Abstract
A variety of antimicrobial peptides against infection have been identified from the skin of amphibians. However, knowledge on amphibian defensins is very limited. A novel anionic defensin designated PopuDef was purified from the skin of tree frog Polypedates puerensis, and the cDNA encoding PopuDef precursor was cloned from the skin cDNA library. The amino acid sequence of PopuDef (net charge: −2, pI: 4.75) shared the highest identity of 57 % (25/44) with the salamander defensin CFBD-1 (net charge: 0, pI: 6.14) from urodela amphibians. PopuDef showed moderate antimicrobial activities against P. aeruginosa and S. aureus (MICs are 19.41 and 17.25 μM, respectively), and relatively weak activities against E. coli and B. subtilis (MICs are 38.82 and 43.14 μM, respectively). Tissue distribution analysis indicated that relatively high expression level of PopuDef mRNA was observed in immune-related tissues including skin, gut, lung and spleen. Furthermore, the expression level of PopuDef was significantly upregulated in these tissues after tree frogs were infected with different bacteria strains mentioned above. Interestingly, the induction of PopuDef challenged with E. coli or B. subtilis, which was less sensitive to PopuDef, was much higher than that did with P. aeruginosa or S. aureus. These findings highlight the key role of PopuDef in innate immunity against infection. To our knowledge, PopuDef is the first anionic defensin characterized from amphibians.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The production of natural antimicrobial peptides (AMPs) is a nonspecific host defense mechanism to fight infection and/or stress in bacteria, fungi, plants and animals (Hassan et al. 2012; Schneider et al. 2010; Barbosa Pelegrini et al. 2011; Yang et al. 2009). Defensins are a family of evolutionarily related AMPs which are stabilized by three disulfide bonds and contain a β hairpin as their principal structural feature (White et al. 1995). In vertebrates, defensins are classified into α-defensins, β-defensins and θ-defensins based on the specific spacing and disulfide bonds of their six cysteines’ residues. α-defensins and θ-defensins are only found in mammals (Yang et al. 2004). The more ancient β-defensins, in which the six cysteines are linked in the 1–5, 2–4, 3–6 pattern, are widely identified from mammals (Yang et al. 2004; Selsted et al. 1985) and other species, such as fishes (Casadei et al. 2009; Ruangsri et al. 2013), birds (Van Dijk et al. 2008), reptiles (Lakshminarayanan et al. 2008). These defensins are commonly cationic peptides, manifesting antimicrobial activity against Gram-positive and Gram-negative bacteria, fungi, and some enveloped viruses (Groot et al. 2008). Although cationic defensins have been abundantly discussed in the literature, anionic defensins have been poorly documented. To date, anionic defensins are restricted to a few animal species and tissues. Most of them are invertebrate defensins and primate β-defensins, such as the first anionic defensins (Amblyomma defensin peptide 2) from tick Amblyomma hebraeum (Lai et al. 2004), the anionic defensin (spliDef) from leafworm Spodoptera littoralis (Seufi et al. 2011), the anionic defensin-like peptide (BmdefA) from silkworm Bombyx mori (Wen et al. 2009), the human β-defensin 118 (DEFB118) and HE2C from the epididymis (Yenugu et al. 2004; von Horsten et al. 2004).
Among vertebrates, amphibians have developed various mechanisms to cope with the stress and/or bacterial infection. Their skins could produce small compounds and gene-coded peptides/proteins which are involved in the host defense (Clarke 1997; Wu et al. 2011). Little research has been done about defensins in amphibians (Anura, Caudata, and Gymnophiona), and currently only two defensins have been identified. One is the CFBD-1 β-defensin which was isolated from the skin secretions of salamander Cynops fudingensis (Caudata: Salamandridae) (Meng et al. 2013), the other is the partial defensin sequence identified by transcriptome analysis from the Chinese brown frog Rana chensinensis (Anura: Ranidae) (Zhang et al. 2009).
In the present study, we described the purification and characterization of a novel anionic defensin (PopuDef) from the skin of tree frog P. puerensis. We also showed the expression profile of PopuDef in different tissues, and the induction of PopuDef mRNA in immune-related tissues after tree frogs were infected with different bacteria.
Materials and methods
Frog collection and tissue preparation
Adult tree frogs of P. puerensis were captured in Pu’er, Yunnan Province, China (24.786°N 101.362°E). Skin secretions were collected as previously reported (Hao et al. 2012). Five randomly selected tree frogs (21–30 g) were killed by destroying their brain and spinal cord with the pithing needle. Tissues (skin, muscle, brain, spleen, heart, kidney, lung, stomach, gut and liver) were quickly removed, and stored in liquid nitrogen until use. Tree frogs used for bacterial infection were housed and randomly presented with mealworm larvae Tenebrio molitor. The large shallow water dishes were provided for nocturnal bathing. The study was approved by the Animal Care and Use Ethics Committee of Kunming Medical University.
Peptide purification
2 g of lyophilized P. puerensis skin secretion was dissolved in 10 mL phosphate-buffered saline (PBS, 0.1 M, pH 6.0) with one tablet of Complete™ Mini Protease Inhibitor Cocktail (Roche, USA). After centrifugation (12,000 × g for 15 min at 4 °C), the supernatant was prepurified through a 10 kDa cutoff Centriprep filter (Millipore, CA, USA). The filtrate was further purified by reversed-phase high performance liquid chromatography (RP-HPLC) on a Wondasil C18 column (25 × 0.46 cm). The linear gradient elution was performed in a 0–60 % acetonitrile containing 0.1 % (v/v) trifluoroacetic acid in 0.1 % (v/v) trifluoroacetic acid/water for 70 min. The eluted peaks were collected and lyophilized for antimicrobial assay, and the eluted peak with antimicrobial activity was subjected to automated Edman degradation analysis on an Applied Biosystems pulsed liquid-phase sequencer (model ABI 491, USA).
MALDI-TOF MS analysis
1 μL of the eluted peak with antimicrobial activity was spotted onto a matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) plate with 1 μL of α-cyano-4-hydroxycinnamic acid matrix (10 mg/mL in 60 % acetonitrile) and analyzed by an UltraFlex I mass spectrometer (Bruker Daltonics, Germany) in a positive ion mode.
cDNA library construction and screening of cDNA encoding defensin
Total RNA extraction from the skin of P. puerensis was performed using TRIzol reagent (Invitrogen, USA). mRNA was purified from the total RNA by affinity chromatography in oligo(dT) cellulose columns (Promega, USA) and then used for cDNA library construction by the In-Fusion SMARTer™ Directional cDNA Library Construction Kit (Takara, Japan) according to the instructions of manufacturer.
The synthesized cDNA was used as template for PCR to screen the cDNAs encoding defensin. Primers used in this research are shown in Table 1. PopuDef-R1 and 5′ PCR primer were used in PCR to screen the 5′ fragments of cDNAs encoding defensin. PopuDef-R1 is designed from the amino acid sequence of PopuDef determined by Edman degradation, and 5′ PCR primer is provided in the kit. The PCR conditions were: 95 °C for 5 min and 30 cycles of 95 °C (30 s), 58 °C (40 s), 72 °C (50 s) followed by an extension step at 72 °C for 10 min. The PCR product was purified by gel electrophoresis, and cloned into pMD19-T vector (Takara, Japan) for sequencing. After the 5′ fragments of cDNA had been obtained, a sense primer (PopuDef-F1) was designed based on the 5′-coding region of defensin cDNA and coupled with 3′ PCR primer based on the adaptor sequence of 3′ In-Fusion SMARTer CDS Primer to screen the full length cDNA encoding defensin. The PCR conditions were: 95 °C for 5 min and 30 cycles of 95 °C (30 s), 56 °C (30 s), 72 °C (1 min) followed by an extension step at 72 °C for 8 min. DNA sequencing was performed on an Applied Biosystems DNA sequencer (model ABI PRISM 377, USA).
Sequence analysis
Deduced defensin sequence was performed with ExPASy Translate Tool (http://web.expasy.org/translate/). Database searches were performed with Blastx (http://www.ncbi.nlm.nih.gov/), and the amino acid sequence identity between defensin sequences was aligned using ClustalW (http://embnet.vital-it.ch/software/ClustalW.html) (Chenna et al. 2003). The theoretical isoelectric point (pI) and molecular weight (Mw) were carried out using ExPASy Compute pI/Mw tool (http://web.expasy.org/compute_pi/) (Bjellqvist et al. 1993). The dendrogram was drawn using the neighbor-joining (NJ) method in Mega 5 package. A total of 1000 bootstrap replicates were used to test the reliability of each branch.
Antimicrobial assay
The effect of PopuDef was evaluated against Gram-positive and Gram-negative bacteria as described in our previous paper (Yang et al. 2009; Hao et al. 2012). Microorganisms including Staphylococcus aureus (ATCC 25923), Bacillus subtilis (ATCC 6633), Escherichia coli (ATCC 25922) and Pseudomonas aeruginosa (ATCC 27853) were cultured in Luria–Bertani broth (LB). The minimal concentrations at which no visible growth of microorganisms occurred were defined as minimal inhibitory concentration (MIC).
Hemolytic assay
Hemolytic assay was conducted as previously reported (Hao et al. 2012). Serial dilutions of PopuDef were incubated with washed human erythrocytes at 37 °C for 30 min and then the cells were centrifuged at 1000 × g for 5 min. The absorbance of supernatant was measured at 540 nm. 1 % (v/v) Triton X-100 was used to determine the maximal hemolysis and 0.9 % saline was used as negative control.
Bacterial challenge
To study the expression of the defensin mRNA following bacterial infection, both sexes of tree frogs (21–30 g, n = 5 in each group) were challenged with E. coli, P. aeruginosa, S. aureus and B. subtilis, respectively. Tree frogs were injected intraperitoneally (50 μL/tree frog) with bacteria (1 × 106 CFU/mL PBS) and killed 24 h later. Tissues (skin, spleen, gut and lung) were collected and stored in liquid nitrogen until use. As controls, five tree frogs reared under the same conditions received the same volume of PBS. The samples were processed to obtain total RNA as mentioned above, and then used for qPCR as described below.
Quantitative PCR (qPCR)
qPCR was performed to analyze the expression profile of the PopuDef mRNA in different tissues (skin, muscle, brain, spleen, heart, kidney, lung, stomach, gut and liver), with the housekeeping gene β-actin as an internal control. To elucidate whether the expression level could be increased further, we used qPCR to detect the inducible expression of PopuDef following different bacterial challenge in immune-related tissues. As listed in Table 1, primers for PopuDef amplification were designed based on the PopuDef cDNA sequence, and β-actin was amplified using primers based on the sequence from tree frog, Hyla japonica. PrimeScript® Reverse Transcriptase (Takara, Japan) and SYBR green master mix (Takara, Japan) were used following the manufacture’s instruction.
q-PCR was performed on a Realplex Mastercycler real-time PCR system (Eppendorf, Germany) with the following parameters: 95 °C for 2 min, and 40 cycles of 95 °C for 30 s, 60 °C for 30 s. PopuDef mRNA expression level was calculated following normalization to β-actin by ΔΔC t method. The accuracy of qPCR was verified by melt curve analysis.
Data and statistical analysis
Statistical analyses were performed using GraphPad Prism 5.0 and Stata 10.0 statistical program. Data were presented as mean ± SEM and compared using two-tailed equal variance Student’s t test. P < 0.05 was considered as statistical significance.
Results
Characterization of PopuDef
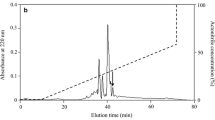
The peptide with antimicrobial activity was purified by C18 RP-HPLC as illustrated in Fig. 1a. After Edman degradation, a primary structure of 15 amino acid residues was identified with the following sequence: GASPALWGCDSFLGY. MALDI-TOF MS analysis (Fig. 1b) indicated that PopuDef had a measured molecular mass of 4636.12 Da, matching well with the calculated molecular mass 4636.37 Da. The complete nucleotide sequence of cDNA (GenBank accession KJ809087) and deduced amino acid sequence of PopuDef precursor are shown in Fig. 2a. The N-terminal deduced sequence of PopuDef precursor is completely consistent with the result of Edman degradation sequencing. The cDNA encoding protein precursor is composed of 66 amino acid (aa) residues, including a predicted 22-aa signal peptide and a 44-aa mature PopuDef. BLAST search indicated that the amino acid sequence of PopuDef (algebraic net charge: −2) shared the highest identity of 57 % (25/44) with the salamander defensin CFBD-1 (net charge: 0) from Cynops fudingensis. It also showed low identities of 38–41 % with fish β-defensin such as β-defensin 4 (net charge: +1) from Oncorhynchus mykiss, defensin-like protein 1 and unnamed protein product (net charge: +2) from Tetraodon nigroviridis. Multi-sequence alignment of β-defensin precursors (Fig. 2b) indicated that the precursor of PopuDef shared low similarity with other defensins. However, twelve amino acids’ residues within the mature peptides are highly conserved, including a signature motif of six conserved cysteines and additional five residues (Trp29, Gly36, Arg39, Glu46 and Gly50).
Isolation of PopuDef from the skin of P. puerensisa and MALDI–TOF MS. a The filtrate of the skin secretion of P. puerensis by 10 kDa cutoff was divided by a Wondasil C18 RP-HPLC column (25 × 0.46 cm) equilibrated with 0.1 % (v/v) trifluoroacetic acid/water. The elution was performed with the indicated gradient of acetonitrile at a flow rate of 1 mL/min. The purified antimicrobial peptide is indicated by an arrow. b MALDI-TOF mass spectrometry analysis of the antimicrobial peptide
cDNA sequence of PopuDef and multiple sequence alignment with known defensins. a The cDNA sequence of PopuDef precursor and the deduced amino acid sequence. Deduced amino acid sequence is shown below the cDNA sequence. The amino acid sequence of mature peptide is underlined and the stop codon is indicated by an asterisk. The putative polyadenylation consensus signal is italicized and gray shaded. Amino acid numbers or nucleotide numbers are shown after the sequences. b Alignment of the amino acid sequence of PopuDef with known defensins. These sequences were based on BLAST search results. The conserved cysteine residues are shaded. The symbols under the alignment indicate: (Asterisk) identical sites; (colon) conserved sites; (dot) less conserved sites. CFBD-1 β-defensin (Cynops fudingensis), β-defensin 4 (Oncorhynchus mykiss), Defensin-like protein1 (Tetraodon nigroviridis), β-defensin-like 3 (Danio rerio). GenBank accession numbers for the analyzed sequences are shown in Fig. 3
Phylogenetic analysis
β-defensins including PopuDef were assembled for evolutionary analysis. As shown in Fig. 3, the constructed phylogenetic tree divided defensins into two distinct clusters. The analysis demonstrated that PopuDef was most closely related to the salamander defensin CFBD-1. In addition, PopuDef also showed a close evolutionary relationship with fish defensins.
Antimicrobial activity
The MICs of the PopuDef against Gram-positive and Gram-negative bacteria were determined. As listed in Table 2, PopuDef showed moderate antimicrobial activities against Gram-negative bacteria P. aeruginosa and Gram-positive bacteria S. aureus with MICs of 19.41 and 17.25 μM, respectively. It also showed relatively weak activities against Gram-negative bacteria E. coli and Gram-positive bacteria B. subtilis with MICs of 38.82 and 43.14 μM, respectively.
Hemolytic activity
Human fresh erythrocytes were used to evaluate the hemolytic activity of PopuDef. The result showed that PopuDef induced 3.6 % human erythrocyte hemolysis at concentration of 43.14 μM, which is the highest MIC value of PopuDef against four tested bacteria.
Expression analysis
Tissue distribution analysis indicated that relatively higher expression of PopuDef mRNA was observed in immune-related tissues including skin, gut, spleen and lung (Fig. 4). Following immune challenge by different bacteria (E. coli, P. aeruginosa, S. aureus and B. subtilis), the expression of PopuDef mRNA in these immune-related tissues was examined. As illustrated in Fig. 5, the mRNA level of PopuDef was apparently higher than naïve tree frogs in skin (31.6, 13.9, 14.8 and 27.2 fold, respectively), in spleen (31.3, 16.3, 13.6 and 39.3 fold, respectively), in gut (53.8, 34.9, 29.5 and 64.2 fold, respectively) and in lung (19.6, 12.3, 14.2 and 19.3 fold, respectively). After different bacterial challenge, the mRNA level of PopuDef showed the highest increase in gut.
Fold increase of PopuDef in immune-related tissues after challenge with different bacteria for 24 h. a Fold increase of PopuDef in skin. b Fold increase of PopuDef in spleen. c Fold increase of PopuDef in gut. d Fold increase of PopuDef in lung. Expression levels in different tissues were calculated relative to the level of PopuDef in corresponding uninfected tissue, which was arbitrarily defined as 1. Values for infection treatment are significantly different from control values. *P < 0.05, **P < 0.01 significantly different compared to the control (n = 5)
Discussion
Defensins are commonly cationic peptides, and abundantly expressed in phagocytes and epithelia in vertebrate (Ganz 2001). The relatively low number of anionic defensin has been reported in the literature; to date, no anionic defensin has been found in amphibians.
Here, a novel anionic defensin, designated PopuDef (the seven letters derive from the first two letters of the genus name and the first two letters from the species name, and followed by the first three letters from the word defensin), was purified from the skin of tree frog Polypedates puerensis (Fig. 1). The cDNA encoding PopuDef precursor was cloned from the skin cDNA library (Fig. 2a). The spacing pattern of PopuDef is X8-C-X6-C-X3-C-X11-C-X5-C-C-X5 (where C is a cysteine, and X is any amino acid except cysteine), which is consistent with the consensus motif of salamander β-defensin and some fish β-defensins (Xn-C-X5-6-C-X3-5-C-X11-12-C-X5-7-C-C-Xn). The result of sequence alignment of β-defensins (Fig. 2b) indicated that besides six absolutely conserved cysteines, there are another five conserved amino acids within the mature peptides, including the glycine residue presented two residues upstream of Cys2, which is absolutely conserved in fish defensins (Casadei et al. 2009; Ruangsri et al. 2013; Nam et al. 2010). However, the role of these conserved residues in the structure and function of these peptides remains to be determined. Phylogenetic analysis (Fig. 3) also showed that PopuDef is closely related to the salamander defensin CFBD-1 and fish defensins. Current identification of amphibian defensin provides a good opportunity to elucidate the evolutionary relationship of lower vertebrate defensins.
Anionic AMPs are different from cationic AMPs in net charge residue composition, secondary structure, hydrophobicity and amphiphilicity. These parameters contribute to the ability of cationic peptides to bind to and disrupt microbial membranes. (Hancock 2001). The cationic AMPs are mainly interacted with microbial membrane components or intracellular targets to exert their highly effective antimicrobial activity (Hancock 2001; Bera et al. 2003). Since too little anionic defensins have been investigated, the structure–function relationships of them are poorly characterized. Generally, anionic defensins possess a relatively narrow spectrum of antimicrobial activities against microorganisms which are closely related to the host (Harris et al. 2009, 2011). In the present study, we observed that PopuDef had an antimicrobial activity against both Gram-positive bacteria (S. aureus and B. subtilis) and Gram-negative bacteria (E. coli and P. aeruginosa) (Table 2). Due to its anionic net charge and anionic computational pI, it seems unlikely that PopuDef is attracted by negatively charged lipid membranes of bacteria. The result suggests that PopuDef might possess some novel mechanisms to exert the antimicrobial effect, and it increases the complexity of the antimicrobial mechanisms of AMPs.
The diverse expression profiles of defensins among different species and even the different isoforms within the same species are still to be unraveled. Generally, the expression of defensins is increased after infection. For instance, anionic defensin (fBDI) gene in teleost paralichehys olivaceus was expressed constitutively in the early developmental stages, and the fBDI gene was dramatically induced in the head kidney of juvenile fish after Edwardsiella tarda challenge (Nam et al. 2010). The relative transcription levels of the anionic insect defensin (SpliDef) from cotton leafworm Spodoptera littoralis were upregulated after bacterial challenge, and the expression of SpliDef in haemocytes peaked at 48 h postinfection (Seufi et al. 2011). Tissue distribution analysis indicated that the relatively higher expression of PopuDef was detected in immune-related tissues (skin, gut, spleen and lung) (Fig. 4). The results suggest that these tissues could response to microbial infection prior to an inflammatory cell influx. qPCR analysis indicated that the relative transcription levels of PopuDef were dramatically upregulated after bacterial challenge (Fig. 5). The induction challenged with sensitive strain (P. aeruginosa or S. aureus, which was inhibited at lower concentration of PopuDef) was much less than that challenged with resistant strain (E. coli or B. subtilis). The results suggest that the regulation of PopuDef is complex, and PopuDef plays an important role in immune responses against bacterial infection in the tree frog P. puerensis.
The current work indicated that besides direct antimicrobial activity, PopuDef may selectively enhance and/or alter host defense mechanisms in protecting against bacterial infection in vivo. The biological activities of PopuDef need to be further investigated to highlight its function in vitro and in vivo.
References
Barbosa Pelegrini P, Del Sarto RP, Silva ON, Franco OL, Grossi-de-Sa MF (2011) Antibacterial peptides from plants: what they are and how they probably work. Biochem Res Int 2011:250349
Bera A, Singh S, Nagaraj R, Vaidya T (2003) Induction of autophagic cell death in Leishmania donovani by antimicrobial peptides. Mol Biochem Parasitol 127:23–35
Bjellqvist B, Hughes GJ, Pasquali Ch, Paquet N, Ravier F et al (1993) The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis 14:1023–1031
Casadei E, Wang TH, Zou J, Gonzalez Vecino JL (2009) Characterization of three novel β-defensin antimicrobial peptides in rainbow trout (oncorhynchus mykiss). Mol Immunol 46(16):3358–3366
Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ et al (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31:3497–3500
Clarke BT (1997) The natural history of amphibian skin secretions, their normal functioning and potential medical applications. Biol Rev Camb Philos Soc 72(3):365–379
Ganz T (2001) Defensins in the Urinary Tract and Other Tissues. J Infect Dis 183(Suppl 1):S41–S42
Groot F, Sanders RW, ter Brake O, Nazmi K, Veerman ECI et al (2008) Histatin 5-derived peptide with improved fungicidal properties enhances human immune deficiency virus type 1 replication by promoting viral entry. J Virol 80:9236–9243
Hancock REW (2001) Cationic peptides: effectors in innate immunity and novel antimicrobials. Lancet Infect Dis 1:156–164
Hao X, Yang HL, Wei L, Yang S, Zhu W et al (2012) Amphibian cathelicidin fills the evolutionary gap of cathelicidin in vertebrate. Amino Acids 43(2):677–685
Harris F, Dennison SR, Phoenix DA (2009) Anionic antimicrobial peptides from eukaryotic organisms. Curr Protein Pept Sci 10:585–606
Harris F, Dennison SR, Phoenix DA (2011) Anionic antimicrobial peptides from eukaryotic organisms and their mechanisms of action. Curr Chem Biol 5:142–153
Hassan M, Kjos M, Nes IF, Diep DB, Lotfipour F (2012) Natural antimicrobial peptides from bacteria: characteristics and potential applications to fight against antibiotic resistance. J Appl Microbiol 113(4):723–736
Lai R, Lomas LO, Jonczy J, Turner PC, Rees HH (2004) Two novel non-cationic defensin-like antimicrobial peptides from haemolymph of the female tick, Amblyomma hebraeum. Biochem J 379:681–685
Lakshminarayanan R, Vivekanandan S, Samy RP, Banerje Y, Chi-Jin EO et al (2008) Structure, self-assembly, and dual role of a beta-defensin-like peptide from the Chinese soft-shelled turtle eggshell matrix. J Am Chem Soc 130:4660–4668
Meng P, Yang S, Shen C, Jiang K, Rong M et al (2013) The first salamander defensin antimicrobial peptide. PLoS One 8(12):e83044
Nam BH, Moon JY, Kim YO, Kong HJ, Kim WJ (2010) Multiple β-defensin isoforms identified in early developmental stages of the teleost Paralichthys olivaceus. Fish Shellfish Immunol 28(2):267–274
Ruangsri J, Kitani Y, Kiron V, Lokesh J, Brinchmann MF et al (2013) A novel beta-defensin antimicrobial peptide in Atlantic cod with stimulatory effect on phagocytic activity. PLoS One 8(4):e62302
Schneider T, Kruse T, Wimmer R, Wiedemann I, Sass V et al (2010) Plectasin, a fungal defensin, targets the bacterial cell wall precursor Lipid II. Science 328(5982):1168–1172
Selsted ME, Harwig SS, Ganz T, Schilling JW, Lehrer RI (1985) Primary structures of three human neutrophil defensins. J Clin Invest 76(4):1436–1439
Seufi AM, Hafez EE, Galal FH (2011) Identification, phylogenetic analysis and expression profile of an anionic insect defensin gene, with antibacterial activity, from bacterial-challenged cotton leafworm Spodoptera littoralis. BMC Mol Biol 12:47
Van Dijk A, Veldhuizen EJA, Haagsman HP (2008) Avian defensins. Vet Immunol Immunopathol 124:1–18
von Horsten HH, Schafer B, Kirchhoff C (2004) SPAG11/isoform HEX, an atypical anionic beta-defensin like peptide. Peptides 25:1223–1233
Wen H, Lan X, Cheng T, He N, Shiomi K et al (2009) Sequence structure and expression pattern of a novel anionic defensin-like gene from silkworm (Bombyx mori). Mol Biol Rep 36(4):711–716
White SH, Wimley WC, Selsted ME (1995) Structure, function, and membrane integration of defensin. Curr Opin Struct Biol 5:521–527
Wu J, Liu H, Yang HL, Yu H, You D et al (2011) Proteomic analysis of skin defensive factors of tree frog Hyla simplex. J Proteome Res 10(9):4230–4240
Yang D, Biragyn A, Hoover DM, Lubkowski J, Openheim JJ (2004) Multiple role of antimicrobial defensins, cathelicidin, and eosinophil-derived neurotoxin in host defense. Annu Rev Immunol 22:181–215
Yang HL, Wang X, Liu XH, Wu J, Liu CB et al (2009) Antioxidant peptidomics reveals novel skin antioxidant system. Mol Cell Proteomics 8(3):571–583
Yenugu S, Hamil KG, Radhakrishnan Y, French FS, Hall SH (2004) The androgen-regulated epididymal sperm-binding protein, human beta-defensin 118 (DEFB118) (formerly ESC42), is an antimicrobial beta-defensin. Endocrinology 145:3165–3173
Zhang Z, Zhang B, Nie X, Liu Q, Xie F et al (2009) Transcriptome analysis and identification of genes related to immune function in skin of the Chinese brown frog. Zoolog Sci 26:80–86
Acknowledgments
This work was supported by Chinese National Natural Science Foundation (81373380, 81360253, 81260258, 81402830) and Jiangsu Province Foundation (BK20140362, 14KJD350003).
Conflict of interest
The authors have declared that no competing interests exist.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Handling Editor: J.-P. Bingham.
L. Wei and H. Che made equal contributions to this work.
Rights and permissions
About this article
Cite this article
Wei, L., Che, H., Han, Y. et al. The first anionic defensin from amphibians. Amino Acids 47, 1301–1308 (2015). https://doi.org/10.1007/s00726-015-1963-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00726-015-1963-8