Abstract
The understanding of physio-biochemical and molecular attributes along with morphological traits contributing to the salinity tolerance is important for developing salt-tolerant rice (Oryza sativa L.) varieties. To explore these facts, rice genotypes CSR10 and MI48 with contrasting salt tolerance were characterized under salt stress (control, 75 and 150 mM NaCl) conditions. CSR10 expressed higher rate of physio-biochemical parameters, maintained lower Na/K ratio in shoots, and restricted Na translocation from roots to shoots than MI48. The higher expression of genes related to the osmotic module (DREB2A and LEA3) and ionic module (HKT2;1 and SOS1) in roots of CSR10 suppresses the stress, enhances electrolyte leakage, promotes the higher compatible solute accumulation, and maintains cellular ionic homeostasis leading to better salt stress tolerance than MI48. This study further adds on the importance of these genes in salt tolerance by comparing their behaviour in contrasting rice genotypes and utilizing specific marker to identify salinity-tolerant accessions/donors among germplasm; overexpression of these genes which accelerate the selection procedure precisely has been shown.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Salinity is one among the major environmental stresses which negatively affects plant growth and productivity across the world. Globally, about one billion hectare area is affected by salt stress (Fageria et al. 2012). Crops are affected by multiple stresses due to salinity, including unbalanced nutrient uptake, accumulation of toxic ions, and oxidative and osmotic stresses (Wang et al. 2012; Singh et al. 2018). Rice (Oryza sativa L.) is an important food crop for billions of people worldwide. Rice-producing areas of the world are still acquiring most of their energy requirement from this crop and its derived products; hence, the demand for food will be increased by 38% in the next 30 years (Joseph et al. 2010). However, its growth and productivity are seriously influenced by soil salinity especially at the seedling stage (Darwish et al. 2009).
Salt tolerance is a complex trait that requires various biochemical and physiological responses by eliciting a gene network when exposed to salt stress (Wu et al. 2013). An elevated intracellular salt content disturbs ionic and osmotic balance which leads to metabolic disruption, growth retardation, and even death of the plant (Pandit et al. 2011). Inter-varietal differences in salt tolerance are mainly attributed to the capability of plants to control the amount of sodium reaching its sensitive leaf tissues or to metabolic reprogramming when roots are subjected to high salt content (Deinlein et al. 2014). Higher tolerance to increased Na is also often linked with these differences (Widodo et al. 2009). The concentrations of numerous metabolites, including proline, and polyols are also increased under salt stress, providing defense against osmotic challenge by serving as compatible solutes (Wu et al. 2013). Root cells mitigate the salinity effects by sequestering Na+ in the vacuole as an osmoticum, and showing a rationalizing nitrogen-containing compounds and sugars in the cytosol for osmotic adjustment and oxidative stress protection to provide plant viability (Annunziata et al. 2017). This induction (increasing concentration) of osmolytes is one of the key physiological markers for salt tolerance traits in rice and is a part of a holistic strategy for multiple abiotic stress defense (Liao et al. 2016). Sodium is absorbed by roots and translocated to shoots mainly through xylem (Deinlein et al. 2014). Salt-tolerant glycophytes have the ability to minimize Na accumulation in a shoot by excluding Na from the cytosol, and compartmentalize it into the vacuoles (Hasegawa 2013). The ability to exclude Na from its shoot and to maintain a lower cellular Na/K ratio are two pivotal traits that correlate highly with the salt tolerance of rice cultivars (Martinez-Atienza et al. 2007). Na transporters such as HKTs and NHXs together with reactive oxygen species (ROS) scavengers play a very important role in maintaining cellular homeostasis under salt stress and positively influence the ability of the plant to cope with increased salinity (Bharathkumar et al. 2016). Salt stress brings about subtle changes in the functioning of the electron transport chain in mitochondria and chloroplasts, disrupting the balance between ROS production and scavenging, thereby causing damage to the membranes, protein oxidation and DNA fragmentation (Mishra et al. 2011). Physio-biochemical mechanisms of salt tolerance have been extensively studied in various crops. Further, several important genes controlling these events are also known. However, their behavior has not been well studied in the contrasting genotypes which would have better defined their roles in salinity tolerance.
Salt stress tolerance is orchestrated by the complex array of molecular events that regulate the induction or repression of a wide range of salt stress-responsive genes (Passaia et al. 2013). Three groups of genes, group I (DREB2A, DREB2B, and LEA3), group II (HKT2;1, HKT1;5, NHX1, and SOS1), and group III (CATA and POX1) pertaining to the osmotic, ionic, and oxidative modules, respectively, play major roles in salt homeostasis and enable the plant to combat with increasing salinity. The expression of most of these genes has not or little been reported for the defensive response of rice to salinity stress. Hence, we selected these nine important genes for our study.
In this paper, we studied the expression of the main genes involved in the ionic, osmotic, and oxidative responses to salinity in two contrasting rice genotypes CSR10 (tolerant) and MI48 (sensitive). This is significant because we can utilize specific marker for these genes for a comprehensive screening and selection of rice germplasm for salinity tolerance and development and/or improvement of new salinity-tolerant accessions.
Materials and methods
Experimental setup and plant material
Two popular rice genotypes showing extremely contrasting salt stress response pattern, i.e., CSR10 (salt tolerant) and MI48 (salt sensitive), were selected for the study and seeds were obtained from the ICAR-Central Soil Salinity Research Institute, Karnal. The seeds of both the genotypes were placed in Petri dishes on moistened filter papers (soaked in 75 and 150 mM NaCl solutions as “salt treatment” and in the distilled water as “control”) for germination in the dark at 28 °C. Four days after sowing (DAS), uniformly germinated seeds were transferred into a hydroponic system with modified Yoshida solution (Yoshida et al. 1976) in the greenhouse. The hydroponic system was covered with perforated Styrofoam floats with a net bottom suspended on buckets that held the plants. The mean air temperature in the greenhouse was 28 and 25 °C during the day and night, respectively, with light intensity 270 μmol m−2 s−1, 14-h photoperiod, and 60% relative humidity.
The nutrient solutions were replaced every 4 days. Plants were grown for 15 days under normal conditions and then salinized with 75 and 150 mM NaCl solutions, which are equivalent to about ECiw 7.5 and 15 dS m−1, respectively. Nutrient solution without NaCl served as the control, whose electrical conductivity was 0 dS m−1. Samples were collected for phenotyping and RNA extraction on the third and sixth days after NaCl treatments. Tissues were subjected to flash freezing by quickly immersing it in liquid nitrogen for further analysis. The electrolyte leakage, chlorophyll content, relative water content, and ROS were measured on fresh weight basis while Na/K determination was done on leaf samples dried in hot air oven at 80 °C for 48 h. These parameters were recorded on both germination and seedling stages. The experiment was carried out in a completely randomized design. All experiments were performed in triplicate.
Germination percent and index
The germination percentage was calculated by the formula GP (%) = (S ∕ T) × 100, where S is the number of germinated seed and T is the total number of seeds sown. The germination index was calculated after complete germination using the following formula: GI = (germination percent in each treatment ∕ germination percent in control) × 100.
Growth measurements
The fresh weight (FW) and dry weight (DW) of leaves, stem, and root of both genotypes grown at ECiw 0, 75, and 150 mM NaCl were determined in three independent experiments. For measurement of DW, shoots and roots were oven dried at 80 °C for 48 h prior to being weighed. The relative reduction in growth caused by imposed salt stress was calculated from the ratio between performances under control and imposed salt stress conditions.
Assay of chlorophyll content
The total chlorophyll content of both genotypes was estimated under control conditions and on the third and sixth days after imposed salt stress (75 and 150 mM NaCl). Fresh leaf samples were accurately weighed and ground in a pestle and mortar using 5 ml of 80% acetone. The homogenate was filtered using Whatman filter paper I and filtrate was collected in a 25-ml volumetric flask. The volumetric flask was covered with aluminum foil to prevent photo-oxidation of pigments. The homogenate was washed three to four times with 80% acetone (5 ml each time) and the final volume was made up to 25 ml. The optical density of the extract was measured at two wavelengths (663 and 645 nm). The amount of chlorophyll (mg g−1 FW) was determined according to the formula given by Arnon (1949).
where
- A :
-
is the optical density at specific wavelengths;
- V :
-
is the final volume of 80% acetone (ml) and
- W :
-
is the fresh weight of tissue (g).
Electrolyte leakage ratio
The electrolyte leakage was estimated as per the protocol developed by Premachandra et al. (1990). Fresh leaf samples were taken from plants grown under control and salt stress condition. These leaves were then immersed in a test tube containing 20 ml of deionized water and kept overnight at room temperature. The electrical conductivity of the solution (EC1) was recorded with an EC meter (LMCM-20, Labman Scientific Instruments Pvt Ltd, Chennai, India). The sample was autoclaved to completely kill the tissues and release all electrolytes; then, final conductivity (EC2) was measured after cooling them to 25 °C. Electrolyte leakage ratio was calculated with the formula: EC1 ∕ EC2.
Proline concentration estimation
Fresh shoot tissues were collected from plants grown under control and salt stress conditions (75 and 150 mM) on third and sixth days. Proline content was determined using the protocol developed by Bates and Waklren (1973). Shoot tissues (100 mg) from both genotypes were ground in 10 ml of 3% sulphosalicylic acid and filtered through Whatman filter I. The clear supernatant (2 ml) was added to glacial acetic acid and acid ninhydrin. The resultant mixture was boiled at 100 °C for 1 h. The reaction was terminated on ice; later, 4 ml toluene was added to the reaction mixture with vigorous vortexing. The chromophore was then aspirated from the aqueous phase and the absorbance was read at 530 nm against toluene as blank. The proline concentration was calculated using a standard curve (l-proline) and expressed as micrograms per gram FW.
Histochemical detection of ROS
The young leaves collected from unstressed and stressed plants were immediately dipped in 0.1% w/v 3,3′-diaminobenzidine (DAB) prepared in 0.1 M sodium citrate buffer (pH 3.8) followed by vacuum infiltration in dark for 30 min. Leaves were then incubated in DAB solution in darkness for 24 h. Upon the appearance of brown/dark spots on leaves, they were bleached in ethanol:acetone (70:30 v/v) solution. Finally, they were observed and photographed (Thordal-Christensen et al. 1997).
Relative water content
Randomly selected shoot tissues from control and each treatment were sampled in polythene bags and sealed properly to minimize water loss. The midvein along with edge sections was cut out and the remaining tissue was weighed immediately and the FW was recorded. Then, the samples were hydrated to full turgidity by floating on deionized water in a closed Petri dish for 4 h at room temperature. After 4 hours, excess moisture was removed from the leaf surface and weighed immediately to obtain turgid weight (TW). The samples were further oven dried at 80 °C for 24 h and the dry weight (DW) was recorded. The RWC was calculated using the following formula: RWC = (FW − DW) ∕ (TW − DW) × 100.
Measurement of ion constituents
The rice plants selected from control and each treatment were separated into roots and shoots and washed repeatedly with tap water and finally with double-distilled water and then oven dried at 80 °C. Four replicates were sampled from each treatment for determining Na and K contents in the root and shoot tissues under control and imposed salt stress conditions. Dry root and shoot tissues were ground and 0.5 g samples were digested in diacid mixture (20 ml) containing NO3 and HClO4 acid (9:4 ratio) using a hot plate digestion system. After proper cooling, the digest was diluted with double-distilled water; filtered and final volume was made up to 50 ml and stored in polypropylene bottles. Elemental analysis of Na and K was performed using inductively coupled plasma emission spectroscopy (ICPE-9000, Shimadzu Europa GmbH, Duisburg, Germany).
RNA isolation and quantitative qRT-PCR
Total RNA was extracted from the root tissues of the control and salt-stressed plants using TRIzol reagent (Invitrogen, Carlsbad, CA). RNA (1 μg) was further digested with DNase I and was reverse transcribed to cDNA using a reverse transcription kit (Applied Biosystems). Expression analysis of salt-responsive genes was carried out using qRT-PCR (Supplementary file-Table S1). The qRT-PCR was performed using Fast SYBR Green Master Mix and an ABI Step-One Plus system (Applied Biosystems). qRT-PCR was conducted with three biological replicates and normalized using ubiquitin5 gene as an internal control. The relative expression level of the transcripts was calculated using the 2−ΔΔCT method. The sequences of the gene specific primers were taken from previous reports (Cai et al. 2015; Wang et al. 2015). All the primers used in this study are listed in supplementary file-Table S1.
Statistical methods
Data were subjected to principal component analysis (PCA) and statistical analysis using analysis of variance (ANOVA) and the means were tested by the Least Significant Difference (LSD) at P ≤ 0.05 using the SAS 9.3 software (SAS Institute Inc., Cary, USA).
Results
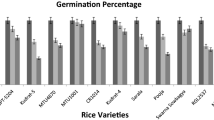
Effect of NaCl on germination percentage and germination index
A significant reduction was marked in germination and germination index of contrasting rice genotypes, i.e., CSR10 and MI48, with increasing salinity (Supplementary file-Table S2). The reduction in germination percent was more in MI48 (79.59%) than in CSR10 (31.63%) at 150 mM NaCl compared to the control conditions (Fig. 1). Similarly, least reduction in germination index was observed in CSR10 (18.55%) than in MI48 (27.55%) compared to control. This shows CSR10 can tolerate a higher degree of salinity than MI48 at seed germination stage.
Growth dynamics under salinity stress
The plant growth was significantly reduced with increasing salt stress (Supplementary file-Table S3). When during the early stages of growth, control plants were compared with plants under higher stress, i.e., 150 mM NaCl, we observed 27.54 and 42.86% reductions in root length and 20.28 and 40.35% reductions in shoot length of CSR10 and MI48, respectively. The deleterious effect of salt stress became more pronounced after a prolonged period of the stress, i.e., sixth day, as 23.74 and 60.47% reductions were observed in root length while 24.80 and 53.93% reductions were observed in shoot length of CSR10 and MI48, respectively. Clearly, the reduction in both root length and shoot length was higher in the sensitive variety MI48 than in the tolerant one CSR10 (Fig. 2(a, d)).
A similar trend was observed in the fresh weight as 37.32 and 51.51% reductions were observed in the root FW while 45.77 and 73.34% reductions were observed in shoot fresh weight during the early stage of stress in CSR10 and MI48, respectively, while we found 41.87 and 51.17% reductions in root FW and 34.02 and 69.53% reductions in shoot FW after a longer period of stress in CSR10 and MI48, respectively under similar conditions (Fig. 2(b, e)).
The salt-sensitive cultivar MI48 recorded greater reduction in root and shoot dry biomass as compared to the tolerant cultivar CSR10 as we found 15.40 and 36.12% reductions in root and 25.85 and 70.64% reductions in shoot dry weight of CSR10 and MI48 seedlings, respectively, on the third day of 150 mM salt stress. After the sixth day of the stress, we observed 40.53 and 72.59% reductions in root and 31.01 and 58.27% reductions in shoot dry weight of tolerant and sensitive cultivar, respectively (Fig. 2(c, f)), as compared to their respective controls.
Ionic profiling in root and shoot under salt stress
In order to observe the effect of salt stress on various ion accumulations, Na and K concentrations were determined in root and shoot of these two varieties under salt stress. Significant differences in ion accumulation were evident in these two contrasting genotypes after the third and sixth days of salt stress as compared to control (Supplementary file-Table S4). With the increasing salinity, Na concentration was increased with a simultaneous reduction in cellular K in root and shoot tissues of both the genotypes. Lower Na concentration was observed in roots of the tolerant cultivar CSR10 as compared to MI48 while in the shoot, a higher Na concentration was observed in CSR10 than in MI48 during normal growth. After mild salt stress (75 mM NaCl) root, Na concentrations were 43.80 and 37.40 mg/g DW in CSR10 and 53.50 and 74.10 mg/g DW in MI48 on the third and sixth days after NaCl stress, respectively. At the higher salt stress, Na concentration increased in both root (73.70, 85.40 mg/g DW) and shoot (58.62, 71.44 mg/g DW) of MI48 than in root (62.10, 73.30 mg/g DW) and shoot (45.60, 59.34 mg/g DW) of CSR10 seedlings, respectively (Fig. 3(a, d)). Although the percentage increase in Na content was higher in roots of CSR10 and in shoot of MI48 seedlings, yet the overall content of Na ions in CSR10 was lower in both roots and shoots.
Effects of salt stress on cellular ion and mineral accumulation in root and shoot Na concentration (a, d), K concentration (b, e), and Na/K ratio (c, f) in contrasting rice genotypes at the 3rd and 6th days of NaCl treatment, respectively. Each bar represents the average value of three biological replicates with standard error
A reduction in cellular K concentration in root and shoot of both genotypes was recorded with increasing levels of salt stress while K concentration was higher in the salt-tolerant genotype CSR10 than in the sensitive genotype MI48 under salt stress. We found initially a higher content of K in MI48 roots (78.90 and 53.36 mg/g DW) on the third and sixth days of salt stress treatment, respectively, as compared to CSR10 roots (56.70 and 46.20 mg/g DW), while no significant difference was observed in shoot K contents of both genotypes under control conditions (Fig. 3(b, e)). However, the percentage decrease in K concentration was higher in MI48 (72.14 and 91.60%) than in CSR10 (54.14 and 82.13%) roots on the third and sixth days of stress, respectively. Similarly, reduction in K concentration was higher in shoot tissues of MI48 (47.65% and 63.50%) than in shoot tissues of CSR10 (18.43 and 40.93%) on the third and sixth days of stress (Fig. 3(b, e)). This resulted in a higher Na/K ratio in MI48 than in CSR10 (Fig. 3(c, e)).
Physiological and biochemical modulations under salt stress
The percentage reduction in total chlorophyll concentration was more at the higher salinity levels compared to control. MI48 showed a significant reduction in chlorophyll concentration under the higher salt stress (55.99, 67.18%) than CSR10 (39.35, 41.63%) as compared to control plants on the third and sixth days of NaCl treatment, respectively (Fig. 4(a, e)). Relative water content was also drastically reduced in the salt-sensitive genotype MI48 (37.11, 65.80%) at 150 mM NaCl than in CSR10 (22.59, 53.65%) after the third and sixth days of stress, respectively (Fig. 4(b, f)). Further, electrolyte leakage was analyzed in order to assess the cell membrane integrity under salinity stress. We found an increase in electrolyte leakage in both CSR10 and MI48; however, the effect of salt stress on electrolyte was more in MI48 (81.81, 92.92%) than in CSR10 (30.91, 54.96%) on the third day and sixth day of stress, respectively. Hence, cell membrane stability was least affected by salinity in CSR10 (Fig. 4(c, g)).
Effects of salt stress on physio-biochemical parameters: total chlorophyll concentration (a, e), relative water content (b, f), electrolyte leakage (c, g), and proline concentration (d, h) in contrasting rice genotypes at third and sixth days of NaCl treatment, respectively. Each bar represents the average value of three biological replicates with standard error
Proline belongs to a very effective class of compatible solutes and a wide range of crop plants have been reported to accumulate this compound under abiotic stress like drought and salinity. Here, CSR10 accumulated lower proline than MI48. A higher increase in proline concentration was found in MI48 (258.18 and 416.13%) than in CSR10 (106.44 and 125.06%) as compared to control plants on the third and sixth days, respectively (Fig. 4(d, h)). The extent of accumulation of proline was higher in the sensitive genotype MI48 to survive against salt stress at higher salinity compared to respective controls.
In vivo deposition of H2O2 in shoot tissues of both the genotypes was studied under control and salt stress conditions. We performed an H2O2-specific histochemical analysis of salt-treated leaves (Fig. 5). No difference in 3,3′-diaminobenzidine staining intensity was observed between the two genotypes under control conditions. While, upon exposure to salt stress (75, 150 mM NaCl) for the third day, an increased staining was observed in leaves of MI48 as compared to CSR10. Further, under the extended duration of stress (6th day), an even stronger staining intensity was observed in the leaf tissues of MI48 than in CSR10.
Expression patterns of genes involved in salt tolerance in CSR10 and MI48
We studied the transcript levels of nine major genes playing significant roles in the salt stress signaling network at different salinity levels (Supplementary file-Table S5). The genes examined were categorized into three groups via group I (DREB2A, DREB2B, and LEA3), group II (HKT2;1, HKT1;5, NHX1, and SOS1), and group III (CATA and POX1) pertaining to the osmotic, ionic, and oxidative modules of salt stress tolerance, respectively (Fig. 6). At moderate salt stress (75 mM NaCl), the expression of group I genes (DREB2A, DREB2B, and LEA3) was higher in CSR10 (tolerant genotype) than in MI48 (susceptible genotype) at the third day of NaCl treatment, but no significant difference was observed after prolonged stress though the level of expression was higher under stress. This suggested MI48 is slow to respond and to maintain homeostasis, while this process is faster in CSR10.
Expression pattern of different salt responsive genes in CSR10 and MI48 rice roots after the 3rd and 6th days of NaCl treatment. qRT-PCR was used for quantification of gene expression. Each bar represents the average value of three independent biological replicates. Relative expression was calculated using MI48-unstressed plants taken as control at both the 3rd and 6th days after stress. Ubiquitin5 gene was taken as endogenous control. Error bar represents std. error among the biological replicates
Group II genes included (NHX1 and SOS1) also showed early and higher induction in CSR10 than in MI48. The expression of HKT1;5 was higher in MI48 during the early and prolonged period of stress. Interestingly, MI48 expressed significantly higher levels of HKT1;5 even in the control conditions as compared to the CSR10, and consequently, the expression of HKT1;5 gene lowers down after imposition of salt stress in CSR10 but it was induced in MI48 roots during early stages of stress. HKT2;1, another member of K transporter family, showed very high levels of expression in CSR10 but the expression levels remain low in MI48 roots during control and all the stress conditions.
Group III genes including CATA and POX1 which conditioned the oxidative module of salt tolerance also showed differential expression behaviour under the salt stress. CATA gene showed the higher expression in MI48 roots under salt stress while the expression levels were lower under control conditions as compared to CSR10 roots. Similarly, the POX1 gene was also exhibited a significantly higher expression in MI48 roots during prolonged stress and in CSR10 during the early phase of the stress. Therefore, it seems to be early salt stress responsive in CSR10 but late salt stress responsive in MI48 as the pattern becomes more differential after the sixth day of stress as compared to the early stage, i.e., third day of the stress.
Principal component analysis
Further data were also analyzed and presented using principal component analysis (PCA). The use of PCA can provide an indication of the most important traits and salt-responsive genes contributing to salinity tolerance in the rice and conditions under study. In Fig. 7, the distribution of the genotypes shows that PCA1 and PCA2 account for 80.65% of the total variability in the set of variables (traits and salt responsive genes) analyzed in each genotype. PCA1 accounts for 50.68% of the variability and it is strongly negatively correlated with the proline content (Shoot_Prol), shoot Na (Shoot_Na), shoot Na/K (Shoot_Na_K), root Na (Root_Na), root Na/K (Root Na_K), electrolyte leakage (Shoot_LE), and salt-responsive genes POX1, DREB2B, CATA, NHX1, CATB, and HKT1;5 (HKT1_5).
The principal component analysis to assess the importance of morpho-physiological traits and salt-responsive genes contributing to salinity tolerance. The traits used here are germination (G), germination index (GI), root length (RL), root fresh weight (RFW), root dry weight (RDW), root Na (Root_Na), root K (Root_K), root Na/K (Root Na_K), shoot length (SL), shoot fresh weight (SFW), shoot dry weight (SDW), shoot Na (Shoot_Na), shoot K (Shoot_K), shoot Na/K (Shoot_Na_K), shoot proline content (Shoot_Prol), electrolyte leakage (Shoot_LE), total chlorophyll (Shoot_TChl), relative water content (Shoot_RWC), and salt-responsive genes used here are DREB2B, SOS1, DREB2A, HKT2;1 (HKT2_1), LEA3, CATA, NHX1, CATB, POX1, and HKT1;5 (HKT1_5)
This is in contrast to germination (G), germination index (GI), root length (RL), root fresh weight (RFW), root dry weight (RDW), root K (Root_K), shoot length (SL), shoot fresh weight (SFW), shoot dry weight (SDW), shoot K (Shoot_K), relative water content (Shoot_RWC), total chlorophyll (Shoot_TChl), and salt-responsive genes SOS1, DREB2A, LEA3, and HKT2;1 (HKT2_1) which are positively correlated with PCA1. Therefore, these traits and salt-responsive genes are positively associated with salinity tolerance in rice. Figure 7 indicates that G, GI, RL, RFW, RDW, Root_K, SL, SFW, SDW, Shoot_K, Shoot_RWC, Shoot_TChl, and salt-responsive gene SOS1, DREB2A, LEA3, and HKT2_1 are the best discriminating parameters for PCA2, explaining 29.97% of the variability.
Discussion
NaCl stress delayed and reduced the germination in both the genotypes. This reduction in germination percentage and germination index was higher in the salt-sensitive genotype MI48 than in the tolerant genotype CSR10. It may be assumed that along with the toxic effects, a higher concentration of salt reduces the water potential in the medium which impedes the water absorption by germinating seeds during imbibition and seed turgescence stages and therefore represses the germination (Jamil et al. 2006). Excess intracellular Na accumulation results in osmotic and oxidative stresses, hence restraining cell elongation in root and shoot (Munns and Tester 2008). Shoot and root growth parameters showed a decreasing trend with increasing salinity level. CSR10 was able to maintain its root and shoot lengths at higher salinity than salt-sensitive genotype with increasing duration and severity of salt stress. Similarly, there was a considerable reduction in fresh and dry weights of shoot and root tissues in both the cultivars, but the extent of reduction was higher in salt-sensitive genotype MI48 than in CSR10. This reduction in fresh and dry weights might be due to poor CO2 fixation efficiency, the toxic effect of NaCl ions, osmotic imbalance, or disturbed metabolic activity under saline conditions (Turhan et al. 2008).
The ability to keep sodium away from the sensitive tissues like leaves is an important determinant of salt tolerance in both monocots and dicots (Munns and Tester 2008). In our study, the lower Na/K ratio in CSR10 under MI48 depicts a controlled uptake mechanism of Na in CSR10. In rice, Na can also enter the plant through the apoplastic pathway (Gong et al. 2006). The differences in this bypass conductance among the genotypes may also lead to the genotypic variations in salt tolerance (Krishnamurthy et al. 2011). Salt stress also reduced the cellular K concentration in root and shoot tissues of MI48 and CSR10, but this reduction was higher in MI48 than in CSR10. This clearly suggests a higher specificity index of K channels together with better Na partitioning ability in CSR10.
Salt stress-mediated decrease in total chlorophyll concentration may be due to toxic ion accumulation which affects the membrane stability and causes damage (Nawaz et al. 2010). This reduction in chlorophyll can be one of the major reasons for the decline in the fresh and dry weights of plant tissues, especially in the sensitive genotype, under saline condition. Physiological analysis revealed a much higher reduction in RWC of MI48 as compared to CSR10 under salt stress. The results suggest that CSR10 had better ability to avoid the salinity-induced osmotic stress than MI48. Such a greater reduction in RWC of salt sensitive has also been reported for salt-sensitive wheat cultivars (El-Bassiouny and Bekheta 2005) which reported a greater reduction in the RWC as compared with the tolerant ones under salt stress.
The cell membrane is another key target of many abiotic stresses and its maintenance of integrity is an important determinant of stress tolerance in plants. Here, CSR10 was found to restrict the amount of electrolyte leakage more efficiently than MI48. It is noteworthy here that protection of membrane damage under salt stress correlated with better salinity tolerance (Djanaguiraman et al. 2010). Osmolytes like proline contribute immensely towards protecting the membrane proteins and other enzymes apart from its basic role of osmoprotection (Ashraf and Foolad 2007). In our study, salt-sensitive genotype, MI48, accumulated higher levels of proline in its tissues under salt stress. Although it is commonly agreed upon the fact that proline accumulation is necessary for plants to tolerate abiotic stresses, a strong correlation between proline accumulation and salt tolerance is not always obvious (Ghars et al. 2008; Chen et al. 2007). However, proline accumulation is a symptom of injury rather than an indicator of stress tolerance in rice (Hoai et al. 2003). Nevertheless, the nature of proline accumulation under stress is still debated. Its higher accumulation is considered as a symptom or response to a stress or an adaptive mechanism, while plant salt tolerance is a complex trait, hence difficult to interpret (Lawlor 2013). However, proline has various crucial roles under stress conditions such as stabilizing membranes and proteins, scavenging ROS, or buffering cellular redox potential in addition to osmolyte. Moreover, it can stimulate the expression of salt stress-responsive genes that have proline-responsive elements (PRE) such as ACTCAT in their promoters (Ashraf and Foolad 2007; Carillo et al. 2011; Woodrow et al. 2017). The γ-aminobutyric acid and proline are accumulated conjointly in response to salt and water stresses (Li et al. 2016). They can be rapidly synthesized for cell protection against stress, mainly as osmolytes and ROS scavengers, and broken down upon relief of stress to provide energy, carbon, and nitrogen to recover and repair stress-induced damages (Hare and Cress 1997; Carillo et al. 2008; Carillo 2018).
The critical balance between production and scavenging of ROS gets disturbed by abiotic stresses like drought and salinity and this leads to a drastic increase in intracellular ROS levels which can cause significant damage to cellular structures. Moreover, higher H2O2 levels found in MI48 further confirm the higher stress level and low ROS scavenging capacity of the sensitive variety than of the tolerant one (Thordal-Christensen et al. 1997).
Salt-responsive transcriptional regulation is a common adaptive strategy of plants that mitigate the undesirable effects of high salt concentration by restoration of cellular ion homeostasis and osmotic balance (Mallikarjuna et al. 2011). Assessing the transcript abundance in response to salt stress can also give a reliable estimate of specific gene activation or downregulation (Liao et al. 2016). The expression patterns of three groups of genes: group I (DREB2A, DREB2B, and LEA3), group II (HKT2;1, HKT1;5, NHX1, and SOS1), and group III (CATA and POX1) pertaining to the osmotic, ionic, and oxidative modules of salt stress tolerance, respectively, were studied. DREB (dehydration-responsive element binding) genes are upregulated under various stresses and their overexpression resulted in greater tolerance to drought, salt, and cold stress treatments. They are thought to impart tolerance against these stresses by regulating the expression of certain stress-responsive genes like rd29A in Arabidopsis (Liu et al. 1998).
The LEA3 is another very important class of proteins that protect plants from damages caused by environmental stress by maintaining the structure of macromolecules and thereby protecting cells from dehydration (Chen et al. 2008; Oh et al. 2009; Matsukura et al. 2010; Hirayama and Shinozaki 2010; Singh et al. 2018). Six different DREB2 transcription factor-encoding genes have been reported in rice. However, only DREB2A and DREB2B genes have been reported to be abiotic stress inducible (Matsukura et al. 2010). Hence, we analyzed the expression pattern of DREB2A, DREB2B, and LEA3 genes in contrasting genotypes under varying levels of salt stress and found that these genes have higher expression levels in the tolerant genotype than in the sensitive one during the early phase of stress, and though they are induced even after a prolonged period of stress, the levels were same in both the genotypes. These results indicated that MI48 takes a longer period in order to activate the stress-responsive genes which might be a factor for late homeostasis and a higher degree of damage due to salt stress (Hirayama and Shinozaki 2010).
Phylogenetic analysis of HKTs (high-affinity K transporters) revealed two major subfamilies of HKTs in plants. These comprise of Na-selective transporters HKT1;x and HKT2;x which are permeable to both Na and K. About seven to nine such HKT transporters have been reported in rice (Hauser and Horie 2010). HKT2;1 which is permeable to both the ions exhibited very high expression in CSR10 as compared to MI48 both under control and stress conditions. The HKT1;5 gene, which belongs to the family-1, found highly induced in MI48 during both early and prolonged stress durations. These HKT transporters effectively regulate ascend of Na along the xylem sap, thus limiting Na levels in the sensitive shoot tissues (Munns et al. 2012). HKT2;1 has been identified as a central transporter mediating cellular ion homeostasis under salt stress and its transcription has been reported to be upregulated in the presence of excess salts (Horie et al. 2007; Hauser and Horie 2010).
The NHX1 is a vacuolar Na/H antiporter which confers Na extrusion and compartmentalization of Na in rice (Fukuda et al. 2004) while SOS1 is reported for sodium extrusion, controlling long-distance Na transport from root to shoot and withdrawing Na from the xylem tissues under higher salt stress (Shi et al. 2002; Martinez-Atienza et al. 2007). SOS1 encodes a Na/H antiporter and its overexpression resulted in the increased salt tolerance (Yang et al. 2009). There was a significant increase in transcript abundance of vacuolar antiporter gene NHX1 after getting subjected to moderate salt stress (75 mM) in both the genotypes and we observed higher expression in CSR10 than in MI48 during the early stage of the stress. Further, extended exposure to stress resulted in higher expression levels of the gene but no significant difference was observed in the contrasting genotypes. These results can be complemented with the K and Na concentrations observed in the tissues. It can be concluded that behaviour of these transporters is modulated by the tolerant genotypes so as to have a minimum accumulation of Na in the tolerant cultivar, while MI48 is not able to regulate these pathways as efficiently as CSR10.
Higher accumulation of Na in MI48 tissues cause more oxidative stress which can be reflected by the expression of CATA and POX1 genes. The expression level of CATA and POX1 was higher in salt-susceptible MI48 plants as compared to the tolerant CSR10 plants which suggest that MI48 plants are experiencing higher levels of ROS accumulation and they need more amount of catalase to establish the homeostasis with the stress (Passaia et al. 2013). It was also evident from the electrolyte leakage and proline accumulation which again confirms that MI48 plants are experiencing higher stress level than CSR10. Moreover, higher H2O2 levels found in MI48 further confirm the higher stress level in the sensitive variety than in the tolerant one.
The principal component analysis (PCA) can provide an idea of the most important traits and salt-responsive genes contributing to salinity tolerance in the rice and conditions under study (Chunthaburee et al. 2016; Negrão et al. 2017). Germination, germination index, root length, root fresh weight, root dry weight, root K, shoot length, shoot fresh weight, shoot dry weight, shoot K, relative water content, total chlorophyll, and salt-responsive genes SOS1, DREB2A, LEA3, and HKT2;1 are positively associated with salinity tolerance in rice. Hence, these should be considered as the best discriminating parameters for selection and breeding of rice for salinity tolerance.
Conclusion
The proportionately higher role of genes pertaining to osmotic and ionic modules of salt stress tolerance is clearly evident in imparting salinity tolerance to CSR 10. Interestingly, HKT 2;1 exhibited constitutive expression pattern coupled with higher induction of osmotic module (DREB2A and LEA3) and ionic module (SOS1) in CSR10 under moderate and very high stresses. The improved salt tolerance in CSR10 might also be attributed to higher RWC, cell membrane stability accompanied by increased quenching of intracellular ROS. Altogether, the specific upregulation of these transcripts might well be the trigger that led to improved growth, cellular ion homeostasis, and salt stress tolerance in CSR10. Further, we can utilize specific marker for these genes for a comprehensive screening and selection of rice germplasm for salinity tolerance showing overexpression of DREB2A and LEA3 (genes pertaining to the osmotic and signaling modules of salt stress tolerance) and HKT 2;1 and SOS1 (genes/sequences emphasize the better protection of cellular membranous network and membrane-bound macromolecules under salt stress), and development and/or improvement of new salinity-tolerant accessions.
Abbreviations
- ROS:
-
Reactive oxygen species
- LEA:
-
Late embryogenesis abundant
- HKT:
-
High-affinity K+ transporter
- NHX:
-
Vacuolar Na+/H+ antiporter
- DREB:
-
Dehydration-responsive element-binding protein
- SOS:
-
Salt overly sensitive
- CATA:
-
Catalase
- POX:
-
Peroxidase
- RWC:
-
Relative water content
- DAB:
-
3,3′-Diaminobenzidine
- FW:
-
Fresh weight
- DW:
-
Dry weight
References
Annunziata MG, Ciarmiello LF, Woodrow P, Maximova E, Fuggi A, Carillo P (2017) Durum wheat roots adapt to salinity remodeling the cellular content of nitrogen metabolites and sucrose. Front Plant Sci 7:2035
Arnon DI (1949) Copper enzymes in intact chloroplast. Polyphenoxidase in Beta vulgaris. Plant Physiol 24:1–15
Ashraf M, Foolad MA (2007) Improving plant abiotic-stress resistance by exogenous application of osmoprotectants glycine betaine and proline. Environ Exp Bot 59:206–216
Bates LS, Waklren RP (1973) Rapid determination of free proline for water stress studies. Plant Soil 39:205–207
Bharathkumar S, Jena PP, Kumar J, Baksh SKY, Samal R, Gouda G, Mukherjee M, Donde R, Vijayan J, Parida M, Reddy JN (2016) Identification of new alleles in salt tolerant rice germplasm lines through phenotypic and genotypic screening. Int J Agric Biol 18:441–448
Cai W, Liu W, Wang WS, Fu ZW, Han TT, Lu YT (2015) Overexpression of rat neurons nitric oxide synthase in rice enhances drought and salt tolerance. PLoS One 10:e0131599
Carillo P (2018) GABA shunt in durum wheat. Front Plant Sci 9:100
Carillo P, Mastrolonardo G, Nacca F, Parisi D, Verlotta A, Fuggi A (2008) Nitrogen metabolism in durum wheat under salinity: accumulation of proline and glycine betaine. Funct Plant Biol 35:412–426
Carillo P, Annunziata MG, Pontecorvo G, Fuggi A, Woodrow P (2011) Salinity stress and salt tolerance. In: Shanker A, Venkateswarlu B (eds) Abiotic stress in plants–mechanisms and adaptations. InTech, Croatia, pp 21–38
Chen Z, Cuin TA, Zhou M, Twomey A, Naidu BP (2007) Compatible solute accumulation and stress-mitigating effects in barley genotypes contrasting in their salt tolerance. J Exp Bot 58(15–16):4245–4255
Chen JQ, Meng XP, Zhang Y, Xia M, Wang XP (2008) Overexpression of OsDREB genes lead to enhanced drought tolerance in rice. Biotechnol Lett 30:2191–2198
Chunthaburee S, Dongsansuk A, Sanitchon J, Pattanagul W, Theerakulpisut P (2016) Physiological and biochemical parameters for evaluation and clustering of rice cultivars differing in salt tolerance at seedling stage. Saudi J Biol Sci 23:467–477
Darwish E, Testerink C, Khalil M, El-Shihy O, Munnik T (2009) Phospholipid signalling responses in salt-stressed rice leaves. Plant Cell Physiol 50(5):986–997
Deinlein U, Stephan AB, Horie T, Luo W, Xu G, Schroeder JI (2014) Plant salt-tolerance mechanisms. Trends Plant Sci 19:371–379
Djanaguiraman M, Prasad PV, Seppanen M (2010) Selenium protects sorghum leaves from oxidative damage under high temperature stress by enhancing antioxidant defense system. Plant Physiol Biochem 48(12):999–1007
El-Bassiouny HMS, Bekheta MA (2005) Effect of salt stress on relative water content, lipid peroxidation, polyamines, amino acids and ethylene of two wheat cultivars. Int J Agric Biol 7(3):363–368
Fageria NK, Stone LF, Santos ABD (2012) Breeding for salinity tolerance. In: Fritsche-Neto R, Borém A (eds) Plant breeding for abiotic stress tolerance. Springer-Verlag, Berlin, pp 103–122
Fukuda A, Nakamura A, Tagiri A, Tanaka H, Miyao A, Hirochika H, Tanaka Y (2004) Function, intracellular localization and the importance in salt tolerance of a vacuolar Na+/H+ antiporter from rice. Plant Cell Physiol 45(2):146–159
Ghars MA, Parre E, Debez A, Bordenave M, Richard L (2008) Comparative salt tolerance analysis between Arabidopsis thaliana and Thellungiella halophila, with special emphasis on K+/Na+ selectivity and proline accumulation. J Plant Physiol 165:588–599
Gong HJ, Randall DP, Flowers TJ (2006) Silicon deposition in the root reduces sodium uptake in rice (Oryza sativa L.) seedlings by reducing bypass flow. Plant Cell Environ 29:1970–1979
Hare PD, Cress WA (1997) Metabolic implications of stress-induced proline accumulation in plants. Plant Growth Regul 21:79–102
Hasegawa PM (2013) Sodium (Na+) homeostasis and salt tolerance of plants. Environ Exp Bot 92:19–31
Hauser F, Horie T (2010) A conserved primary salt tolerance mechanism mediated by HKT transporters: a mechanism for sodium exclusion and maintenance of high K+/Na+ ratio in leaves during salinity stress. Plant Cell Environ 33:552–565
Hirayama T, Shinozaki K (2010) Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant J 61:1041–1052
Hoai NTT, Shim IS, Kobayashi K, Kenji U (2003) Accumulation of some nitrogen compounds in response to salt stress and their relationships with salt tolerance in rice (Oryza sativa L.) seedlings. Plant Growth Regul 41:159–164
Horie T, Costa A, Kim TH, Han MJ, Horie R, Leung HY, Miyao A, Hirochika H, An G, Schroeder JI (2007) Rice OsHKT2;1 transporter mediates large Na+ influx component into K+-starved roots for growth. EMBO J 26:3003–3014
Jamil M, Lee DB, Jung KY, Ashraf M, Lee SC, Rhal ES (2006) Effect of salt (NaCl) stress on germination and early seedling growth of four vegetables species. J Cent Eur Agric 7:273–282
Joseph B, Jini D, Sujatha S (2010) Biological and physiological perspectives of specificity in abiotic salt stress response from various rice plants. Asian J Agric Sci 2:99–105
Krishnamurthy P, Ranathunge R, Nayak S, Schreiber L, Mathew MK (2011) Root barriers block Na+ traffic to shoots in rice (Oryza sativa L.). J Exp Bot 62:4215–4228
Lawlor D (2013) Genetic engineering to improve plant performance under drought. J Exp Bot 64:83–108
Li Z, Yu J, Peng Y, Huang B (2016) Metabolic pathways regulated by γ-aminobutyric acid (GABA) contributing to heat tolerance in creeping bentgrass (Agrostis stolonifera). Sci Rep 6:30338
Liao YD, Lin KH, Chen CC (2016) Oryza sativa protein phosphatase 1a (OsPP1a) involved in salt stress tolerance in transgenic rice. Mol Breed 36:22
Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought-and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10:1391–1406
Mallikarjuna G, Mallikarjuna K, Reddy MK, Kaul T (2011) Expression of OsDREB2A transcription factor confers enhanced dehydration and salt stress tolerance in rice (Oryza sativa L.). Biotechnol Lett 33:1689–1697
Martinez-Atienza J, Jiang X, Garciadeblas B, Mendoza I, Zhu JK, Pardo JM (2007) Conservation of the salt overly sensitive pathway in rice. Plant Physiol 143:1001–1012
Matsukura S, Mizoi J, Yoshida T, Todaka D, Ito Y, Maruyama K, Shinozaki K, Yamaguchi-Shinozaki K (2010) Comprehensive analysis of rice DREB2-type genes that encode transcription factors involved in the expression of abiotic stress-responsive genes. Mol Gen Genomics 283:185–196
Mishra P, Bhoomika K, Dubey RS (2011) Differential responses of antioxidative defense system to prolonged salinity stress in salt-tolerant and salt-sensitive Indica rice (Oryza sativa L.) seedlings. Protoplasma 250:3–19
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59:651–681
Munns R, James RA, Xu B, Athman A, Conn SJ, Jordans C, Byrt CS, Hare RA, Tyerman SD, Tester M (2012) Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nat Biotechnol 30:360–364
Nawaz K, Talat AI, Hussain K, Majeed A (2010) Induction of salt tolerance in two cultivars of sorghum (Sorghum bicolor L.) by exogenous application of proline at seedling stage. World Appl Sci J 10:93–99
Negrão S, Schmöckel SM, Tester M (2017) Evaluating physiological responses of plants to salinity stress. Ann Bot 119:1–11
Oh SJ, Kim YS, Kwon CW, Park HK, Jeong JS, Kim JK (2009) Overexpression of the transcription factor AP37 in rice improves grain yield under drought conditions. Plant Physiol 150:1368–1379
Pandit A, Rai V, Sharma TR, Sharma PC, Singh NK (2011) Differentially expressed genes in sensitive and tolerant rice varieties in response to salt stress. J Plant Biochem Biotechnol 20:149–154
Passaia G, Spagnolo FL, Caverzan A, Jardim-Messeder D, Christoff AP, Gaeta ML, de Araujo Mariath JE, Margis R, Margis-Pinheiro M (2013) The mitochondrial glutathione peroxidase GPX3 is essential for H2O2 homeostasis and root and shoot development in rice. Plant Sci 208:93–101
Premachandra GS, Saneoka H, Ogata S (1990) Cell membrane stability an indicator of drought tolerance as affected by applied N in soybean. J Agric Sci 115:63–66
Shi H, Quintero FJ, Pardo JM, Zhu JK (2002) The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 14:465–477
Singh J, Singh V, Sharma PC (2018) Elucidating the role of osmotic, ionic and major salt responsive transcript components towards salinity tolerance in contrasting chickpea (Cicer arietinum L.) genotypes. Physiol Mol Biol Plants 24(3):441–453. https://doi.org/10.1007/s12298-018-0517-4
Thordal-Christensen H, Zhang Z, Wei Y, Collinge DB (1997) Subcellular localization of H2O2 in plants, H2O2 accumulation in papillae and hypersensitive response during barley-powdery mildew interaction. Plant J 11:1187–1194
Turhan H, Genc L, Smith SE, Bostanci YB, Turkmen OS (2008) Assessment of the effect of salinity on the early growth stage of the common sunflower using spectral discrimination techniques. Afr J Biotechnol 7(6):750–756
Wang H, Zhang M, Guo R, Shi D, Liu B, Lin X, Yang C (2012) Effects of salt stress on ion balance and nitrogen metabolism of old and young leaves in rice (Oryza sativa L.). BMC Plant Biol 12:194
Wang R, Jing W, Xiao L, Jin Y, Shen L, Zhang W (2015) The rice high-affinity potassium transporter1;1 is involved in salt tolerance and regulated by an MYB-type transcription factor. Plant Physiol 168:1076–1090
Widodo JHP, Newbigin E, Tester M, Bacic A (2009) Metabolic responses to salt stress of barley (Hordeum vulgare L.) cultivars, Sahara and Clipper, which differ in salt tolerance. J Exp Bot 60:4089–4103
Woodrow P, Ciarmiello L, Annunziata MG, Pacifico S, Iannuzzi F, Mirto A, D’Amelia L, Dell’Aversana E, Piccolella S, Fuggi A, Carillo P (2017) Durum wheat seedling responses to simultaneous high light and salinity involve a fine reconfiguration of amino acids and carbohydrate metabolism. Physiol Plant 159(3):290–312
Wu D, Cai S, Chen M, Ye L, Chen Z, Zhang H (2013) Tissue metabolic responses to salt stress in wild and cultivated barley. PLoS One 8:e55431
Yang Q, Chen ZZ, Zhou XF, Yin HB, Li X, Xin XF, Hong XH, Zhu JK, Gong Z (2009) Overexpression of SOS (salt overly sensitive) genes increases salt tolerance in transgenic Arabidopsis. Mol Plant 2:22–31
Yoshida S, Forno DA, Cock JH, Gomez KA (1976) Laboratory manual for physiological studies of rice. International Rice Research Institute, Laos Banos
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Handling Editor: Néstor Carrillo
Electronic supplementary material
ESM 1
(DOCX 38 kb)
Rights and permissions
About this article
Cite this article
Singh, V., Singh, A.P., Bhadoria, J. et al. Differential expression of salt-responsive genes to salinity stress in salt-tolerant and salt-sensitive rice (Oryza sativa L.) at seedling stage. Protoplasma 255, 1667–1681 (2018). https://doi.org/10.1007/s00709-018-1257-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00709-018-1257-6