Abstract
Hepatitis B virus (HBV) infection affects more than 2 billion people throughout the world. Among them, more than 240 million have chronic infection. Every year, 0.5–1.2 million people die of chronic hepatitis B virus infection (CHBVI), and approximately 60 % of liver cancers are related to CHBI and subsequent liver cirrhosis (LC). These HBVI-related diseases impose a considerable economic burden as well as morbidity on patients, families, and society. Family and twin studies have indicated that the host genetic constitution greatly influences the clinical outcomes of HBV infection. During the past several years, genome-wide association studies (GWAS) have identified susceptibility variants for various HBVI-related diseases. Of these variants, SNPs rs3077 and rs9277535 in HLA-DP on chromosome 6 show the strongest evidence for association with CHBVI and with viral clearance. However, whether there exists an association between HLA-DP variants and the progression of CHBVI remains to be determined. Thus, further study should focus not only on identifying more variants in HLA-DP that are associated with various HBVI-related diseases but also on characterizing any newly discovered functional variants at the molecular level. Further, given the complexity of CHBV infection and its progression, gene–gene and gene–environment interactions should also be taken into consideration. Moreover, because both smoking and alcohol affect HBV infection and progression, it is important to understand how these factors interact with genetics to influence HBV-related diseases.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hepatitis B virus (HBV) is one of the world’s most serious health problems, especially in Asian counties. Although a highly effective hepatitis B vaccine was introduced in 1982, chronic hepatitis B infection (CHBI) remains common, with more than 240 million people now affected (World Health Organization, 2013; http://www.who.int/mediacentre/factsheets/fs204/en/index.html).
The prevalence CHBI is about 5 % worldwide (Lai et al. 2003), but it shows large regional divergences, as well as population differences. Thus, such infection is endemic in sub-Saharan Africa and East Asia, with most people becoming infected during childhood and 5–10 % of the adult population being chronically infected. High CHBI rates also are found in the Amazon Basin and the southern parts of eastern and central Europe. About 2–5 % of the general population is chronically infected in the Middle East and the Indian subcontinent. In contrast, less than 1 % of the population of Western Europe and North America is affected (World Health Organization, 2013; http://www.who.int/mediacentre/factsheets/fs204/en/index.html). In the high-endemic regions, most affected people become infected in the perinatal or preschool period (Lok 2002; Gust 1996), which is associated with a very high probability of chronic infection (Chen et al. 2000). Most infections are acquired by horizontal transmission in early adult life in low-endemic areas (Gust 1996).
It is estimated that the worldwide mortality of CHBI is 0.5–1.2 million per year (Liaw and Chu 2009). Approximately 60 % of liver cancers are considered to be related to CHBI and subsequent liver cirrhosis (LC) (Lai et al. 2003). About 25 % of persons who acquire HBV as children develop cirrhosis or primary liver cancer as adults (Hsieh et al. 1992; Mahoney 1999). The typical progression of cirrhosis is from compensated to decompensated if treatment is not provided. Patients with decompensated cirrhosis would die without liver transplantation. Because of the high HBV-related morbidity and mortality rates, the economic burden of HBVI is severe. A survey conducted on patients with HBVI-related diseases who were hospitalized for 7 or more days in 13 representative hospitals in Shandong Province of China from April 2010 to November 2010 (Lu et al. 2013) showed, as expected, that the cost escalated with increasing severity of liver disease. It has been estimated that the direct cost in 2010 for CHBI, compensated cirrhosis, decompensated cirrhosis, and primary liver cancer was US$4,552, $7,400, $6,936, and $10,635, respectively. These costs ranged from 78.79 % (for CHBI) to 297.85 % (for primary liver cancer) of the average annual household income of the population, a huge economic burden for the family.
The clinical outcomes of HBVI, from spontaneous clearance to chronic infection that may go on LC and liver cancer, are influenced by viral, environmental, and host factors (Ganem and Prince 2004; Frodsham 2005). Family and twin studies indicate that host genetic constitution is an important factor (Lin et al. 1989). The human leukocyte antigen (HLA) allele DRB1*1302 was first reported to be associated with a protective effect for CHBI in Gambians (Thursz et al. 1995). Subsequent studies confirmed the involvement of other HLA genes in HBV clearance or persistence (Ahn et al. 2000; Thio et al. 1999, 2000). In addition, non-HLA genes involved in cytokine production, receptor binding, and immune regulation are also significantly associated with HBV infection. These factors include interferon-gamma and tumor necrosis factor (Ben-Ari et al. 2003), estrogen receptor alpha (Deng et al. 2004), vitamin D receptor (Bellamy et al. 1999), mannose-binding protein (Thomas et al. 1996), cytotoxic T-lymphocyte antigen 4 (Thio et al. 2004), and macrophage migration inhibitory factor (Zhang et al. 2013). However, the results of these candidate gene-based association studies are less conclusive in many cases.
In the past several years, genome-wide association study (GWAS) has been used increasingly to identify common genetic variants for complex human disorders such as HBV infection (Wellcome Trust Case Control Consortium 2007). To identify disease-predisposing variants, Kamatani et al. (2009) carried out a two-stage GWAS for CHB, which revealed that several single nucleotide polymorphisms (SNPs) in HLA-DP are associated with risk. Since then, many laboratories independently confirmed the association of HLA-DP variants with hepatitis B. Together, these studies have led to a new insight and substantial advancement in our understanding of HBV infection in human beings. Thus, the primary objective of this communication is to provide an updated view of recent progress in genetic association studies on the association of variants in HLA-DP with various HBV-related diseases.
Association of HLA-DP variants with chronic HBV infection and clearance
Using a two-stage GWAS approach, Kamatani et al. (2009) identified an association between CHB and 11 SNPs in HLA-DP loci in 786 Japanese CHB cases and 2,201 control individuals in the first discovery stage: rs2395309, rs3077, rs2301220, rs9277535, rs3117222, rs3128917, rs3135021, rs9380343, rs9277341, rs10484569, and rs2281388. Two of these SNPs, namely rs3077 in HLA-DPA1 and rs9277535 in HLA-DPB1, were the most significantly associated SNPs identified from the first discovery stage and were then selected for subsequent validation in three Japanese and Thai samples, which included two independent Japanese case–control samples comprising 274 cases and 274 controls as well as 718 cases and 1,280 controls and one Thai sample comprising 308 cases and 546 controls. A meta-analysis of these samples from both the discovery and replication stages revealed a P value of 2.31 × 10−38 for rs3077 with an odds ratios (OR) of 0.56, 95 % confidence interval (CI) of 0.51, 0.61; and of 6.34 × 10−39 for rs9277535 with OR of 0.57 and 95 % CI of 0.52, 0.62. In addition, after genotyping SNPs in the highly polymorphic exon 2 of HLA-DP, it was found that HLA-DPA1*0103, DPA1*0202, DPB1*0402, and DPB1*0501 were significantly associated with CHBI (P = 2.93 × 10−11, 4.45 × 10−8, 2.27 × 10−7, and 6.98 × 10−7, respectively) (Kamatani et al. 2009). Considering that haplotype and haplotype-based association analysis not only show how these alleles are organized along a chromosomal region but also provide more information than individual SNPs, especially when genotyped SNPs for a region of interest are limited, the authors performed haplotype analysis for 11 SNPs and variants in exon 2. Their analysis revealed a strong linkage disequilibrium (LD) among some of these 11 SNPs, and haplotypes DPA1*0103-DPB1*0402 and DPA1*0103-DPB1*0401 showed protective effects for CHBI (P = 6.00 × 10−8; OR = 0.52; 95 % CI 0.35, 0.75; and P = 0.002; OR = 0.57; 95 % CI 0.33, 0.96, respectively). In contrast, they found that haplotypes DPA1*0202-DPB1*0501 and DPA1*0202-DPB1*0301 were risk factors for CHBI (P = 5.79 × 10−6; OR = 1.45; 95 % CI 1.16, 1.81 and P = 0.002; OR = 2.31; 95 % CI 1.39, 3.84, respectively).
Since the first GWAS, which revealed a significant association of genetic variants in HLA-DP with CHBI, association of this genomic region on chromosome 6 with CHBVI has been replicated in a number of studies (Wong et al. 2013; Seto et al. 2013; Kim et al. 2013; Hu et al. 2013; Vermehren et al. 2012; Hu et al. 2012; Nishida et al. 2012; Mbarek et al. 2011; Li et al. 2011; Wang et al. 2011; Guo et al. 2011; An et al. 2011; Al-Qahtani et al. 2014). For example, after genotyping 11 SNPs in 521 chronic HBV carriers and 819 controls (571 individuals with HBV natural clearance and 248 healthy individuals), Guo et al. (2011) reported that this region was significantly associated with chronic HBV infection and clearance in the Han Chinese population from northern China. Moreover, they found that haplotype AACT (defined as block 1 in the original report), formed by the protective alleles of SNPs rs2395309, rs3077, rs2301220, and rs9277341 in HLA-DPA1, was significantly associated with a lower risk of chronic HBV infection (OR = 0.54; P = 8.73 × 10−7). In addition, they found that haplotypes GAGATT, all formed by the susceptibility minor alleles of SNPs rs9277535, rs10484569, rs3128917, rs2281388, rs3117222, and rs9380343, and GGGGTC, formed by the same SNPs with the minor alleles for rs9277535, rs3128917 and rs3117222 (defined as block 2), were significantly associated with a higher risk of chronic HBV infection (OR = 1.98, P = 1.37 × 10−10 and OR = 1.7, P = 0.002; respectively). Further, these two blocks were tested for their joint effects, which showed that the joint protective haplotypes AACT and AGTGCC from each block exerted a significant protective effect against chronic HBV infection. In addition, the joint haplotypes that included the protective haplotype AGTGCC from block 2 showed significant protective effects. From these results, we can infer that individuals with protective haplotypes should have a lower risk of chronic HBV infection.
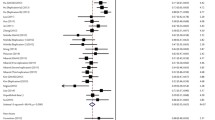
In addition, a number of studies have been conducted to investigate the association of rs3077 in HLA-DPA1 and rs9277535 in HLA-DPB1 with chronic HBV infection and spontaneous clearance in different populations (Table 1). Generally speaking, the “A” alleles of both rs3077 and rs9277535 have protective effects against CHBI and encourage HBV clearance in the Han Chinese population (Wang et al. 2011; Li et al. 2011), as does the “A” allele of rs2395309 (Li et al. 2011). Moreover, An et al. (2011) reported that the protective effect of the rs3077/A allele was mainly attributable to more efficient viral clearance (OR = 2.41; 95 % CI 1.83, 3.18; P = 4.65 × 10−10) rather than reduction of the possibility of infection (OR = 0.62; 95 % CI 0.47, 0.83; P = 0.0013). Additionally, Seto et al. (2013) reported that the probability of viral clearance was further reduced in younger patients; among patients aged ≤50 years, the OR of HBV clearance was 0.56 (P = 0.027) (Fig. 1), whereas rs3077 had no association with HBV clearance among patients aged ≥50 years (P = 0.617). Also, there was no difference between male and female patients in the association of rs3077 with HBV clearance (P = 0.091 and P = 0.190, respectively). In Asians, rs3077/A was the minor allele. On the contrary, in one study with 201 chronic HBV carriers and 235 healthy people, it was found that the rs3077/A allele was the major allele in a Caucasian population (Vermehren et al. 2012). In the HapMap project, frequencies of rs3077/A and rs9277535/A were higher in European populations than in the corresponding population from Asia. One possible reason for such a discrepancy is that the higher prevalence of HBV infection in Asia may be secondary to fewer protective alleles in HLA-DP. Nevertheless, Wang et al. (2011) reported that rs3077 was not significantly associated with CHBI in the Chinese Zhuang ethnic population (OR = 0.8; P = 0.206). A meta-analysis conducted by Yan et al. (2012) showed that rs3077 was significantly associated with CHBI (OR = 0.57; 95 % CI = 0.44, 0.75; P < 0.001). However, the sample size in the study of Wang et al. (2011) was small, with only 177 CHBI and 208 spontaneous clearance subjects included, so the study might not have had sufficient power to detect significance.
Odds ratios (OR) of HLA-DP rs3077/A in association with hepatitis B virus (HBV) clearance at different ages [Adopted from Seto et al. (2013)]
Association of HLA-DP variants with LC and hepatocellular carcinoma (HCC)
Although chronic HBV infection is an important factor in LC and HCC (Lai et al. 2003) and contributes to 70 % of the HCC in Asian and African populations (Llovet et al. 2003), only 0.06 % of persistent carriers develop HCC per year (Chen et al. 2010). So far, only four studies reported the relation between HLA-DP variants (rs3077, rs9277535, and rs2395309) and LC and HCC (An et al. 2011; Hu et al. 2012; Li et al. 2011; Nishida et al. 2012). Most of these studies reported that there is no association of HLA-DP variants with LC and HCC (An et al. 2011; Li et al. 2011; Nishida et al. 2012). Even though rs9277535 and rs2395309 were initially reported to be strongly associated with CHBI and HBV clearance in the initial GWAS (Li et al. 2011; Kamatani et al. 2009), when compared with asymptomatic HBV carriers, Li et al. (2011) revealed that these two SNPs were not associated with CHBI, LC, and HCC, indicating there was no association of these two SNPs in HLA-DP with progression of chronic HBV infection. In an independent study, Zhang et al. (2010) conducted GWAS for HCC in a Chinese population and identified a new susceptibility locus in the UBE4B-KIF1B-PGD region on 1p36.22. This region involves in numerous cellular functions such as proliferation, apoptosis, DNA repair, and other intracellular actions (Hoeller et al. 2006). Based on this work, it is likely that different genes play distinct roles in chronic HBV infection and progression in different populations. HLA-DP or other genes might be the primary cause of chronic HBV infection, but an intracellular pathway, such as ubiquitin or others, is mainly involved in the progression of chronic infection (Li et al. 2011). Unfortunately, such association of the genomic region on chromosome 1 has not been replicated in other studies (Li et al. 2012; Hu et al. 2012), and the exact reason for this discrepancy remains to be examined.
Regarding the variants on chromosome 6 reported by Kamatani et al. (2009), Hu et al. (2012) found that SNP rs3077 AG and AA genotypes significantly decreased host HCC risk (OR = 0.78; 95 % CI 0.67, 0.92; P = 3.63 × 10−3) when compared with persistent HBV carriers with GG genotype in dominant genetic models. But after meta-analysis by combining the results from An et al. (2011), this association becomes non-significant (Hu et al. 2012). Thus, further research, with a larger sample and in the same or different populations, needs to be conducted in order to demonstrate whether there exists any association of HLA-DP variants with the susceptibility and progression of chronic HBV infection.
Association of HLA-DP variants with the immune system
The outcomes of HBV infection depend mainly on the immune response. HLA class II molecules (HLA-DR, -DQ and -DP) encode proteins expressed on the surface of antigen-presenting cells and are crucial in that they present peptides to CD4+ helper T-cells (Pieters 2000). Although HLA-DP has a structure similar to that of other classical HLA class II molecules, the roles of HLA-DP molecules in the immune response have not been well characterized. Using single amino acid substitutions in HLA-DPB1*02012 to analyze the role of polymorphic residues of the HLA-DPB1 domain on peptide binding and T-cell allorecognition, Diaz et al. (2003) demonstrated that HLA-DPβ peptide residues 9, 11, 35, 55, 56, 69, and 84–87 play crucial roles in peptide binding and T-cell allorecognition, probably by influencing the binding of peptide in the groove of HLA-DPB and altering the conformation of the MHC–peptide complex which is recognized by T-cell receptor. However, there is still very limited information about peptide interactions and the roles of variants in HLA-DP both in peptide binding and in T-cell recognition. Because none of the identified 11 SNPs in HLA-DP on chromosome 6 leads to a change of amino acids in the encoded protein, it is likely that these 11 SNPs influence binding or presentation of viral peptides, resulting in either chronic HBV infection or viral clearance.
Up to now, the most convincing and well-investigated SNPs regarding chronic HBV infection and clearance are rs3077 and rs9277535, which are about 22 kb away from each other and located in the 3′-untranslated regions (UTRs) of HLA-DPA1 and HLA-DPB1 (Fig. 2), respectively. Because they are not located in the HLA-DP coding region, their biological effects could be realized through influencing the antigen-binding site indirectly, changing the expression of HLA-DP genes through altering some microRNA-binding sites, which leads to changes in the stability and translation of mRNA, and/or through altering transcription factor binding (An et al. 2011; O’Brien et al. 2011). These SNPs upregulate HLA-DPA1 and HLA-DPB1 expression in normal liver tissues (Figs. 3, 4) (O’Brien et al. 2011). High HLA-DPA1 and HLA-DPB1 expression might be more effective in presenting viral antigens to CD4+ helper T-cells, increasing the immune response to encourage HBV clearance and aiding in the clearance of chronic HBV infection. The reason these SNPs had no obvious impact on disease progression to cirrhosis or HCC remains to be explored.
Relative locations of SNPs rs3077 and rs9277535 in HLA-DPA1 and HLA-DPB1 loci on chromosome 6. The names of two SNPs are shown on the top, and their chromosomal positions are marked on the ruler in the middle. The HLA-DPA1 and HLA-DPB1 genes are shown as arrows at the bottom, with the exons shown as black boxes, introns as open boxes, and untranslated regions as gray boxes/arrows
Relationship of SNP rs3077 genotypes in HLA-DPA1 with HLA-DPA1 RNA expression level (a) and odds ratios (OR) for its association with chronic hepatitis B virus infection (b). Gene expression values represent the arithmetic increase in mean-log gene expression compared with the risk GG genotype, which has been set to zero as a reference [Adopted from O’Brien et al. (2011)]
Relationship of SNP rs9277535 in HLA-DPB1 with HLA-DPB1 RNA expression level (a) and odds ratios (OR) for its association with chronic hepatitis B virus infection (b). Gene expression values represent the arithmetic increase in mean-log gene expression compared with the risk GG genotype, which has been set to zero as a reference [adopted from O’Brien et al. (2011)]
Conclusion and future research directions
Both GWAS and candidate gene-based association studies have revealed significant association of variants in HLA-DP with viral clearance and chronic HBV infection. In addition, GWAS demonstrated that rs9277535 has a great impact in the response to booster hepatitis B vaccination in adolescents who received postnatal hepatitis B vaccination. The concordance of alleles associated with protective effects for both chronic infection and vaccine response indicates that these two HBV-related phenotypes share some major aspects of genetic etiologies, and the lesser effectiveness of the vaccine in those individuals carrying those risk alleles may attributable to their high likelihood of being infected with HBV (Wu et al. 2013). But whether there exists any association of HLA-DP variants with the progression of chronic HBV infection remains to be confirmed with larger samples and different origins of samples.
As reviewed here, most current researches focus mainly on SNPs rs3077 and rs9277535 in HLA-DP on chromosome 6. Because none of them can lead to an amino acid change, future study should focus on identifying more variants in this genomic region on chromosome 6 and illustrate how these variants affect gene expression and biological function of the product. A detailed elucidation of the molecular mechanism of HLA-DP variants and HBV infection will shed light on its pathogenesis and promote development of new therapies for HBV infection and prevention of disease progression.
Moreover, smoking and alcohol drinking are risk factors for HBV infection and progression (Ohnishi et al. 1987). How they contribute to such processes is rarely addressed in the reported studies; thus, if possible, these factors should be considered when one designs a new genetic study on HBV infection and analyzes this type of data, given the information available for the dataset of interest. Besides, gene–gene and gene–environment interactions should be taken into account. It is our hope that such comprehensive studies will lead to a better understanding of the involvement of variants within this genomic region on chromosome 6 in chronic HBV infection and progression and eventually lead to targeted and effective epidemic control strategies.
References
Ahn SH, Han KH, Park JY, Lee CK, Kang SW, Chon CY, Kim YS, Park K, Kim DK, Moon YM (2000) Association between hepatitis B virus infection and HLA-DR type in Korea. Hepatology 31(6):1371–1373. doi:10.1053/jhep.2000.7988
Al-Qahtani AA, Al-Anazi MR, Abdo AA, Sanai FM, Al-Hamoudi W, Alswat KA, Al-Ashgar HI, Khalaf NZ, Eldali AM, Viswan NA, Al-Ahdal MN (2014) Association between HLA variations and chronic hepatitis B virus infection in Saudi Arabian patients. PLoS ONE 9(1):e80445. doi:10.1371/journal.pone.0080445
An P, Winkler C, Guan L, O’Brien SJ, Zeng Z, Consortium HBVS (2011) A common HLA-DPA1 variant is a major determinant of hepatitis B virus clearance in Han Chinese. J Infect Dis 203(7):943–947. doi:10.1093/infdis/jiq154
Bellamy R, Ruwende C, Corrah T, McAdam KP, Thursz M, Whittle HC, Hill AV (1999) Tuberculosis and chronic hepatitis B virus infection in Africans and variation in the vitamin D receptor gene. J Infect Dis 179(3):721–724. doi:10.1086/314614
Ben-Ari Z, Mor E, Papo O, Kfir B, Sulkes J, Tambur AR, Tur-Kaspa R, Klein T (2003) Cytokine gene polymorphisms in patients infected with hepatitis B virus. Am J Gastroenterol 98(1):144–150. doi:10.1111/j.1572-0241.2003.07179.x
Chen CJ, Wang LY, Yu MW (2000) Epidemiology of hepatitis B virus infection in the Asia-Pacific region. J Gastroenterol Hepatol 15(Suppl):E3–E6
Chen JD, Yang HI, Iloeje UH, You SL, Lu SN, Wang LY, Su J, Sun CA, Liaw YF, Chen CJ, Risk Evaluation of Viral Load E, Associated Liver Disease/Cancer in HBVSG (2010) Carriers of inactive hepatitis B virus are still at risk for hepatocellular carcinoma and liver-related death. Gastroenterology 138(5):1747–1754. doi:10.1053/j.gastro.2010.01.042
Deng G, Zhou G, Zhai Y, Li S, Li X, Li Y, Zhang R, Yao Z, Shen Y, Qiang B, Wang Y, He F (2004) Association of estrogen receptor alpha polymorphisms with susceptibility to chronic hepatitis B virus infection. Hepatology 40(2):318–326. doi:10.1002/hep.20318
Diaz G, Amicosante M, Jaraquemada D, Butler RH, Guillen MV, Sanchez M, Nombela C, Arroyo J (2003) Functional analysis of HLA-DP polymorphism: a crucial role for DPbeta residues 9, 11, 35, 55, 56, 69 and 84-87 in T cell allorecognition and peptide binding. Int Immunol 15(5):565–576
Frodsham AJ (2005) Host genetics and the outcome of hepatitis B viral infection. Transpl Immunol 14(3–4):183–186. doi:10.1016/j.trim.2005.03.006
Ganem D, Prince AM (2004) Hepatitis B virus infection—natural history and clinical consequences. N Engl J Med 350(11):1118–1129. doi:10.1056/NEJMra031087
Guo X, Zhang Y, Li J, Ma J, Wei Z, Tan W, O’Brien SJ (2011) Strong influence of human leukocyte antigen (HLA)-DP gene variants on development of persistent chronic hepatitis B virus carriers in the Han Chinese population. Hepatology 53(2):422–428. doi:10.1002/hep.24048
Gust ID (1996) Epidemiology of hepatitis B infection in the Western Pacific and South East Asia. Gut 38(Suppl 2):S18–S23
Hoeller D, Hecker CM, Dikic I (2006) Ubiquitin and ubiquitin-like proteins in cancer pathogenesis. Nat Rev Cancer 6(10):776–788. doi:10.1038/nrc1994
Hsieh CC, Tzonou A, Zavitsanos X, Kaklamani E, Lan SJ, Trichopoulos D (1992) Age at first establishment of chronic hepatitis B virus infection and hepatocellular carcinoma risk. A birth order study. Am J Epidemiol 136(9):1115–1121
Hu L, Zhai X, Liu J, Chu M, Pan S, Jiang J, Zhang Y, Wang H, Chen J, Shen H, Hu Z (2012) Genetic variants in human leukocyte antigen/DP-DQ influence both hepatitis B virus clearance and hepatocellular carcinoma development. Hepatology 55(5):1426–1431. doi:10.1002/hep.24799
Hu Z, Liu Y, Zhai X, Dai J, Jin G, Wang L, Zhu L, Yang Y, Liu J, Chu M, Wen J, Xie K, Du G, Wang Q, Zhou Y, Cao M, Liu L, He Y, Wang Y, Zhou G, Jia W, Lu J, Li S, Liu J, Yang H, Shi Y, Zhou W, Shen H (2013) New loci associated with chronic hepatitis B virus infection in Han Chinese. Nat Genet 45(12):1499–1503. doi:10.1038/ng.2809
Kamatani Y, Wattanapokayakit S, Ochi H, Kawaguchi T, Takahashi A, Hosono N, Kubo M, Tsunoda T, Kamatani N, Kumada H, Puseenam A, Sura T, Daigo Y, Chayama K, Chantratita W, Nakamura Y, Matsuda K (2009) A genome-wide association study identifies variants in the HLA-DP locus associated with chronic hepatitis B in Asians. Nat Genet 41(5):591–595. doi:10.1038/ng.348
Kim YJ, Kim HY, Lee JH, Yu SJ, Yoon JH, Lee HS, Kim CY, Cheong JY, Cho SW, Park NH, Park BL, Namgoong S, Kim LH, Cheong HS, Shin HD (2013) A genome-wide association study identified new variants associated with the risk of chronic hepatitis B. Hum Mol Genet 22(20):4233–4238. doi:10.1093/hmg/ddt266
Lai CL, Ratziu V, Yuen MF, Poynard T (2003) Viral hepatitis B. Lancet 362(9401):2089–2094. doi:10.1016/S0140-6736(03)15108-2
Li J, Yang D, He Y, Wang M, Wen Z, Liu L, Yao J, Matsuda K, Nakamura Y, Yu J, Jiang X, Sun S, Liu Q, Jiang X, Song Q, Chen M, Yang H, Tang F, Hu X, Wang J, Chang Y, He X, Chen Y, Lin J (2011) Associations of HLA-DP variants with hepatitis B virus infection in southern and northern Han Chinese populations: a multicenter case–control study. PLoS ONE 6(8):e24221. doi:10.1371/journal.pone.0024221
Li S, Qian J, Yang Y, Zhao W, Dai J, Bei JX, Foo JN, McLaren PJ, Li Z, Yang J, Shen F, Liu L, Yang J, Li S, Pan S, Wang Y, Li W, Zhai X, Zhou B, Shi L, Chen X, Chu M, Yan Y, Wang J, Cheng S, Shen J, Jia W, Liu J, Yang J, Wen Z, Li A, Zhang Y, Zhang G, Luo X, Qin H, Chen M, Wang H, Jin L, Lin D, Shen H, He L, de Bakker PI, Wang H, Zeng YX, Wu M, Hu Z, Shi Y, Liu J, Zhou W (2012) GWAS identifies novel susceptibility loci on 6p21.32 and 21q21.3 for hepatocellular carcinoma in chronic hepatitis B virus carriers. PLoS Genet 8(7):e1002791. doi:10.1371/journal.pgen.1002791
Liaw YF, Chu CM (2009) Hepatitis B virus infection. Lancet 373(9663):582–592. doi:10.1016/S0140-6736(09)60207-5
Lin TM, Chen CJ, Wu MM, Yang CS, Chen JS, Lin CC, Kwang TY, Hsu ST, Lin SY, Hsu LC (1989) Hepatitis B virus markers in Chinese twins. Anticancer Res 9(3):737–741
Llovet JM, Burroughs A, Bruix J (2003) Hepatocellular carcinoma. Lancet 362(9399):1907–1917. doi:10.1016/S0140-6736(03)14964-1
Lok AS (2002) Chronic hepatitis B. N Engl J Med 346(22):1682–1683. doi:10.1056/NEJM200205303462202
Lu J, Xu A, Wang J, Zhang L, Song L, Li R, Zhang S, Zhuang G, Lu M (2013) Direct economic burden of hepatitis B virus related diseases: evidence from Shandong. China. BMC Health Serv Res 13:37. doi:10.1186/1472-6963-13-37
Mahoney FJ (1999) Update on diagnosis, management, and prevention of hepatitis B virus infection. Clin Microbiol Rev 12(2):351–366
Mbarek H, Ochi H, Urabe Y, Kumar V, Kubo M, Hosono N, Takahashi A, Kamatani Y, Miki D, Abe H, Tsunoda T, Kamatani N, Chayama K, Nakamura Y, Matsuda K (2011) A genome-wide association study of chronic hepatitis B identified novel risk locus in a Japanese population. Hum Mol Genet 20(19):3884–3892. doi:10.1093/hmg/ddr301
Nishida N, Sawai H, Matsuura K, Sugiyama M, Ahn SH, Park JY, Hige S, Kang JH, Suzuki K, Kurosaki M, Asahina Y, Mochida S, Watanabe M, Tanaka E, Honda M, Kaneko S, Orito E, Itoh Y, Mita E, Tamori A, Murawaki Y, Hiasa Y, Sakaida I, Korenaga M, Hino K, Ide T, Kawashima M, Mawatari Y, Sageshima M, Ogasawara Y, Koike A, Izumi N, Han KH, Tanaka Y, Tokunaga K, Mizokami M (2012) Genome-wide association study confirming association of HLA-DP with protection against chronic hepatitis B and viral clearance in Japanese and Korean. PLoS ONE 7(6):e39175. doi:10.1371/journal.pone.0039175
O’Brien TR, Kohaar I, Pfeiffer RM, Maeder D, Yeager M, Schadt EE, Prokunina-Olsson L (2011) Risk alleles for chronic hepatitis B are associated with decreased mRNA expression of HLA-DPA1 and HLA-DPB1 in normal human liver. Genes Immun 12(6):428–433. doi:10.1038/gene.2011.11
Ohnishi K, Terabayashi H, Unuma T, Takahashi A, Okuda K (1987) Effects of habitual alcohol intake and cigarette smoking on the development of hepatocellular carcinoma. Alcohol Clin Exp Res 11(1):45–48
Pieters J (2000) MHC class II-restricted antigen processing and presentation. Adv Immunol 75:159–208
Seto WK, Wong DK, Kopaniszen M, Proitsi P, Sham PC, Hung IF, Fung J, Lai CL, Yuen MF (2013) HLA-DP and IL28B polymorphisms: influence of host genome on hepatitis B surface antigen seroclearance in chronic hepatitis B. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America 56(12):1695–1703. doi:10.1093/cid/cit121
Thio CL, Carrington M, Marti D, O’Brien SJ, Vlahov D, Nelson KE, Astemborski J, Thomas DL (1999) Class II HLA alleles and hepatitis B virus persistence in African Americans. J Infect Dis 179(4):1004–1006. doi:10.1086/314684
Thio CL, Thomas DL, Carrington M (2000) Chronic viral hepatitis and the human genome. Hepatology 31(4):819–827. doi:10.1053/he.2000.4316
Thio CL, Mosbruger TL, Kaslow RA, Karp CL, Strathdee SA, Vlahov D, O’Brien SJ, Astemborski J, Thomas DL (2004) Cytotoxic T-lymphocyte antigen 4 gene and recovery from hepatitis B virus infection. J Virol 78(20):11258–11262. doi:10.1128/JVI.78.20.11258-11262.2004
Thomas HC, Foster GR, Sumiya M, McIntosh D, Jack DL, Turner MW, Summerfield JA (1996) Mutation of gene of mannose-binding protein associated with chronic hepatitis B viral infection. Lancet 348(9039):1417–1419
Thursz MR, Kwiatkowski D, Allsopp CE, Greenwood BM, Thomas HC, Hill AV (1995) Association between an MHC class II allele and clearance of hepatitis B virus in the Gambia. N Engl J Med 332(16):1065–1069. doi:10.1056/NEJM199504203321604
Vermehren J, Lotsch J, Susser S, Wicker S, Berger A, Zeuzem S, Sarrazin C, Doehring A (2012) A common HLA-DPA1 variant is associated with hepatitis B virus infection but fails to distinguish active from inactive Caucasian carriers. PLoS ONE 7(3):e32605. doi:10.1371/journal.pone.0032605
Wang L, Wu XP, Zhang W, Zhu DH, Wang Y, Li YP, Tian Y, Li RC, Li Z, Zhu X, Li JH, Cai J, Liu L, Miao XP, Liu Y, Li H (2011) Evaluation of genetic susceptibility loci for chronic hepatitis B in Chinese: two independent case–control studies. PLoS ONE 6(3):e17608. doi:10.1371/journal.pone.0017608
Wellcome Trust Case Control Consortium (2007) Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447(7145):661–678. doi:10.1038/nature05911
Wong DK, Watanabe T, Tanaka Y, Seto WK, Lee CK, Fung J, Lin CK, Huang FY, Lai CL, Yuen MF (2013) Role of HLA-DP polymorphisms on chronicity and disease activity of hepatitis B infection in Southern Chinese. PLoS ONE 8(6):e66920. doi:10.1371/journal.pone.0066920
Wu TW, Chu CC, Ho TY, Chang Liao HW, Lin SK, Lin M, Lin HH, Wang LY (2013) Responses to booster hepatitis B vaccination are significantly correlated with genotypes of human leukocyte antigen (HLA)-DPB1 in neonatally vaccinated adolescents. Hum Genet 132(10):1131–1139. doi:10.1007/s00439-013-1320-5
Yan Z, Tan S, Dan Y, Sun X, Deng G, Wang Y (2012) Relationship between HLA-DP gene polymorphisms and clearance of chronic hepatitis B virus infections: case–control study and meta-analysis. Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases 12(6):1222–1228. doi:10.1016/j.meegid.2012.03.026
Zhang H, Zhai Y, Hu Z, Wu C, Qian J, Jia W, Ma F, Huang W, Yu L, Yue W, Wang Z, Li P, Zhang Y, Liang R, Wei Z, Cui Y, Xie W, Cai M, Yu X, Yuan Y, Xia X, Zhang X, Yang H, Qiu W, Yang J, Gong F, Chen M, Shen H, Lin D, Zeng YX, He F, Zhou G (2010) Genome-wide association study identifies 1p36.22 as a new susceptibility locus for hepatocellular carcinoma in chronic hepatitis B virus carriers. Nat Genet 42(9):755–758. doi:10.1038/ng.638
Zhang K, Pan X, Shu X, Cao H, Chen L, Zou Y, Deng H, Li G, Xu Q (2013) Relationship between MIF-173 G/C polymorphism and susceptibility to chronic hepatitis B and HBV-induced liver cirrhosis. Cell Immunol 282(2):113–116. doi:10.1016/j.cellimm.2013.04.013
Acknowledgments
The preparation of this review was supported in part by the Ministry of Science and Technology of China (2012AA020405) and the National Natural Science Foundation of China grant 81273223.
Conflict of interest
The authors declare that they have no conflict of interest on this report.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jiang, X., Ma, Y., Cui, W. et al. Association of variants in HLA-DP on chromosome 6 with chronic hepatitis B virus infection and related phenotypes. Amino Acids 46, 1819–1826 (2014). https://doi.org/10.1007/s00726-014-1767-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00726-014-1767-2