Abstract
Protein phosphatase 2Cs (PP2Cs) belong to the largest protein phosphatase family in plants. Some members have been described as being negative modulators of plant growth and development, as well as responses to hormones and environmental stimuli. However, little is known about the members of PP2C clade D, which may be involved in the regulation of signaling pathways, especially in response to saline and alkali stresses. Here, we identified 13 PP2C orthologs from the wild soybean (Glycine soja) genome. We examined the sequence characteristics, chromosome locations and duplications, gene structures, and promoter cis-elements of the PP2C clade D genes in Arabidopsis and wild soybean. Our results showed that GsPP2C clade D (GsAPD) genes exhibit more gene duplications than AtPP2C clade D genes. Plant hormone and abiotic stress-responsive elements were identified in the promoter regions of most PP2C genes. Moreover, we investigated their expression patterns in roots, stems, and leaves. Quantitative real-time PCR analyses revealed that the expression levels of representative GsPP2C and AtPP2C clade D genes were significantly influenced by alkali and salt stresses, suggesting that these genes might be associated with or directly involved in the relevant stress signaling pathways. Our results established a foundation for further functional characterization of PP2C clade D genes in the future.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Saline and alkali stresses are considered major threats to plant growth and development. Numerous studies have focused on the mechanisms of salt stress (Zhu 2001; Phang et al. 2008; Huang et al. 2012; Sanchita et al. 2014). However, only a few studies on alkali stress have been reported so far. Salt stress affects plant growth mainly by ionic poisoning and osmotic stress (Tuteja 2007). Alkaline stress, which is characterized by HCO3 −, CO3 2−, Na+, and high pH, imposes detrimental effects on plant cells (Alhendawi et al. 1997; Wang et al. 2007; Zhang et al. 2013). We previously identified some alkaline stress-responsive genes, such as GsTIFY10, using a high throughput RNA sequencing technique, and described their functions under alkaline stress (Zhu et al. 2011). Some PP2C genes were also found to be significantly upregulated under alkaline stress and may function as putative alkaline stress-responsive genes.
Protein phosphatase 2C proteins are Mg2+⁄Mn2+-dependent enzymes (MacKintosh et al. 1991). In the genomes of Arabidopsis thaliana and Oryza sativa, a total of 80 and 78 putative PP2C genes were identified, respectively (Singh et al. 2010). Based on multiple sequences alignments, the AtPP2C family can be divided into 10 or 13 subfamilies (Schweighofer et al. 2004; Xue et al. 2008). Some members have been described as modulators in response to developmental signaling cascades or environmental stimuli. For example, PP2Cs of clade A function as co-receptors of ABA signaling and play significant roles in ABA-dependent stress responses, including drought, cold, and salt (Christmann et al. 2006; Santiago et al. 2009; Park et al. 2009; Banerjee and Roychoudhury 2017). Some members of clade B have been demonstrated to be negative regulators of MAPK activities (Schweighofer et al. 2007). Members of clade C, including POLTERGEIST and POLTERGEIST-LIKE, participate in stem-cell identity and shoot and root meristem (Song and Clark 2005; Song et al. 2006). Whereas in clade E, AtPP2C52 can interact with UMP1 (proteasome maturation factor) and RD21a (cysteine proteinase), and AtPP2C6-6 can interact with a histone acetyltransferase GCN5 to regulate stress-responsive genes (Benhamed et al. 2006; Servet et al. 2008; Liu et al. 2013). However, besides a single report (Tovar-Mendez et al. 2014), little is known about PP2C clade D in Arabidopsis or other plants, especially in respond to stress responses until now.
Wild soybean (Glycine soja) exhibits very high adaptability to saline-alkali soil compared to cultivated soybean (Glycine max) (Ying et al. 2009). It can germinate and grow well in the saline-alkali soil (pH 9.02) and can survive in the Hoagland nutrient solution containing 50 mmol/L NaHCO3 at pH 8.5 (Ge et al. 2010). Previously, we identified some PP2C clade D genes in wild soybean as bicarbonate stress-responsive genes based on transcriptome sequencing data (DuanMu et al. 2015). In this study, we highlighted the important features of PP2C clade D genes in wild soybean and Arabidopsis, as well as the differences in evolution and divergence. In addition, we also examine the tissue-specific and alkali-salt stress-inducible expression patterns of the PP2C clade D genes in wild soybean and Arabidopsis.
Materials and methods
Sequence retrieval and identification
To assemble all members of the PP2C gene family in soybean, we performed a BLAST search on NCBI (http://www.ncbi.nlm.nih.gov/) and PHYTOZOME (http://phytozome.Jgi. doe.gov/pz/portal.html) using the G. max database. All putative non-redundant PP2C gene sequences were assessed with Pfam database and SMART (http://smart.embl-heidelberg.de/) analyses (Sonnhammer et al. 1997; Schultz et al. 2000). The sequences of Arabidopsis PP2C family genes were also obtained from PHYTOZOME.
Information regarding amino acid sequences and chromosome locations was obtained from PHYTOZOME. The isoelectric point (pI) and molecular weight (MW) were predicted by online softwares (http://au.expasy.org).
Phylogenetic analysis of PP2C family in wild soybean and Arabidopsis
Multiple sequence alignments of the full-length protein sequences were performed using MEGA 5.0. Phylogenetic trees were constructed by the neighbor-joining (NJ) method, with 1000 replicates of bootstrap analysis to support the values for each branch (Tamura et al. 2011).
Chromosomal location and gene duplication
Information about the chromosome locations of PP2C clade D genes was extracted from the PHYTOZOME database and NCBI. Gene locations were mapped on chromosomes using the MapInspect tool. Gene duplications were investigated according to two criteria: (1) the shorter sequence covers at least 80% of the longer sequence, and (2) the similarity of two genes was at least 70% (Gu et al. 2002; Yang et al. 2008).
Analyses of genetic structures and cis-acting elements
Genomic sequences and CDS sequences were obtained from the PHYTOZOME database and NCBI. Genetic structure maps were produced using Gene Structure Display Server (GSDS) online tools (Hu et al. 2015). The 3000-bp promoter sequences of GsPP2C clade D genes and 1500-bp promoter sequences of AtPP2C clade D genes were obtained from the G. soja sequence database (http://www.ncbi.nlm.nih.gov/assembly/ GCA) and the A. thaliana database (http://phytozome.Jgi. doe.gov/pz/portal.html) (Sun et al. 2014), respectively. Cis-acting elements were analyzed using PLANT CARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
Plant materials, growth conditions, and stress treatments
Wild soybean (07256) seeds were obtained from Jilin Academy of Sciences (Changchun, China). To promote germination, the seeds were treated with 98% sulfuric acid and shaken for 12–15 min, and then, were washed with distilled water for three times. The seeds were incubated in the dark for 1 day. Subsequently, the seeds were transferred to Hoagland’s nutrient solution at 23–28 °C and germinated under 16-h light/8-h dark environmental conditions. To explore the gene expression patterns, the roots, stems, and leaves of 21-day-old seedlings were harvested, respectively. For the alkali and salt treatments, the 21-day-old seedlings were transferred into Hoagland’s nutrient solution containing 50 mM NaHCO3 or 200 mM NaCl. Equal amounts of seedling roots were harvested at six time points (0, 1, 3, 6, 12, and 24 h), and the samples were snap-frozen in liquid nitrogen and stored at −80 °C.
The soybean (DN50) seeds were incubated and germinated in the dark for 2 days. Subsequently, the seeds were transferred to Hoagland’s nutrient solution at 23–28 °C with 16-h light/8-h dark environmental conditions. For the alkali and salt stress treatments, 15-day-old seedlings were transferred into Hoagland’s nutrient solution containing 50 mM NaHCO3 or 200 mM NaCl. Equal amounts of seedling roots were harvested at six time points (0, 1, 3, 6, 12, and 24 h), and the samples were snap-frozen in liquid nitrogen and stored at −80 °C.
Arabidopsis thaliana (Col-0) seedlings were grown in a greenhouse at 21–23 °C and with a 16-h light/8-h dark cycle. To investigate gene expression patterns, the roots, stems, and leaves of 18-day-old seedlings were harvested, respectively. For the gene expression analyses under alkali and salt stresses, the 12-day-old seedlings were treated with 10 mM NaHCO3 or 150 mM NaCl. Equal amounts of seedlings were harvested at six time points (0, 1, 3, 6, 12, and 24 h), and the samples were frozen in liquid nitrogen and stored at −80 °C.
Quantitative real-time PCR
Total RNA was isolated from wild soybean, soybean, and Arabidopsis materials using the EasyPure™ Plant Kit (TransGen, Beijing, China), and cDNAs was synthesized using the GoldScript cDNA kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. qRT-PCR assays were performed with SYBR Green Master Mix on an ABI 7500 sequencer according to the manufacturer’s protocols. ACTIN2 in Arabidopsis and GAPDH in wild soybean and soybean were used as internal controls (Czechowski et al. 2005; Huis et al. 2010). All experiments were performed with three independent biological replicates for statistical analyses. Statistical analyses were performed by one-way ANOVA with Bonferroni post-test or one-way ANOVA with Tukey’s pairwise comparison tests. The primers used for quantitative RT-PCR are listed in Supplemental Table S1.
Results
Identification of PP2C genes in cultivated soybean and PP2C clade D genes in wild soybean
Although the genome sequence data for wild soybean are available now, the genome annotation is still in progress. Because the wild and cultivated soybean species have a close genetic relationship, and their genomes share a high level of sequence similarity (Zeng et al. 2012; Chung et al. 2014), we employed the G. max database from NCBI and PHYTOZOME databases to identify the soybean PP2C genes. As a result, a total of 131 putative PP2C genes were obtained. Subsequently, 103 non-redundant soybean PP2C genes were extracted using the Pfam database and SMART online tool analysis. Simultaneously, 80 PP2C genes were obtained from Arabidopsis.
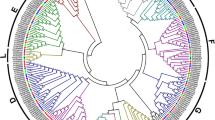
Using 103 GmPP2C and 80 AtPP2C full-length amino acid sequences, we performed phylogenetic analysis to construct a neighbor-joining phylogenetic tree. According to a previous study, AtPP2Cs were divided into 13 subfamilies. In this study, 10 soybean PP2C subfamilies (A, B, C, D, E, F1, F2, G, H, I) were classified, which is consistent with the previous study (Fig. 1) (Schweighofer et al. 2004). The soybean and Arabidopsis PP2Cs genes are closely related to each other within each subfamily. The GmPP2C clade D subfamily contains 14 genes according to the PP2C clade D nomenclature in Arabidopsis. Moreover, PP2C clade D in soybean and Arabidopsis has high bootstrap support, indicating that the genes in the same subfamily have a close evolutionary relationship. However, eight genes could not be clustered according to the nomenclature of the AtPP2C subfamily.
Phylogenetic relationship of PP2C families of soybean and Arabidopsis. Multiple sequence alignments were generated by using MEGA 5.0 with full-length protein sequences. The phylogenetic tree was constructed by the neighbor-joining (NJ) method, and the bootstrap values were based on 1000 replications for each branch. The frequency values > 50 (%) are shown. GsPP2Cs were divided into 10 subfamilies (A, B, C, D, E, F1, F2, G, H, and I) and marked by different shapes
We also obtained PP2C clade D gene sequences of wild soybean from www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi and http://www.ncbi.nlm.nih.gov/assembly/ GCA (Kim et al. 2010; Qi et al. 2014), by comparing to the sequences of soybean PP2C clade D genes. We found GsPP2C and GmPP2C clade D proteins share high sequence similarity (Fig. S1). Additionally, phylogenetic and gene structure analyses showed that these genes have reasonably high bootstrap support and share analogous exon/intron structures (Fig. S2). These results indicated that GsPP2C and GmPP2C clade D genes share a high level of sequence similarity.
Identification and analysis of the physical locations and duplications of PP2C clade D genes in wild soybean and Arabidopsis
To explore the basic properties of PP2C clade D proteins, the polypeptide lengths, theoretical MW, and pI were predicted (Table 1). The polypeptide lengths of GsPP2C clade D proteins range from 344 to 397aa, shorter than those of AtPP2C clade D proteins, which range from 370 to 400aa. Moreover, the theoretical MWs of GsPP2C clade D proteins are in the range of 38,198.54 to 44,137.56 Da, also smaller than those of AtPP2C clade D proteins, which range from 41,334.32 to 44,140.21 Da. The average pI of GsPP2C clade D proteins is 7.37, which is lower than that of AtPP2C clade D with an average pI at 7.91.
To determine the genomic distribution and duplication of GsPP2C clade D genes, their physical locations on the wild soybean chromosomes were determined using the soybean genome database (http://phytozome.jgi.doe.gov/pz/portal.html). The results demonstrated that 14 PP2C clade D genes were mapped to 10 chromosomes, and each chromosome was assigned to one or two genes (Fig. 2a). Moreover, 12 genes were mainly located on the edges of the chromosomes except for GsAPD4. Additionally, nine AtPP2C clade D genes were unevenly distributed among three chromosomes, with the exception of chromosomes 1 and 2. All genes were found to be located near the edges of the chromosomes, which is similar to the locations of GsPP2C clade D genes (Fig. 2b). In addition, we investigated gene duplication events. The results of this analysis showed that all GsPP2C clade D genes were linked to each other, whereas four paralogous genes were identified in AtPP2C clade D.
Physical locations and duplications of PP2C clade D genes in wild soybean (a) and Arabidopsis (b). The black bars represent the chromosomes and the scales represent megabases (Mb). The chromosome numbers are shown on the top of each chromosome representation. GsPP2C (a) and AtPP2C (b) genes are distributed on 10 and 3 chromosomes, respectively. The paralogous genes are identified and connected by purple lines
Phylogenetic and structure analyses of PP2C clade D genes in wild soybean and Arabidopsis
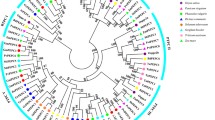
To gain insights into the evolutionary relationships and structural diversity of GsPP2C and AtPP2C clade D genes, we constructed a neighbor-joining phylogenetic tree and compared the gene structure between these two clades (Fig. 3a–b). The results suggested that these two clades have a close evolutionary relationship. For example, GsPP2C and AtPP2C clade D genes have a high bootstrap support, and they shared 48 to 76% sequence similarity. GsAPD6 and AtAPD3, AtAPD6 and AtAPD9, GsAPD12 and AtAPD1 and AtAPD4, GsAPD5 and AtAPD5, and GsAPD4 and AtAPD8 share high sequence similarities. Furthermore, almost all Arabidopsis clade D genes contain four exons and three introns, except for AtAPD4. However, GsPP2C clade D genes have longer sequences than AtPP2C clade D genes, although they share similar protein sequence lengths. Moreover, more paralogous pairs of GsPP2C clade D genes have high bootstrap support and share analogous exon/intron structures than AtPP2C clade D genes, which is consistent with the characteristics of genomic distribution and duplication.
Phylogenetic relationships and exon-intron structure analysis of PP2C clade D genes in wild soybean and Arabidopsis. a The phylogenetic tree was produced using MEGA 5.0 by the N-J method. Bootstrap values are based on 1000 replications for each branch. Pair of genes with bootstrap values higher than 98% were considered to be paralogues. b Structure analysis of PP2C clade D genes was performed using GSDS online tools. The untranslated regions (UTRs), CDSs, and introns are indicated by light blue boxes, yellow boxes, and black lines, respectively
Analyses of cis-acting elements of PP2C clade D gene promoters in wild soybean and Arabidopsis
Previous studies have suggested that hormone and abiotic stress-related cis-acting elements play active roles in response to stresses (Narusaka et al. 2003; Hernandez-Garcia and Finer 2014; Passricha et al. 2017). To elucidate whether PP2C clade D genes contain such cis-acting elements, we analyzed the PP2C clade D gene promoters in wild soybean (3 kb) and Arabidopsis (1.5 kb) using the PLANTCARE database. Our analyses demonstrated that almost all members carried putative stress-related cis-acting elements in their promoters, including ABA (ABRE), MeJA, ERE (ethylene), SA (Salicylic acid), GA (Gibberellin), Defense and Stress, HSE (heat), and MBS (drought)-responsive elements (Table 2). For example, all GsPP2C clade D genes contain HSE-responsive elements. All GsPP2C clade D genes contain defense and stress- and GA-responsive elements except for GsAPD12. In contrast, only 61.53% of GsPP2C clade D and 33.33% of AtPP2C clade D members have ERE-responsive elements. In all, these results suggested that both PP2C clade D genes may be responsive to abiotic stresses and hormone stimuli.
Expression patterns of PP2C clade D genes in wild soybean and Arabidopsis
Expression patterns are useful to assess the potential functions of a gene in certain developmental stages or under certain environmental conditions. To further understand the tissue-specific expression patterns of PP2C clade D genes, we examined the spatial expression levels of these genes in roots, stems, and leaves of Arabidopsis and wild soybean. Transcripts of all the clade D genes were detectible in these three organs of Arabidopsis and wild soybean (Fig. 4). Five GsPP2C clade D genes, including GsAPD5, -6, -11, -12, and -13, displayed high expression levels in roots. Eight AtPP2C clade D genes, AtAPD1, -2, -3, -4, -5, -6, -8, and -9, showed higher expression levels in roots than the other organs. The high expression of these genes in roots or in all three organs suggested that these genes may significantly play functional roles in developmental processes or under specific conditions. In addition, we identified some PP2C clade D genes shared highly correlated expression pattern in wild soybean or Arabidopsis using correlation analysis. We found 15 pairs of GsPP2C clade D genes showed a highly correlated expression pattern (r > 8), such as GsAPD2 and GsAPD8, GsAPD5 and GsAPD13, and GsAPD6 and GsAPD12 (Fig. S3a). Sixteen pairs of PP2C clade D genes showed a highly correlated expression pattern (r > 8) in Arabidopsis, such as AtAPD1 and AtAPD2, AtAPD1 and AtAPD3, and AtAPD2 and AtAPD8 (Fig. S3b). These results showed that these genes with highly correlated expression patterns may play similar roles in plant developmental processes.
Tissue-specific expression of PP2C clade D genes in wild soybean (a) and Arabidopsis (b). Total RNA samples were extracted from root, stems, and leaves of 21-day-old wild soybean and 18-day-old Arabidopsis plants. The relative expression levels were detected by qRT-PCR. GAPDH and ACTIN2 were used as internal controls in wild soybean and Arabidopsis, respectively. Statistical analyses were performed by one-way ANOVA followed by Tukey’s test. *P < 0.05, **P < 0.01
Expression analysis of PP2C clade D genes in wild soybean and Arabidopsis under alkaline treatment
Previously, we identified six GsPP2C clade D genes (GsAPD1, -5, -7, -10, -11, and -13) by transcriptome sequencing data and revealed that these PP2C genes were rapidly and significantly upregulated under alkaline treatment. To verify the data from the transcriptome sequencing analysis, we examined the expression patterns of these genes in wild soybean under alkaline treatment using qRT-PCR analysis (Fig. 5a). The gene expression patterns were roughly consistent with the transcriptome sequencing data, except that GsAPD7 had contrary results at 12 and 24 h. In addition, the expression of GsAPD1, -5, -10, and -13 increased and reached a maximum point at 1 h. GsAPD7 and GsAPD11 were obviously induced at 12 and 24 h, respectively.
Expression analysis of PP2C clade D genes in wild soybean (a) and Arabidopsis (b) under alkaline treatment. Total RNA samples were extracted from 21-day-old wild soybean seedlings under 50 mM NaHCO3 treatment and 12-day-old Arabidopsis seedlings under 10 mM NaHCO3 treatment, respectively. The relative expression levels were detected using qRT-PCR. GAPDH and ACTIN2 were used as internal controls in wild soybean and Arabidopsis, respectively. All experiments were performed in three independent biological replicates to enable statistical analyses. Statistical analyses were performed by one-way ANOVA with Bonferroni post-test. *P < 0.05, **P < 0.01, as compared with the control before treatment (0 h)
Similarly, the expression profiles of all AtPP2C clade D genes in Arabidopsis under alkali stress were also examined using qRT-PCR (Fig. 5b). Four genes (AtAPD5, -6, -7, and -9) showed differential expression. Interestingly, AtAPD6 and AtAPD7 were downregulated in response to alkali stress across all time points, and AtAPD5 and AtAPD9 were upregulated at different time points. In conclusion, our results showed that most of GsPP2C and AtPP2C clade D genes showed significant responses to alkali stress, providing the novel clues for us to better understand and to further dissect the mechanisms of alkali-related stresses at the cellular and molecular levels in plant.
Expression analyses of PP2C clade D in wild soybean and Arabidopsis under salt treatment
Plants growing on saline-alkali soils suffer from not only sodium toxicity but also from HCO3 − and CO3 2− toxicity. To determine whether PP2C clade D genes are responsive to salt stress, we assessed the expression levels of all clade D genes under salt stress (Fig. 6). All eight genes (GsAPD1, -2, -5, -7, -8, -10, -11, and -13) were upregulated at different time points under salt stress in wild soybean. However, only two genes (AtAPD4 and AtAPD7) appeared to be upregulated in Arabidopsis, and AtAPD7 was downregulated at 1, 3, 6, and 12 h, and then was upregulated at 24 h. These results indicate that more GsPP2C genes than AtPP2C genes in the clade D subfamily may participate in salt resistance and play important roles in response to salt stress.
Expression analysis of PP2C clade D genes in wild soybean (a) and Arabidopsis (b) under salt treatment. Total RNA samples were extracted from 21-day-old wild soybean seedlings and 12-day-old Arabidopsis seedlings under 200 mM NaCl and 150 mM NaCl treatment. qRT-PCR was performed to detect the relative expression levels. All experiments were performed in three independent biological replicates to enable statistical analysis. Statistical analyses were performed by one-way ANOVA with Bonferroni post-test. *P < 0.05, **P < 0.01, as compared with the control before treatment (0 h)
Discussion
Phosphatase 2C family proteins play key regulatory roles in multiple signaling transduction pathways related to diverse biological processes, especially abiotic stress responses (Fuchs et al. 2013). Previously, we found that six GsPP2C clade D genes were significantly upregulated under alkaline treatment by transcriptome sequencing data (DuanMu et al. 2015). However, little is known about members of PP2C clade D, except for their localization and possible roles in regulating PM H+-ATPase activity (Tovar-Mendez et al. 2014). In this study, we identified soybean PP2C clade D genes by investigating the relationship between the soybean and Arabidopsis PP2C protein families by phylogenetic analysis (Fig. 1). Then, we compared the sequences of GsPP2C clade D genes using GmPP2C clade D genes as references in the absence of wild soybean genome annotation.
In this study, to explore a genome-wide overview of the PP2C clade D subfamily, we compared the chromosome locations and duplications and gene structures in wild soybean and Arabidopsis and found that AtPP2C clade D genes underwent fewer gene duplications than GsPP2C clade D (Fig. 2), although a number of studies have showed that the Arabidopsis genome has undergone large-scale gene duplications and contains numerous large duplicated chromosomal segments (Blanc et al. 2003). First, all GsPP2C clade D genes had gene paralogs, compared with only four pairs of AtPP2C clade D gene paralogs. Second, more paralogous gene pairs of GsPP2C clade D genes have relatively high bootstrap support and share similar exon/intron structures compared to AtPP2C clade D (Fig. 3a–b). In plants, the gene duplication is an important evolutionary mechanism, resulting in novel gene generation, gene loss, and gene rearrangement, which helps plants to adapt to different and diverse environments (Bowers et al. 2003; Shang et al. 2013). Thus, our results indicated that the PP2C clade D genes expanded differently in wild soybean and Arabidopsis, and suggested that the wild soybean genome is a result of large-scale gene duplications to adapt to different environmental conditions.
There are 15 and 16 pairs of PP2C clade D genes which exhibited similar expression patterns in wild soybean and Arabidopsis, respectively (Fig. S3a–b). Some paralogous pairs of PP2C clade D genes also exhibited similar expression patterns in wild soybean and Arabidopsis, such as GsAPD5 and GsAPD13, GsAPD6 and GsAPD12, and AtAPD3 and AtAPD9, indicating that they may play similar roles in plant developmental processes. However, eight of the AtPP2C clade D genes displayed higher expression in roots than the other organs, whereas only five GsPP2C clade D genes showed observable expression in roots. In particular, GsAPD10 displayed high expression in stem, indicating that it may play special roles in stem. In total, these results clearly suggested that AtPP2C clade D genes may mainly play functional roles in roots under specific environmental conditions. Additionally, GsPP2C and AtPP2C clade D genes may play different functional roles at certain developmental processes or under different stresses based on their different expression patterns.
Previous reports have illustrated that SAUR proteins can interact with some AtPP2C clade D proteins to regulate PM H+-ATPase activity, which plays regulatory roles in cell expansion, intracellular pH, salinity, and alkalinity (Rober-Kleber et al. 2003; Gevaudant et al. 2007; Spartz et al. 2014; Li et al. 2016). We screened putative cis-acting elements in the promoter regions of wild soybean and Arabidopsis PP2C clade D genes and found that most of them contain plant hormone-responsive elements and stress-responsive elements (Table 2 and Table S2). Subsequently, we determined the expression patterns PP2C clade D genes under salt and alkali stresses in wild soybean and Arabidopsis. The results demonstrated that six (GsAPD1, -5, -7, -10, -11, and -13) and four (AtAPD5, -6, -7, and -9) genes responded to alkali stress in wild soybean and Arabidopsis, respectively (Fig. 5). And eight (GsAPD1, -2, -5, -7, -8, -10, -11, and -13) and two (AtAPD4 and AtAPD7) genes responded to salt stress in wild soybean and Arabidopsis, respectively (Fig. 6). These results indicate that both GsPP2C and AtPP2C clade D subfamily genes may play important roles in response to alkali and salt stresses.
In addition, the data show that much more PP2C clade D genes from wild soybean than Arabidopsis were significantly regulated under salt and alkali stresses, suggesting that the wild soybean genome has undergone more large-scale gene duplications than the Arabidopsis genome, which might result in better adaptability of wild soybean to environmental stresses. Moreover, AtPP2C clade D genes displayed more variable expression patterns compared to GsPP2C clade genes, which were all upregulated. For example, AtAPD5 and AtAPD9 showed significant upregulation, whereas, AtAPD6 and AtAPD7 were downregulated under alkali stress, indicating AtPP2C clade D genes may participate in different signaling pathways compared with GsPP2C clade D genes. In our previous studies, we found that GsCML27 plays opposite roles in plant responses to alkali and salt stresses, even though it is upregulated under both alkali and salt treatments (Chen et al. 2015). GsACA1 overexpression in alfalfa increased plant tolerance to both alkaline and salt stresses (Sun et al. 2016). In this study, we also found that some genes were induced by both alkali and salt stresses, such as GsAPD1, GsAPD5, GsAPD7, GsAPD10, and AtAPD7, and these genes may have important functions under alkali and salt stresses. However, further functional characterization of PP2C clade D genes is required to understand the molecular mechanisms of their responses to saline and alkali stresses.
Wild soybean is the ancestor of the cultivated soybean, and wild soybean exhibits high adaptability to saline-alkali soil compared to the cultivated species (Ying et al. 2009). In this study, we extracted the sequences of GsPP2C clade D genes using GmPP2C clade D genes as references because they have high sequence homology. Therefore, it is worthy to discuss the PP2C clade D genes in soybean and wild soybean. We found that GsPP2C and GmPP2C clade D genes not only shared high protein sequence similarity but also had high bootstrap support and shared analogous exon/intron structures (Figs. S1-S2). These results are consistent with the previous studies, indicating that there is only 0.31% genomic difference between wild soybean and domesticated soybean (Kim et al. 2010).
We also assessed the expression levels of four GmPP2C clade D genes (GmAPD1, 5, 7, and 10) under alkali and salt stresses (Fig. S4), and found that all were upregulated in wild soybean. Although these genes were upregulated under alkali and salt stresses, they have some different expression patterns compared with GsPP2C clade D genes (GsAPD1, 5, 7, and 10). For example, GmAPD1 showed no response to alkali stress, whereas GsAPD1 was obviously induced at 1, 3, 6, and 24 h under alkali stress. GsAPD7 was induced and reached a maximum point at 6 h, whereas GmAPD7 was induced and reached a maximum point at 12 h. We demonstrated that all GmPP2C clade D genes harbored putative stress-related cis-acting elements in their promoters, but there were some different stress-related cis-acting elements in GsPP2C clade D genes (Table S3). Overall, these results indicated that some GsPP2C and GmPP2C clade D genes may participate in different signaling pathways. This may be consistent with the differences in adaptability to stress tolerance mechanisms between wild soybean and cultivated soybean (Zhang et al. 2016).
References
Alhendawi R, Romheld V, Krikby E, Marschner H (1997) Influence of increasing bicarbonate concentrations on plant growth, organic acid accumulation in roots and iron uptake by barley, sorghum, and maize. J Plant Nutr 20(12):1731–1753
Banerjee A, Roychoudhury A (2017) Abscisic-acid-dependent basic leucine zipper (bZIP) transcription factors in plant abiotic stress. Protoplasma 254(1):3–16. https://doi.org/10.1007/s00709-015-0920-4
Benhamed M, Bertrand C, Servet C, Zhou DX (2006) Arabidopsis GCN5, HD1, and TAF1/HAF2 interact to regulate histone acetylation required for light-responsive gene expression. Plant Cell 18(11):2893–2903. https://doi.org/10.1105/tpc.106.043489
Blanc G, Hokamp K, Wolfe KH (2003) A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res 13(2):137–144. https://doi.org/10.1101/gr.751803
Bowers JE, Chapman BA, Rong JK, Paterson AH (2003) Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422(6930):433–438. https://doi.org/10.1038/nature01521
Chen C, Sun X, Duanmu H, Zhu D, Yu Y, Cao L, Liu A, Jia B, Xiao J, Zhu Y (2015) GsCML27, a gene encoding a calcium-binding Ef-hand protein from Glycine soja, plays differential roles in plant responses to bicarbonate, salt and osmotic stresses. PLoS One 10(11):e0141888. https://doi.org/10.1371/journal.pone.0141888
Christmann A, Moes D, Himmelbach A, Yang Y, Tang Y, Grill E (2006) Integration of abscisic acid signalling into plant responses. Plant Biol 8(3):314–325. https://doi.org/10.1055/s-2006-924120
Chung WH, Jeong N, Kim J, Lee WK, Lee YG, Lee SH, Yoon W, Kim JH, Choi IY, Choi HK, Moon JK, Kim N, Jeong SC (2014) Population structure and domestication revealed by high-depth resequencing of Korean cultivated and wild soybean genomes. DNA Res 21(2):153–167. https://doi.org/10.1093/dnares/dst047
Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139(1):5–17. https://doi.org/10.1104/pp.105.063743
DuanMu H, Wang Y, Bai X, Cheng S, Deyholos MK, Wong GK, Li D, Zhu D, Li R, Yu Y, Cao L, Chen C, Zhu Y (2015) Wild soybean roots depend on specific transcription factors and oxidation reduction related genesin response to alkaline stress. Funct Integr Genomics 15(6):651–660. https://doi.org/10.1007/s10142-015-0439-y
Fuchs S, Grill E, Meskiene I, Schweighofer A (2013) Type 2C protein phosphatases in plants. FEBS J 280(2):681–693. https://doi.org/10.1111/j.1742-4658.2012.08670.x
Ge Y, Li Y, Zhu YM, Bai X, Lv DK, Guo DJ, Ji W, Cai H (2010) Global transcriptome profiling of wild soybean (Glycine soja) roots under NaHCO3 treatment. BMC Plant Biol 10:153. https://doi.org/10.1186/1471–2229–10-153
Gevaudant F, Duby G, von Stedingk E, Zhao R, Morsomme P, Boutry M (2007) Expression of a constitutively activated plasma membrane H+-ATPase alters plant development and increases salt tolerance. Plant Physiol 144(4):1763–1776. https://doi.org/10.1104/pp.107.103762
Gu Z, Cavalcanti A, Chen FC, Bouman P, Li WH (2002) Extent of gene duplication in the genomes of Drosophila, nematode, and yeast. Mol Biol Evol 19(3):256–262
Hernandez-Garcia CM, Finer JJ (2014) Identification and validation of promoters and cis-acting regulatory elements. Plant Sci 217-218:109–119. https://doi.org/10.1016/j.plantsci.2013.12.007
Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G (2015) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31(8):1296–1297. https://doi.org/10.1093/bioinformatics/btu817
Huang GT, Ma SL, Bai LP, Zhang L, Ma H, Jia P, Liu J, Zhong M, Guo ZF (2012) Signal transduction during cold, salt, and drought stresses in plants. Mol Biol Rep 39(2):969–987. https://doi.org/10.1007/s11033-011-0823-1
Huis R, Hawkins S, Neutelings G (2010) Selection of reference genes for quantitative gene expression normalization in flax (Linum usitatissimum L.) BMC Plant Biol 10:71. https://doi.org/10.1186/1471–2229–10-71
Kim MY, Lee S, Van K, Kim TH, Jeong SC, Choi IY, Kim DS, Lee YS, Park D, Ma J, Kim WY, Kim BC, Park S, Lee KA, Kim DH, Kim KH, Shin JH, Jang YE, Kim KD, Liu WX, Chaisan T, Kang YJ, Lee YH, Kim KH, Moon JK, Schmutz J, Jackson SA, Bhak J, Lee SH (2010) Whole-genome sequencing and intensive analysis of the undomesticated soybean (Glycine soja Sieb. and Zucc.) genome. Proc Natl Acad Sci U S A 107(51):22032–22037. https://doi.org/10.1073/pnas.1009526107
Li Z, Zhen Z, Guo K, Harvey P, Li J, Yang H (2016) MAPK-mediated enhanced expression of vacuolar H(+)-ATPase confers the improved adaption to NaCl stress in a halotolerate peppermint (Mentha piperita L.) Protoplasma 253(2):553–569. https://doi.org/10.1007/s00709-015-0834-1
Liu H, Tsugama D, Liu S, Takano T (2013) Functional analysis of a type-2C protein phosphatase (AtPP2C52) in Arabidopsis thaliana genomics and applied. Biology 4(1):1–7
MacKintosh C, Coggins J, Cohen P (1991) Plant protein phosphatases. Subcellular distribution, detection of protein phosphatase 2C and identification of protein phosphatase 2A as the major quinate dehydrogenase phosphatase. Biochem J 273(Pt 3):733–738
Narusaka Y, Nakashima K, Shinwari ZK, Sakuma Y, Furihata T, Abe H, Narusaka M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J 34(2):137–148. https://doi.org/10.1046/j.1365-313X.2003.01708.x
Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, Alfred SE, Bonetta D, Finkelstein R, Provart NJ, Desveaux D, Rodriguez PL, McCourt P, Zhu JK, Schroeder JI, Volkman BF, Cutler SR (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324(5930):1068–1071. https://doi.org/10.1126/science.1173041
Passricha N, Saifi S, Ansari MW, Tuteja N (2017) Prediction and validation of cis-regulatory elements in 5' upstream regulatory regions of lectin receptor-like kinase gene family in rice. Protoplasma 254(2):669–684. https://doi.org/10.1007/s00709-016-0979-6
Phang TH, Shao G, Lam HM (2008) Salt tolerance in soybean. J Integr Plant Biol 50(10):1196–1212. https://doi.org/10.1111/j.1744-7909.2008.00760.x
Qi X, Li MW, Xie M, Liu X, Ni M, Shao G, Song C, Kay-Yuen Yim A, Tao Y, Wong FL, Isobe S, Wong CF, Wong KS, Xu C, Li C, Wang Y, Guan R, Sun F, Fan G, Xiao Z, Zhou F, Phang TH, Liu X, Tong SW, Chan TF, Yiu SM, Tabata S, Wang J, Xu X, Lam HM (2014) Identification of a novel salt tolerance gene in wild soybean by whole-genome sequencing. Nat Commun 5:4340. https://doi.org/10.1038/ncomms5340
Rober-Kleber N, Albrechtova JTP, Fleig S, Huck N, Michalke W, Wagner E, Speth V, Neuhaus G, Fischer-Iglesias C (2003) Plasma membrane H+-ATPase is involved in auxin-mediated cell elongation during wheat embryo development. Plant Physiol 131(3):1302–1312. https://doi.org/10.1104/pp.013466
Sanchita, Dhawan SS, Sharma A (2014) Analysis of differentially expressed genes in abiotic stress response and their role in signal transduction pathways. Protoplasma 251(1):81–91. https://doi.org/10.1007/s00709-013-0528-5
Santiago J, Dupeux F, Round A, Antoni R, Park SY, Jamin M, Cutler SR, Rodriguez PL, Marquez JA (2009) The abscisic acid receptor PYR1 in complex with abscisic acid. Nature 462(7273):665–668. https://doi.org/10.1038/nature08591
Schultz J, Copley RR, Doerks T, Ponting CP, Bork P (2000) SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res 28(1):231–234
Schweighofer A, Hirt H, Meskiene L (2004) Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci 9(5):236–243. https://doi.org/10.1016/j.tplants.2004.03.007
Schweighofer A, Kazanaviciute V, Scheikl E, Teige M, Doczi R, Hirt H, Schwanninger M, Kant M, Schuurink R, Mauch F, Buchala A, Cardinale F, Meskiene I (2007) The PP2C-type phosphatase AP2C1, which negatively regulates MPK4 and MPK6, modulates innate immunity, jasmonic acid, and ethylene levels in Arabidopsis. Plant Cell 19(7):2213–2224. https://doi.org/10.1105/tpc.106.049585
Servet C, Benhamed M, Latrasse D, Kim W, Delarue M, Zhou DX (2008) Characterization of a phosphatase 2C protein as an interacting partner of the histone acetyltransferase GCN5 in Arabidopsis. BBA-Gene Regul Mech 1779(6–7):376–382. https://doi.org/10.1016/j.bbagrm.2008.04.007
Shang H, Li W, Zou C, Yuan Y (2013) Analyses of the NAC transcription factor gene family in Gossypium raimondii Ulbr.: chromosomal location, structure, phylogeny, and expression patterns. J Integr Plant Biol 55(7):663–676. https://doi.org/10.1111/jipb.12085
Singh A, Giri J, Kapoor S, Tyagi AK, Pandey GK (2010) Protein phosphatase complement in rice: genome-wide identification and transcriptional analysis under abiotic stress conditions and reproductive development. BMC Genomics 11:435. https://doi.org/10.1186/1471–2164–11-435
Song SK, Clark SE (2005) POL and related phosphatases are dosage-sensitive regulators of meristem and organ development in Arabidopsis. Dev Biol 285(1):272–284. https://doi.org/10.1016/j.ydbio.2005.06.020
Song SK, Lee MM, Clark SE (2006) POL and PLL1 phosphatases are CLAVATA1 signaling intermediates required for Arabidopsis shoot and floral stem cells. Development 133(23):4691–4698. https://doi.org/10.1242/dev.02652
Sonnhammer EL, Eddy SR, Durbin R (1997) Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins 28(3):405–420
Spartz AK, Ren H, Park MY, Grandt KN, Lee SH, Murphy AS, Sussman MR, Overvoorde PJ, Gray WM (2014) SAUR inhibition of PP2C-D phosphatases activates plasma membrane H+-ATPases to promote cell expansion in Arabidopsis. Plant Cell 26(5):2129–2142. https://doi.org/10.1105/tpc.114.126037
Sun X, Luo X, Sun M, Chen C, Ding X, Wang X, Yang S, Yu Q, Jia B, Ji W, Cai H, Zhu Y (2014) A Glycine soja 14-3-3 protein GsGF14o participates in stomatal and root hair development and drought tolerance in Arabidopsis thaliana. Plant Cell Physiol 55(1):99–118. https://doi.org/10.1093/pcp/pct161
Sun M, Jia B, Cui N, Wen Y, Duanmu H, Yu Q, Xiao J, Sun X, Zhu Y (2016) Functional characterization of a Glycine soja Ca(2+)ATPase in salt-alkaline stress responses. Plant Mol Biol 90(4–5):419–434. https://doi.org/10.1007/s11103-015-0426-7
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739. https://doi.org/10.1093/molbev/msr121
Tovar-Mendez A, Miernyk JA, Hoyos E, Randall DD (2014) A functional genomic analysis of Arabidopsis thaliana PP2C clade D. Protoplasma 251(1):265–271. https://doi.org/10.1007/s00709-013-0526-7
Tuteja N (2007) Mechanisms of high salinity tolerance in plants. Methods Enzymol 428:419–438. https://doi.org/10.1016/S0076-6879(07)28024-3
Wang Y, Yang C, Liu G, Jiang J (2007) Development of a cDNA microarray to identify gene expression of Puccinellia tenuiflora under saline-alkali stress. Plant Physiol Biochem 45(8):567–576. https://doi.org/10.1016/j.plaphy.2007.05.006
Xue T, Wang D, Zhang S, Ehlting J, Ni F, Jakab S, Zheng C, Zhong Y (2008) Genome-wide and expression analysis of protein phosphatase 2C in rice and Arabidopsis. BMC Genomics 9:550. https://doi.org/10.1186/1471-2164-9-550
Yang SH, Zhang XH, Yue JX, Tian DC, Chen JQ (2008) Recent duplications dominate NBS-encoding gene expansion in two woody species. Mol Gen Genomics 280(3):187–198. https://doi.org/10.1007/s00438-008-0355-0
Ying G, Zhu Y, De-Kang L, Dong T, Wang W (2009) Research on responses of wild soybean to alkaline stress. Pratacultural Sci 26:47–52
Zeng QY, Yang CY, Ma QB, Li XP, Dong WW, Nian H (2012) Identification of wild soybean miRNAs and their target genes responsive to aluminum stress. BMC Plant Biol 12:182. https://doi.org/10.1186/1471–2229–12-182
Zhang LM, Liu XG, XN Q, Yu Y, Han SP, Dou Y, YY X, Jing HC, Hao DY (2013) Early transcriptomic adaptation to Na(2)CO(3) stress altered the expression of a quarter of the total genes in the maize genome and exhibited shared and distinctive profiles with NaCl and high pH stresses. J Integr Plant Biol 55(11):1147–1165. https://doi.org/10.1111/jipb.12100
Zhang J, Yang D, Li M, Shi L (2016) Metabolic profiles reveal changes in wild and cultivated soybean seedling leaves under salt stress. PLoS One 11(7):e0159622. https://doi.org/10.1371/journal.pone.0159622
Zhu JK (2001) Plant salt tolerance. Trends Plant Sci 6(2):66–71
Zhu D, Bai X, Chen C, Chen Q, Cai H, Li Y, Ji W, Zhai H, Lv D, Luo X, Zhu Y (2011) GsTIFY10, a novel positive regulator of plant tolerance to bicarbonate stress and a repressor of jasmonate signaling. Plant Mol Biol 77(3):285–297. https://doi.org/10.1007/s11103-011-9810-0
Acknowledgements
This work was supported by the Heilongjiang Provincial Higher School Science and Technology Innovation Team Building Program (2011TD005), National Natural Science Foundation of China (31501331), and Advanced Talents Foundation of QAU (6631115032).
Author contribution statements
Chao Chen, Yang Yu, Huizi Duanmu, and Lei Cao performed the experiment, analyzed data; Chao Chen, Yu Yang, and Dan Zhu wrote the manuscript; Beidong Liu, Xiaoli Sun, and Zaib-un-Nisa edited the manuscript; and Qiang Li, Yanming Zhu, and Xiaodong Ding provided ideas, designed the research; all authors read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no conflict of interest.
Additional information
Handling Editor: Bhumi Nath Tripathi
Electronic supplementary material
Supplemental Table 1
Gene-specific primers used for quantitative RT-PCR assays (DOCX 16 kb)
Supplemental Table 2
DNA sequence of cis-acting regulatory elements (DOCX 14 kb)
Supplemental Table 3
Distribution of cis-acting elements on the promoters of PP2C clade D genes in soybean (DOCX 14 kb)
Supplemental Fig. 1
Sequence analysis of GmPP2C and GsPP2C clade D proteins. Multiple sequence alignments of the full-length amino acid sequences of GmPP2C and GsPP2C clade D. Sequences were aligned using clustalx1.83. (JPEG 3492 kb)
High resolution image
(TIFF 19293 kb)
Supplemental Fig. 2
Phylogenetic relationships and exon-intron structure analysis of PP2C clade D genes in cultivated and wild soybean. The phylogenetic tree was produced using MEGA 5.0 by N-J method. Bootstrap values based on 1000 replications for each branch. Gene structure analysis of PP2C clade D was performed by using GSDS online tools. (JPEG 142 kb)
High resolution image
(TIFF 6364 kb)
Supplemental Fig. 3
Correlation with PP2C clade D genes in wild soybean (a) and Arabidopsis (b). The correlation of the gene expression patterns measured from root, stems and leaves in wild soybean and Arabidopsis plants. The relative expression levels were detected by qRT-PCR. r ≥ 8: two variables are highly correlation. (JPEG 225 kb)
High resolution image
(TIFF 247 kb)
Supplemental Fig. 4
Expression analysis of four PP2C clade D genes in soybean under alkaline and salt treatments. Total RNA samples were extracted from 15-day-old soybean seedlings under 50 mM NaHCO3 and 200 mM NaCl treatments. The relative expression levels were detected using qRT-PCR. GAPDH was used as an internal control. Statistical analyses were performed by one-way ANOVA with Bonferroni post-test. *P < 0.05, **P < 0.01, as compared with the control before treatment (0 h). (JPEG 120 kb)
High resolution image
(TIFF 236 kb)
Rights and permissions
About this article
Cite this article
Chen, C., Yu, Y., Ding, X. et al. Genome-wide analysis and expression profiling of PP2C clade D under saline and alkali stresses in wild soybean and Arabidopsis . Protoplasma 255, 643–654 (2018). https://doi.org/10.1007/s00709-017-1172-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00709-017-1172-2