Abstract
Breast cancer susceptibility gene 2 (BRCA2) is an important tumor suppressor, which is participated in repair of damaged DNA by its highly conserved BRC repeat motifs regulating RAD51 protein homologous recombination and thereby preventing cell carcinogenesis. In this study, the BRCA2(1524–1548)–RAD51(241–260) complex structure was obtained based on PDB bank data 1N0W, which provided the basis for site-specific mutation of BRCA2(1524–1548). The BRC4 and BRC4 analogous peptides were synthesized, and the interaction between BRC peptide and RAD51(241–260) was studied by fluorescence spectroscopy, circular dichroism spectroscopy and microscale thermophoresis (MST). The results of circular dichroism showed that the changes in secondary structures of RAD51(241–260) occurred after adding BRC4 analogous peptides, and the α-helix content increased significantly. Fluorescence spectral data demonstrated that the model of BRC peptide binding to RAD51(241–260) was static quenching, and the binding constants of BRC4, P1, P2, P4 with RAD51(241–260) were 1.647 × 10−4 L mol−1, 2.532 × 10−4 L mol−1, 3.161 × 10−4 L mol−1, 1.705 × 10−4 L mol−1, respectively. The results of MST indicated that P2 and RAD51(241–260) have better affinity for dissociation constant 44.286 μM. The strongest affinity between P2 and RAD51(241–260) indicated that the mutation of amino acid residue constituting BRC α-helix affects the structure and interaction of BRC peptide and RAD51(241–260).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer has always been the most prevalent malignant tumor among women, with the growing numbers of the disease at a young age (Pan et al. 2017). The occurrence of breast cancer involves changes in a variety of genes and their products that cause the proliferation of cells to become cancerous. In recent years, the susceptibility gene such as P53 and RAD51 are regulated by changing breast cancer susceptibility gene 2 (BRCA2) existing in the human body to participate in the repair of damaged DNA, which has caused more and more attention (Walsh and King 2007).
BRCA2, a tumor suppressor gene closely related to breast cancer, is located on human chromosome 13. The interaction between BRCA2 and RAD51 plays a key role in the process of homologous recombination and double-strand DNA repair (Esashi et al. 2007). Mutations in BRCA2 can affect gene regulation and DNA repair, which lead to the accumulation of damaged DNA and the disorder of homologous recombination repair function in the cells, thereby increasing cancer incidence such as breast cancer and ovarian cancer (Antoniou et al. 2008; Gao et al. 2011). BRCA2 protein contains eight highly conserved repeat motifs (BRC1–BRC8) in its 1003–2085 sequence. The presence of these mutational conserved repeat motifs in various cancer cells indicates that BRC is an important functional region of BRCA2 protein and involved in a variety of cancers’ pathological processes (Marmorstein et al. 1998). The BRC4(1519–1551) has the strongest affinity with RAD51 among eight highly conservative motifs and includes a key peptide fragment—BRCA2(1524–1548). BRCA2(1524–1548), which contains two fragments 1524-FxxA-1527 and 1545-LFDE-1548 that are essential for binding to RAD51, is chosen as the mutated template (Rajendra and Venkitaraman 2010).
RAD51 is an important DNA repair protein and also is a widely studied tumor suppressor gene product (Smith et al. 2003). In the pathway of maintaining genomic integrity and stability, homologous recombination can accurately repair the broken DNA double strand, and RAD51 is one of the important proteins involved in homologous recombination (Carreira et al. 2009). The human RAD51 gene is located on chromosome 15 and encodes a protein consisting of 339 amino acids. The amino acid sequence of RAD51 from 96 to 314 is the functional core region, in which the sequence of RAD51 from 191 to 260 is a key region binding to BRCA2 protein (Baumann and West 1998; Benson et al. 1994). The RAD51–BRC4 crystal structure shows that the key amino acids on RAD51 such as 247Arg and 251Met have stronger interactions with Phe and Glu on BRC4 peptide (Pellegrini et al. 2002).
In this work, RAD51(241–260), that is 241–260 peptide segment, which contains 247Arg, 251Met, etc., key amino acid residues are indispensable for interaction with RAD51, is selected as the target peptide. The alanine scan approach is performed based on the structure of BRCA2(1524–1548)–RAD51(241–260) to obtained active sites, then site-directed mutations were performed to achieve better binding. The preliminary study on the interaction between BRC4 analogous peptides and RAD51(241–260) provides ideas and basis for understanding the structure of BRC repeat motifs and the design of polypeptide drugs.
Materials and methods
Materials
Fmoc-Rink-Amide-MBHA resin, Fmoc-AA-OH,1-[Bis(dimethyllamino)methylenee]-1H-1,2,3-triazolo[4,5-b] pyridinium 3-oxid hexafluorophosphate (HATU), N, N-diisopropylethylamine (DIPEA), and 1-hydroxybenzotriazole (HOBT) were purchased from GL Biochem Ltd. (Shanghai, China). N, N-dimethylformamide (DMF), dichloromethane (DCM) and trifluoroacetic acid (TFA) were purchased from Tianjin Kemiou Chemical Reagent Co., Ltd. Ethanethiol (EDT), thioanisole and phenol were obtained from Aladdin reagent Co., Ltd. (Shanghai, China). Piperidine, acetonitrile and methanol were provided from Tianjin Siyou Fine Chemicals Company (Tianjin, China). The acetonitrile was chromatographic reagent and other reagents were analytical grade.
Design of BRC4 analogous peptides
Our experimental team in order to obtain peptides that bind better to RAD51 and the structure of BRCA2 was optimized. A shorter peptide BRCA2(1524–1548) was chosen from the existing literature. The crystal structure of RAD51(241–260) and BRCA2(1524–1548) was obtained from 1N0W provided by PDB bank (Pellegrini et al. 2002). The alanine scanning was performed on the crystal structure of RAD51(241–260) and BRCA2(1524–1548) to identify the active sites. The 1542Val, 1545Leu were selected to undergo mutation and assessed the binding free energy for complex conformations (Bradshaw et al. 2011). In this work, all peptides were treated with N-terminal acetylation and C-terminal amidation to simulate the structure of peptides in human body (Kitamura et al. 2006).
Synthesis of peptides
The peptides were synthesized by solid-phase peptide synthesis (SPPS) (Zhao and Lu 2015; Zheng et al. 2013). The synthesis step of RAD51(241–260) (Ac-RQMHIARFLRMLLRLADEFG-NH2) was detailed as below. Fmoc-Rink-amide-MBHA Resin (1.5 g, 0.34 mmol g−1) and DMF (10 mL) were added to a 100 mL solid-phase synthesis tube and swelled with nitrogen for 1 h, then treated with 10 ml of 20% piperidine/DMF resin for 20 min (2 × 20 min) to deprotect group Fmoc. The solution was filtered off with a vacuum pump, then rinsed with DMF (3 × 10 ml, each time for 1 min), methanol (3 × 10 ml, each time for 1 min), and DMF (3 × 10 ml, each time for 1 min). Determining whether the amino protecting group is completely removed by Kaiser test. DIPEA (481.77 μl), Fmoc-Gly-OH (0.5780 g) and HOBT (0.2626 g), HATU (0.7391 g) were dissolved in 10 ml of DMF to prepare a coupling reagent. The coupling reagent was added to the synthesis tube for nitrogen bubbling reaction for 4 h. The resin was washed successively with DMF (3 × 10 ml, each time for 1 min), methanol (3 × 10 ml, each time for 1 min), DMF (3 × 10 ml, each minute), respectively and the Kaiser test Proved that the coupling reaction is completely. The experimental procedure was repeated for the remaining amino acids until RAD51(241–260) was synthesized, and finally blocked with 20% acetic anhydride/DMF (10 ml), DIPEA (481.77 μl) to form a stable amide bond. The RAD51(241–260) was cleaved from the Fmoc-Rink-Amide-MBHA resin using TFA/thioanisole/thioethanol/H2O/phenol (82.5:5:5:2.5:5, v/v) cutting reagent at room temperature for 4 h. The RAD51(241–260) was suction filtered into ice ether for refrigeration and placed in a centrifuge at 6500 r min−1 for 5 min (5 × 5 min) repeatedly rinsed with ether, dried and dissolved in water. Then, freeze-drying in a freeze dryer to obtain a crude peptide, and stored at − 20 °C. The other peptides were synthesized by the same method.
Separation and purification of peptides by RP-HPLC
The lyophilized crude peptide was passed through a reversed-phase high-performance liquid chromatography (RP-HPLC) on an Agilent Zorbax 300SB-C18 column (4.6 mm × 250 mm) using 5 μm silica gel as the stationary phase. The gradient elution with eluent A (water containing 0.1% TFA), eluent B (acetonitrile) was used separated and purified for peptides at a flow rate of 1 ml min−1. The scan measurement wavelength was set at 220 nm. Gradient: 0–10–20 min corresponds to the range 40–60–40% of eluent B.
Characterization of peptides by mass spectrometry
The solutions of RP-HPLC main peak were collected, freeze dried and characterized by Thermo-LCQ Advantage Mass Spectrometer (Thermo Fisher Scientific, American). The mass spectrometer was equipped with ion jet source, working in positive ion mode and methanol as solvent.
Circular dichroism (CD) spectroscopy
To study the effect of BRC4 peptide on the secondary structure of RAD51(241–260), a MOS–500 spectrometer (France biologic company, France) was used to perform circular dichroism under the conditions of scanning range 190–320 nm, step size 1 nm, scanning speed 60 nm min−1 and at room temperature. The concentration of BRC peptides and RAD51(241–260) was 1 × 103 μM, 10 μM, respectively, and all dissolved in 50 mM (pH = 7.4) Tris–HCl buffer solution. The BRC4 peptide solution was mixed with RAD51(241–260) in different volumes to obtain a series of samples, which were heated for 30 min at 37 °C. The 3 ml Tris–HCl contains different volumes of BRC4 peptide solution was prepared for the blank control groups, then the corresponding background absorption of each blank reagent was subtracted from the absorption spectrum to eliminate the effects of BRC4 peptide autofluorescence. Each concentration of the sample is measured three times to take the average value.
Fluorescence spectroscopy studies
Fluorescence spectra were obtained on a Cary Eclipse spectrofluorimeter (Agilent, Australia) using a buffer solution as the blank control groups, which the (50 mM, pH = 7.4) Tris–HCl was added different volumes of 1 × 103 μM BRC4 peptide solution. A series of mixed solutions were obtained with the different volumes of BRC4 peptide solution but a constant concentration of RAD51(241–260), which was heated at 37 °C for 30 min and measured in the range of 280–400 nm at an excitation wavelength of 275 nm. The entrance of the fluorescence spectrum is 5 nm, and the exit slit is 10 nm. Each concentration of the sample is measured two times to take the average value.
Microscale thermophoresis (MST)
The MST measurements for the binding of BRC4 peptide to RAD51(241–260) were carried out in a Nano Temper Monolith NT.115 apparatus with blue channels at 298 K. The buffer solution was 20 mM PBS (pH = 7.4) with 50 mM NaCl, 1 mM MgCl2 (Nomme et al. 2010; Wienken et al. 2010). Adding the corresponding amount of BRC4 sample to 1 ml PBS (pH = 7.4) to prepare analogous peptide solution at a concentration of 2 mM and taking 1.416 mg sample dissolved in 100 ml PBS (pH = 7.4) and added 5% (vol:vol) DMSO to prepare target peptide solution at a concentration of 5 μM using RAD51(241–260) labeled by 5-FAM. The prepared BRC4 peptide solution was serially diluted in 10 μl centrifuge tube using the buffer PBS (pH = 7.4), then the same amount of RAD51(241–260) solution was added to each diluted BRC4 peptide solution. Finally, the samples were heated for 30 min at 37 °C and measured at 40% LED power and 60% MST power. Laser off/on time was 5 s and 30 s, respectively.
Results and discussion
Design and analysis of peptides
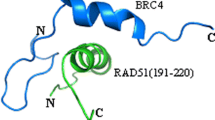
The interaction between BRCA2 and RAD51 was mainly achieved via eight highly conservative BRC repeat motifs (BRC1–BRC8) in BRCA2. BRC4(1519–1551) was the BRC motif strongest binding to RAD51 (Wong et al. 1997). It had been reported that the two motifs, denoted 1524-FxxA-1527 and 1545-LFDE-1548, were essential for BRC4(1519–1551) binding to RAD51(Rajendra and Venkitaraman 2010). Therefore, the sequence of amino acid residue 1524–1548, that is BRCA2(1524–1548), containing the two essential motifs was chosen from BRC4(1519–1551) as P1 to mutated. RAD51(241–260), which contains key amino acid residues such as 247Arg, 251Met had better interaction with BRC4, was selected as target peptide. The RAD51(241–260) and BRCA2(1524–1548) complex structure was intercepted from the PDB 1N0W. The alanine scanning approach was performed on the complex structure and revealed one active site, 1542Val. Moreover, in the complex structure of BRCA2(1524–1548)–RAD51(241–260), 1545Leu was closer to RAD51(241–260) as shown in Fig. 1 and its side chain group extends to RAD51(241–260). Mutated at 1545Leu may alter the α-helix structure of BRCA2(1524–1548), thereby affecting the interaction with RAD51. Therefore, 1542Val and 1545Leu were selected as the mutated sites.
The virtual scanning mutagenesis method was applied to calculate the binding free energy (ΔΔGbind) between peptides (Martins et al. 2013; Huo et al. 2002). There were six scanning results that can be chosen for us (Table 1) and we picked and synthesized three mutated peptides with low energy. The ΔΔG bind value − 0.82 kcal mol−1 of P3 was lower than that of P1, which indicated the mutation Leu1545Trp in P3 may improve the stability of complex structure P3–RAD51(241–260). The ΔΔG bind values − 0.44 kcal mol−1 of P2 and − 0.41 kcal mol−1 of P4 revealed the effect of mutation Val1542Arg in P2 or Leu1545Arg in P4 had some binding disadvantage compared with P3, but there was a certain degree of enhancement contrast in P1 (Zhao et al. 2018).
Based on calculations and structure, the 1542Val and 1545Leu in BRCA2(1524–1548) were selected to undergo mutation. The 1542Val being an amino acid residue that constitutes an α-helix structure and its position between the conservative amino acid residues 1541Lys and 1543Lys. Analogous peptide P2 obtained by the mutation of 1542Val to Arg may affect α-helix structure of BRCA2(1524–1548), thereby affecting the interaction with RAD51. Since 1545Leu was closer to RAD51(241–260) in complex structure, the mutation of Leu to Arg in P4 for enhancing hydrophilicity on one hand and increasing activity of side chain group of the polypeptide on the other hand, which may improve the affinity of P4 with RAD51(241–260). Replacing the 1545Leu with Trp investigates whether the flexible structure of the C-terminal in BRCA2(1524–1548) significantly affects the structure and function of the polypeptide. In addition, the side chain group of 1545Leu in complex structure towards RAD51(241–260), the increase of polypeptide side chain activity after the mutation of 1545Leu to Trp may enhance binding between BRC peptide and RAD51(241–260), P3 was designed. The sequences of peptides and their simple physicochemical properties are shown in Table 2.
Characterization of peptides
The seven peptides were synthesized by SPPS with a ratio of resin, amino acid and coupling agent 1:4:6. The target peptide RAD51(241–260) used in the MST assay was labeled with the fluorophore 5-FAM to enhance the fluorescence intensity of the system. The aqueous solution of synthesized crude peptide was filtered, then separated and purified by RP-HPLC with a purity of more than 90%. All peptides were characterized by ESI–MS, and the results are shown in Table 3 (RP-HPLC and ESI–MS spectra of all peptides are shown in Figs. S1–S7). The multiple charge peaks of the measured polypeptide are basically consistent with the calculated theoretical values, indicating the successful synthesis of target polypeptides.
Analysis of CD spectroscopy for the interaction between BRC4 analogous peptides and RAD51(241–260)
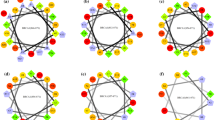
Circular dichroism is a common method for studying proteins due to its sensitivity to protein secondary structure and protein conformational changes. The intensity and position of the circular dichroic bands produced by their secondary structure are also different due to the different amino acid sequences that make up the polypeptide. In the near-ultraviolet range of 200–250 nm, the peak shape is “W”, the positive absorption peak at 192 nm and negative absorption peak at 208 nm and 222 nm, which is considered to be an α-helical structure. The β-sheet presents a positive peak at 195 nm, the β-turn shows a weak negative peak at 220–230 nm, and the negative peak of 200 nm indicates an irregular rolling structure (Wang et al. 2019; Jasim et al. 2018). Therefore, the interaction between polypeptides and RAD51(241–260) can be reflected by change in spectral band displacement and intensity.
As shown in Fig. 2, the negative absorption peaks of RAD51(241–260) at 208 nm and 222 nm had changed after adding different BRC4 analogous peptides. The absorption intensity of RAD51(241–260) was slightly reduced after adding different contents of BRC peptides, and accompanied by red shift (P2: from 212 to 219 nm, P1: from 212 to 220 nm). The changes of RAD51(241–260) absorption intensity after adding other three peptides are shown in Fig. S8. This phenomenon may be due to increase of positive charge produced electrostatic effect under physiological conditions (pH = 7.0) after adding BRC4 analogous peptides (Zheng et al. 2011). After the addition of different concentrations of BRC4 peptide interacted with RAD51(241–260), the microenvironment changed to make the structure of RAD51(241–260) more compact and ordered, and the increase of α-helix, which resulted in a slight decrease in peak intensity and accompanied by red shift. The results indicated that the presence of BRC peptides exerts an influence on the structure of RAD51(241–260).
The α-helix content of the secondary structure of RAD51(241–260) in the presence and absence of BRC peptide was calculated by CDpro as shown in Table 4 (Li et al. 2018). These date show that the α-helix content of RAD51(241–260) has significantly changed with the increase of BRC4 analogous peptide concentration. In particular, BRC4 analogs P2–P4, which, respectively, increased the proportion of the α-helical structure of RAD51(241–260) from 53.67 to 82.2%, 53.67 to 79.61%, and 53.67 to 79.64%. The result shows that the mutation of amino acid residues in the structure BRCA2(1524–1548) exerts a marked influence on the interaction between BRCA2(1524–1548) mutated peptides and RAD51(241–260). This phenomenon may be due to electrostatic effect between BRC4 analogous peptides and RAD51(241–260) under physiological conditions making the structure of RAD51(241–260) compact and orderly. In addition, the basic amino acid residue Arg in BRC analogous peptides can charge–charge bind to the acidic amino acid residues Asp/Glu in RAD51(241–260). Then, the structure of RAD51(241-260) is tightened. The order of the effects of BRC4 analogous peptides on the secondary structure of RAD51(241–260) were P2 > P4 ≈ P3 > P1 > BRC4. Next, the intensity of the interaction between the BRC peptide and RAD51(241–260) is measured by fluorescence spectrometer.
Fluorescence quenching studies
Fluorescence spectroscopy is one of the most useful methods for studying peptide–protein interaction mechanisms. The spectrum of the interaction between BRC4 peptide and RAD51(241–260) was obtained by fluorescence quenching of the BRC–RAD51(241–260) system shown in Fig. S9, then the binding mode was derived based on the spectral. As observed from the spectral that the emission peak intensity of RAD51(241–260) was reduced slightly and the emission peak position was without shifted at 313 nm (maximum emission wavelength) after adding BRC4, P1, P2, P4. The quenching of fluorescence intensity of BRC–RAD51(241–260) was analyzed by the Stern–Volmer equation (Rehman et al. 2015).
where F0 and F were the fluorescence intensities of RAD51(241–260) and BRC–RAD51(241–260) complexes, respectively. [Q] is the concentration of quencher BRC peptides, τ0 is the average lifetime of fluorophore without quencher (10−8 s), and Kq is the bimolecular quenching rate constant (Lv et al. 2018; Tang et al. 2012). If Kq > 2.0 × 1010 L mol−1 s−1, proving that the non-fluorescent substance was produced during the interaction between BRC peptides and RAD51(241–260), then the quenching process is static. The binding constant (K) and the number of binding site (n) were calculated by the following equation (Shahabadi and Maghsudi 2014):
The fluorescent quenching fitting curve and values of Kq, K and n are shown in Fig. 3 and Table 5, respectively.
The results in Table 4 show that the Kq values of the BRC peptides except P3 were greater than the maximum collision constant of diffusion regulation, 2 × 1010 L mol−1 s−1, then the binding model was static quenching. Compared with BRC4, the affinity of BRC4 analogous peptides binding to RAD51(241–260) has enhanced, in which P1 and P2 are more strongly combined, but the combine affinity of the P4–RAD51(241–260) has some disadvantage. This phenomenon may be due to the mutation of Leu to Arg, thus minor changing the isoelectric point of the polypeptide to 10.6, and the P4 with the positive charge under physiological conditions interacted with RAD51(241–260) and resulted in electrostatic effects (Zheng et al. 2011). The side chain groups of 1542Arg, conservative amino acid residues 1541Lys and 1543Lys in P2 have stronger hydrophilicity and the ability to form hydrogen bonds. The strongest affinity of P2 with RAD51(241–260) may be due to synergistic relationship of 1541Lys, 1542Arg and 1543Lys side chain groups. It can be seen from Fig. S9 that the spectral of P3–RAD51(241–260) does not conform to the Stern–Volmer equation. The fluorescence intensity has a slight quenching first and then enhanced dramatically with increasing concentration of P3 during the fluorescence titration experiment. The phenomenon may be due to the 1545Trp itself has a relatively strong fluorescence, and changes of structure in P3 C-terminal after interaction between P3 and RAD51(241–260) led to more exposure of the aromatic ring of the 1545Trp side chain, thereby making the fluorescence intensity of the system enhanced greatly. The maximum absorption wavelength has a significant changed from 315 to 356 nm, which may be attributed to increase of positive charge generate electrostatic repulsion after the mutation of 1545Leu into Trp, thus the structure of RAD51(241–260) was tighten. Furthermore, the mutated amino acid residue Trp itself has a strong fluorescence, so that the complex as a whole exhibits an excitation wavelength similar to the amino acid residue Trp (355 nm). Therefore, there is a significant red shift after the interaction of P3 and RAD51(241–260) compared with other BRC4 peptides.
Precise detection of high affinity interaction by MST
MST is currently the best technique for analyzing the size range of objects and the range of detection dynamics. MST is highly adaptable, suitable for different environmental requirements, different biomolecules, and analyzes biomolecular interactions by measuring intermolecular affinity (Kd value) (Wienken et al. 2010; Zhang et al. 2016). The results of the measurement described that the affinity of P2 with RAD51(241–260) is the strongest among the five peptides, Kd = 38 ± 5 μM. The MST spectrum of the BRC peptides combined with RAD51(241–260) is shown in Fig. S10. This result may be due to Val1542Arg forms more intermolecular hydrogen bond ratio than other peptides, and then these hydrogen bonds stabilize their conformation during P3 interacted with RAD51(241–260) and increase the affinity between them. In addition, the structures in Fig. 4 also imply that noncovalent interactions are the major driving forces in this peptide complex. However, the specific mechanism of action between them still needs further experimental research.
Conclusion
The crystal structure of BRC4 and RAD51 (PDB bank: 1N0W) laid the foundation for specific mutations in BRC4 analogous peptides. The binding mechanism between BRC peptide and RAD51(241–260) was analyzed by CD spectroscopy, fluorescence spectroscopy and MST. CD spectroscopic measurements showed that the secondary structure of RAD51(241–260) changed and the α-helix content increased significantly after adding BRC4 analogous peptides. Fluorescence and MST results showed that the affinity had obviously enhanced between RAD51(241–260) and BRC4 analogous peptides, and the P2 had the strongest affinity binding to RAD51(241–260) among the five peptides. This study proved that the mutations of amino acid residues composing the BRC4 α-helix, uncommonly affect the structure and interaction of BRC with RAD51(241–260). Although the interaction between the fragment peptides does not represent the role of the protein, this work provides idea and theoretical basis to further study the interaction mechanism of their spatial structure and the design of breast cancer suppression drugs.
References
Antoniou AC et al (2008) Common breast cancer-predisposition alleles are associated with breast cancer risk in BRCA1 and BRCA2 mutation carriers. Am J Hum Genet 82:937–948. https://doi.org/10.1016/j.ajhg.2008.02.008
Baumann P, West SC (1998) Role of the human RAD51 protein in homologous recombination and double-stranded break repair. Trends Biochem Sci 23:247–251. https://doi.org/10.1016/s0968-0004(98)01232-8
Benson FE, Stasiak A, West SC (1994) Purification and characterization of the human Rad51 protein, an analogue of E. coli RecA. EMBO J 13:5764–5771. https://doi.org/10.1002/j.1460-2075.1994.tb06914.x
Bradshaw RT, Patel BH, Tate EW, Leatherbarrow RJ, Gould IR (2011) Comparing experimental and computational alanine scanning techniques for probing a prototypical protein-protein interaction. Protein Eng Des Sel 24:197–207. https://doi.org/10.1093/protein/gzq047
Carreira A, Hilario J, Amitani I, Baskin RJ, Shivji MKK, Venkitaraman AR, Kowalczykowski SC (2009) The BRC repeats of BRCA2 modulate the DNA-binding selectivity of RAD51. Cell 136:1032–1043. https://doi.org/10.1016/j.cell.2009.02.019
Esashi F, Galkin VE, Yu X, Egelman EH, West SC (2007) Stabilization of RAD51 nucleoprotein filaments by the C-terminal region of BRCA2. Nat Struct Mol Biol 14:468–474. https://doi.org/10.1038/nsmb1245
Gao LB et al (2011) RAD51 135G/C polymorphism and breast cancer risk: a meta-analysis from 21 studies. Breast Cancer Res Treat 125:827–835. https://doi.org/10.1007/s10549-010-0995-8
Huo S, Massova I, Kollman PA (2002) Computational alanine scanning of the 1:1 human growth hormone-receptor complex. J Comput Chem 23:15–27. https://doi.org/10.1002/jcc.1153
Jasim SB, Li Z, Guest EE, Hirst JD (2018) DichroCalc: improvements in computing protein circular dichroism spectroscopy in the near-ultraviolet. J Mol Biol 430:2196–2202. https://doi.org/10.1016/j.jmb.2017.12.009
Kitamura A, Kiyota T, Lee S, Sugihara G (2006) N- and C-terminal effect of Amphiphilic.ALPHA.Helical peptides on the interaction with model and bio-membranes. Bull Chem Soc Japan 71:1151–1158
Li LL, Lv MX, Lu K, Liu GB, Peng L (2018) Design and synthesis of breast cancer susceptibility gene BRCA1 analogs peptides and the interaction of analogs peptides with breast cancer suppressor gene protein RAD51. Chin J Org Chem 38:246–252. https://doi.org/10.6023/cjoc201708041
Lv MX, Wang MW, Lu K, Duan BC, Zhao YF (2018) Non-covalent interaction between CA-TAT and calf thymus DNA: deciphering the binding mode by in vitro studies. Int J Biol Macromol 114:1354–1360. https://doi.org/10.1016/j.ijbiomac.2017.11.158
Marmorstein LY, Ouchi T, Aaronson SA (1998) The BRCA2 gene product functionally interacts with p53 and RAD51. Proc Natl Acad Sci USA 95:13869–13874. https://doi.org/10.1091/jb/mvt040
Martins SA, Perez MAS, Moreira IS, Sousa SF, Ramos MJ, Fernandes PA (2013) Computational alanine scanning mutagenesis: MM-PBSA vs TI. J Chem Theory Comput 9:1311–1319. https://doi.org/10.1021/ct4000372
Nomme J et al (2010) Design of potent inhibitors of human RAD51 recombinase based on BRC motifs of BRCA2 protein: modeling and experimental validation of a chimera peptide. J Med Chem 53:5782–5791. https://doi.org/10.1021/jm1002974
Pan YB, Liu GH, Yuan YF, Zhao J, Yang Y, Li YR (2017) Analysis of differential gene expression profile identifies novel biomarkers for breast cancer. Oncotarget 8:114613–114625. https://doi.org/10.18632/oncotarget.23061
Pellegrini L, Yu DS, Lo T, Anand S, Lee M, Blundell TL, Venkitaraman AR (2002) Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature 420:287–293. https://doi.org/10.1038/nature01230
Rajendra E, Venkitaraman AR (2010) Two modules in the BRC repeats of BRCA2 mediate structural and functional interactions with the RAD51 recombinase. Nucleic Acids Res 38:82–96. https://doi.org/10.1093/nar/gkp873
Rehman SU, Sarwar T, Ishqi HM, Husain MA, Hasan Z, Tabish M (2015) Deciphering the interactions between chlorambucil and calf thymus DNA: a multi-spectroscopic and molecular docking study. Arch Biochem Biophys 566:7–14. https://doi.org/10.1016/j.abb.2014.12.013
Shahabadi N, Maghsudi M (2014) Multi-spectroscopic and molecular modeling studies on the interaction of antihypertensive drug; methyldopa with calf thymus DNA. Mol Biosyst 10:338–347. https://doi.org/10.1039/c3mb70340a
Smith DD et al (2003) Modifications to the N-terminus but not the C-terminus of calcitonin gene-related peptide (8-37) produce antagonists with increased affinity. J Med Chem 46:2427–2435. https://doi.org/10.1021/jm020507f
Tang J et al (2012) Studies on the binding behavior of prodigiosin with bovine hemoglobin by multi-spectroscopic techniques. Spectrochim Acta Part A 96:461–467. https://doi.org/10.1016/j.saa.2012.05.059
Walsh T, King MC (2007) Ten genes for inherited breast cancer. Cancer Cell 11:103–105. https://doi.org/10.1016/j.ccr.2007.01.010
Wang M, Lv M, Lu K, Liu G, Mai W, Yu B, Lou Y (2019) Design, synthesis and interaction of BRCA1 peptide fragments with RAD51(181-200). Int J Pept Res Ther. https://doi.org/10.1007/s10989-019-09821-7
Wienken CJ, Baaske P, Rothbauer U, Braun D, Duhr S (2010) Protein-binding assays in biological liquids using microscale thermophoresis. Nat Commun. https://doi.org/10.1038/ncomms1093
Wong AKC, Pero R, Ormonde PA, Tavtigian SV, Bartel PL (1997) RAD51 interacts with the evolutionarily conserved BRC motifs in the human breast cancer susceptibility gene brca2. J Biol Chem 272:31941–31944. https://doi.org/10.1074/jbc.272.51.31941
Zhang G, Mendez BL, Sedgwick GG, Nilsson J (2016) Two functionally distinct kinetochore pools of BubR1 ensure accurate chromosome segregation. Nat Commun. https://doi.org/10.1038/ncomms12256
Zhao DX, Lu K (2015) Design, synthesis, and characterization of BRC4 mutants based on the crystal structure of BRC4-RAD51(191-220). J Mol Model. https://doi.org/10.1007/s00894-015-2831-x
Zhao DX, Lu K, Liu GB, Ma L, Zhu HJ, He J (2018) Design and synthesis of BRC analogous peptides and their interactions with a key p53 peptide. FEBS Lett 592:3438–3445. https://doi.org/10.1002/1873-3468.13256
Zheng P, Cao Y, Bu TJ, Straus SK, Li HB (2011) Single molecule force spectroscopy reveals that electrostatic interactions affect the mechanical stability of proteins. Biophys J 100:1534–1541. https://doi.org/10.1016/j.bpj.2011.01.062
Zheng JS, Tang S, Qi YK, Wang ZP, Liu L (2013) Chemical synthesis of proteins using peptide hydrazides as thioester surrogates. Nat Protoc 8:2483–2495. https://doi.org/10.1038/nprot.2013.152
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 21572046).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
There are no conflicts of interest in this work.
Research involving human and animal participants
This article does not contain any studies with human participants performed by any of the authors.
Additional information
Handling Editor: K. W. Li.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, J., Lu, K., Lv, M. et al. Design, synthesis and interaction of BRC4 analogous peptides with RAD51(241–260) . Amino Acids 52, 361–369 (2020). https://doi.org/10.1007/s00726-019-02813-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00726-019-02813-3