Abstract
HBsAg point mutations within the major hydrophilic region (MHR) have frequently been reported to be associated with diagnostic failure, vaccine escape and immunotherapy escape. However, the prevalence of escape mutations in chronic hepatitis B (CHB) patients has not been systematically studied in patients from southern China within the past decade. This study aimed to determine the prevalence of escape mutations within the MHR of hepatitis B virus in patients in Dongguan, southern China. Between June 2015 and May 2016, 391 patients who were chronically infected with HBV were enrolled in the study, including 240 patients with the genotype B strain and 151 with the genotype C strain. The most frequent mutated position was s126 (4.3%), followed by s100 (3.3%), s101 (2.8%), s133 (2.8%), s145 (2.3%), s120 (2.0%) and s129 (1.8%). Furthermore, the mutations sY100C, sQ101R/K, sS114A, sP120T, sT/I126A/N/S, sQ129R, sM133L/T/S and sG145R/A were prevalent in at least one genotype, with a frequency higher than 1%, which indicated that these mutations were relatively common. In addition, sQ101K/R was found only in genotype C isolates (P < 0.05), and sT126A was only discovered in genotype B isolates (P = 0.047), indicating that such mutations were genotype-associated mutations. Notably, combinations of escape mutations within the MHR were also frequently discovered in genotypes B (5.0%) and C (6.6%), with no significant difference (P = 0.498). These results indicated that we should increase the surveillance HBsAg mutations among HBV-infected patients in China.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hepatitis B virus (HBV) infection is still a serious public health problem, and more than 350 million people are chronic carriers of HBV [23]. In the Global Burden of Disease study in 2013, HBV infection caused 686,000 deaths per year [30]. Universal vaccination for HBV is currently the most cost-effective way to protect against HBV infection and can significantly decrease the prevalence of HBV infection and the occurrence of childhood HCC and fulminant hepatitis [22, 26].
Recent reports have indicated that escape mutations within the major hydrophilic region (MHR) of hepatitis B surface antigen (HBsAg) are related to detection failure, vaccine failure and hepatitis B hyperimmune globulin (HBIG) escape [5, 46, 53], leading to occult HBV infection (OBI) [4, 6, 31, 35, 36, 41, 47, 53] or the breakthrough of HBV in liver transplant patients and in children born to HBsAg carriers despite prophylaxis with hepatitis B vaccination and HBIG [9, 11, 43, 46, 51].
Continued monitoring is necessary to determine whether the prevalence of these escape mutants is increasing. However, information on the prevalence of point mutations in the MHR of HBV strains in southern China over the past decade is limited. Therefore, in this study, we investigated the prevalence and clinical characteristics of escape mutations within the MHR of hepatitis B virus in patients in Dongguan, southern China.
Materials and methods
Patients and serum samples
Between June 2015 and May 2016, 391 eligible serum samples were obtained from HBV patients in Dongguan Eighth People’s Hospital. Participants were included in the study if they met all of the following inclusion criteria: 1) subjects had HBsAg for at least six months; 2) patients were not infected with hepatitis A virus, hepatitis C virus, hepatitis D virus, syphilis or human immunodeficiency virus; and 3) all participating subjects or their guardians gave signed informed consent for their samples to be used in the study. The study protocol conformed to the Declaration of Helsinki, and it was approved by the Ethics Committee of Dongguan Eighth People’s Hospital.
Serologic test for HBV markers
HBV serological markers were detected using the chemiluminescent microparticle immunoassay (CMIA) technique with an ARCHITECT i2000 automatic analyzer (Abbot Laboratories, IL, USA). Commercially available kits were purchased from Abbott Laboratories. Samples with HBsAg >0.05 IU/ml and anti-HBs >10 mIU/ml were defined as positive. In the current study, all 391 samples were positive only for HBsAg. Serum alanine aminotransferase (ALT) levels were detected using a Beckman Coulter AU5800 analyzer (Beckman Coulter, CA, USA), using commercial kits purchased from Beckman Coulter.
HBV DNA quantification
Viral DNA was extracted and quantified using commercial kits (Biotgene, Wuhan, China) according to the manufacturer’s instructions.
PCR amplification and sequencing
PCR fragment amplification and sequencing of the full-length S gene were performed as follows: The primer sequences were synthesized by Sangon Biotech Company (Shanghai, China). PCR was carried out in a 50-μL reaction mixture containing 2 μL of HBV DNA template, 0.3 μM each primer, 0.4 mM each dNTP, 25 μL of PCR buffer and 1 μL of KOD FX polymerase (TOYOBO, Japan). The PCR reaction was performed at 94 °C for 2 min, followed by 35 cycles at 98 °C for 10 s, at 56 °C for 1 min, and at 68 °C for 1 min 45 s for outer primers or 50 s for inner primers, and incubation at 72 °C for 5 min. The outer upstream primer was 5’-GGGTCACCATATTCTTGGGAAC-3’ (nt 2814-2835), and the outer downstream primer was 5’-GGGGGTTGCGTCAGCAAACAC-3’ (nt 1180-1200). The inner upstream primer, which was also a sequencing primer, was 5’-ACTTTCCTGCTGGTGGCTCC-3’ (nt 51-70), and the downstream primer was 5’-CATATCCCATGAAGTTAAGG -3’ (nt 867–886). The PCR products were purified and sequenced directly on an ABI 3730xl DNA Analyzer (Applied Biosystems, Foster City, CA) after reaction with BigDye Terminator v3.1 (Applied Biosystems, Foster City, CA). Sequence analysis was conducted using MEGA6.0 [44]. HBV genotyping was conducted by phylogenetic analysis of the sequence and was confirmed by >96% similarity of the surface gene sequences with the reference sequences [2]. The HBV reference sequences in the study were D00330 for genotype B and AB033556 for genotype C. The escape mutations associated with detection failure, occult hepatitis B virus infection (OBI), or vaccination and hepatitis B hyperimmune globulin (HBIG) breakthrough were analyzed as described previously by Echevarria et al. [8], Zhu et al. [53] and Lazarevic [20]. HBV quasispecies were defined as a mixture of wild-type and variant strains in the viral population, resulting in two peaks, including “inner small peaks” in the sequencing results.
Statistical analysis
Statistical analysis was performed using SPSS 22.0 software (Chicago, IL, USA). χ2 or Fisher’s exact test was used to compare categorical data. Quantitative data were compared using Student’s t-test. All tests were two-tailed, and a P-value <0.05 was considered statistically significant.
Results
Characteristics of patients
Overall, 391 hepatitis B patients were enrolled in this study. The mean age of the patients was 44.1 ± 15.0 (range, 4 to 85) years, and 227 (58.1%) patients were male (Table 1). In total, 240 cases (61.4%) were grouped in genotype B, which indicated that genotype B was more prevalent in Dongguan, southern China. Interestingly, a significant difference in gender distribution was discovered when comparing genotypes B and C, and there were more male patients in the genotype B group (P = 0.025) (Table 1). For 12 samples, a negative qPCR result was obtained. HBV DNA was quantified in 379 cases (Table 1), and no difference was discovered between genotypes B and C (P = 0.792). The median serum ALT level of all patients was 36.0 (23.0-70.9) U/L in the study group, and there was no difference between genotypes B and C (P = 0.434).
Genetic diversity within the MHR among genotype B and C strains
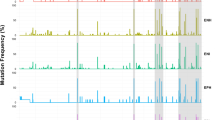
Among the 391 HBsAg-positive patients, 96 were infected with viruses with (24.6%) harbored aa substitutions within the MHR, and the prevalence of mutants within the “a” determinant (124-147) was 48 out of 391 (12.3%). No significant aa substitution diversity was observed within the MHR (99-169) of the surface gene between genotype B and C isolates (0.42 vs. 0.49 substitutions per 100 aa, the same as below, P = 0.345), within the “a” determinant (aa 124-147) (0.59 vs. 0.66, P = 0.665), in MHR3 (aa 124-137) (0.83 vs. 0.85, P = 0.943), in MHR4 (aa 138-147) (0.25 vs. 0.40, P = 0.607), or in MHR5 (aa 148-169) (0.25 vs. 0.12, P = 0.201) (Table 1). Mutations were discovered more frequently in MHR1 (aa 99-119) (0.24 vs. 0.76, P = 0.001) in genotype C strains. However, variants were more frequently found in MHR2 (aa 120-123) (1.25 vs. 0.17, P = 0.021) in genotype B isolates.
The most frequently mutated position was at s126 (17/391; 4.3%), followed by s100 (13/391; 3.3%), s101(11/391; 2.8%), s133 (11/391; 2.8%), s145 (9/391; 2.3%), s120 (8/391; 2.0%) and s129 (7/391; 1.8%) (Table 1). Mutations at position s120 were found only in genotype B strains (8/240; 3.3%; P = 0.026) (Table 1), and mutations at position s101 were found only in genotype C isolates (11/151; 7.3%; P < 0.001) (Table 2). This explains the significant difference within MHR1 and 2 between genotypes B and C.
In addition, when considering the prevalence of specific point mutations in genotypes B and C, the mutations sY100C, sQ101R/K, sS114A, sP120T, sT/I126A/N/S, sQ129R, sM133L/T/S and sG145R/A were prevalent in at least one genotype, with a frequency higher than 1%, which indicated that these mutations were relatively common. Furthermore, sQ101K/R was found only in genotype C (P < 0.05), and sT/I126A was found only in genotype B (P = 0.047), which indicated that such mutations were genotype-associated mutations.
Interestingly, HBV quasispecies variants were found in two genotype B isolates (2/240; 0.8%) and three genotype C isolates (3/151; 2.0%). All of these sites are marked in yellow in Fig. 1. These strains contained variants in coexistence with the wild-type strains, resulting in two peaks including “inner small peaks” in the sequencing results (Fig. 2).
Amino acid substitutions within the MHR (aa 99-169) in CHB patients. The reference amino acid sequences of genotypes B and C are deduced from D00330 and AB033556, respectively. The capital letters represent amino acids, and letters highlighted in yellow represent variants in combination with the wild-type sequence. Letters highlighted in blue represent genotype-related amino acid variations, which are not considered mutations. Letters highlighted in red represent insertions (color figure online)
Combinations of two or more mutations within the MHR were also found in genotype B (5.0%) and C (6.6%) strains (Table 1), with no significant difference (P = 0.498). In genotype B strains, sY100C/Q129R/G145A, sY100C/E164G, sY100S/M133L, sM103I/T123A, sS114K/T116N/S132Y, sP120T/M133L, sP120S/K160N, sP120T/A157G, sP120Q/T126A, sK122I/P127T/G130N and sD144A/A157G were found (Fig. 1). In genotype C isolates, sY100C/Q101K, sY100C/I126N/M133T, sL104F/ G145R, sL109P/A128V/Q129K, sS114A/G145A, sG130K/A166G, sT131N/Q101K/I126S/M133T, sT131N/sM133T/F134L and sM133S/G145A and sT131N/M133S were found (Fig. 1).
Additionally, we found two strains of HBV genotype B with an insertion of “GATCTA” between nucleotide positions 341 and 346, which affected the coding of all amino acids after s113 and were thus not counted in the number of point mutations.
Discussion
Escape mutations located in the major hydrophilic region (MHR) of HBsAg have been widely reported worldwide. Previous studies demonstrated that the prevalence of point mutations within the MHR among hepatitis B patients was 14.8% in Argentina [32], 15% in Morocco [17], 15.4% in Poland [12], 14%-32.8% in Iran [28, 29, 33], 22.6% in Serbia [19], 27.8% in France [37], and 44.1% in South Korea [21]. MHR mutations could lead to diagnostic problems, occult HBV infection, the emergence of vaccine-escape mutants, and failure of HBIG therapy [1, 10, 13, 20, 39].
In China, the overall frequency of MHR mutations among HBV genotype C strains was 46.6% in Beijing, northern China [40]. However, limited information on the prevalence of point mutations within the MHR has been reported in southern China over the past decade. We examined the sequences of HBsAg variants within the MHR from patients with hepatitis B in Dongguan, southern China.
In the current study, 24.6% and 12.3% of the strains harbored aa substitutions within the MHR and “a” determinant, respectively, which indicated that the presence of HBsAg escape mutations among HBV-infected patients in China should be monitored.
Our results demonstrated that the position s126T/I was the most frequent mutation site, with the amino acid replaced by Ala (A) (7/391; 1.8%), Ser (S) (7/391; 1.8%) and Asn (N) (3/391; 0.8%). Previous reports demonstrated that the binding reactivity to anti-HBs of HBsAg with s126I, s126T and s126A was comparable, but HBsAg with sT126S had a significantly lower antigenicity, which might lead to immune escape in chronic HBV infection [34], and result in the occurrence of OBI [53] or vaccine escape [20]. In addition, a previous study showed that sT126S impaired the secretion of subviral particles and virions, both in vitro and in vivo [14]. In the current study, sT/I126S mutations were prevalent in both genotype B (2.1%) and C (1.3%), indicating that this might be a hot spot for immune selection.
Mutations were also frequently found at position s100, including sY100C (10/391; 2.6%), sY100S (2/391; 0.5%) and sY100H (1/391; 0.3%). An in vitro study concluded that the sY100C substitution alone did not play a role in reducing the amounts of HBsAg or the HBsAg affinity in a commercial ELISA assay [27]. sY100S was reported to correlate with occult infection in genotype D strains [20, 42], but not in genotypes B and C [14, 20]. Furthermore, sY100S was relatively uncommon in genotypes B (0.4%) and C (0.7%). The clinical implications of the sY100H mutation and the impact on HBsAg antigenicity are unknown and require further investigation.
Another mutational hot spot was at s101, including sQ101R (7/391; 1.8%) and sQ101K (4/391; 1%), which was only discovered in genotype C strains. sQ101R has been reported to correlate with occult infection in genotype D strains [20, 42], but not in genotypes B and C [14, 20]. sQ101R was associated with lower reactivity in HBsAg assays when combined with some mutations [39]. As far as we know, the clinical implications of sQ101K are still unknown.
Mutations at positions s120 and s129 were mainly sP120T (5/391; 1.3%) and sQ129R (5/391; 1.3%), both of which were only discovered in genotype B strains and are considered “immune escape” mutations [20, 53]. Previous studies demonstrated that amino acids within the region between 120 and 123 are crucial for the antigenicity of HBsAg [45], and that sP120T mutants exhibit impaired binding to MAbs [14], thus indicating its association with immune escape. A previous study also showed that sQ129R decreased the secretion of surface proteins and virions [14]. This mutation was frequently discovered in chronic hepatitis B patients with coexistence of HBsAg and anti-HBs [7, 18, 24]. Therefore, sP120T and sQ129R are important escape mutations in genotype B strains in China.
The prevalence of mutations at position s133 among all patients was 2.8%, including sM133L (4/391; 1%), sM133T (5/391; 1.3%) and sM133S (2/391; 0.5%). Interestingly, sM133L was only discovered in genotype B strains (4/240; 1.7%), and sM133S was only found in genotype C strains (2/151; 2.0%). Various studies have shown that sM133L is associated with escape from vaccine-induced immunity [20, 46]. sM133T showed great clinical significance in the following two aspects. First, the sM133T mutation could create an additional glycosylation site, 131NST133, in the presence of the sT131N mutation, which contributes to immune escape [46, 52]. In the current study, we discovered one strain of genotype C harboring the sT131N/M133T double mutation. Second, the sM133T mutation could rescue virion secretion for a wide range of mutants (sI110M, sG145R, sN146Q, sN146S, sR169H, and, to a lesser extent, sG119E) [15]. In our study, combinations of mutations, sY100C/I126N/M133T, sI126S/M133T and sM133T/F134L, were discovered in genotype C strains. Thus, sM133L/T should be considered an important escape mutation and studied, especially when combined with other mutations. The clinical implications of sM133S, which is relatively uncommon and rarely reported, require further study.
The prevalence of mutations at position s145 was 2.3%, including sG145R (4/391; 1%) and sG145A (5/391; 1.3%). Both of these mutations were primarily discovered in genotype C variants. Over the past several decades, sG145R has become the most widely reported vaccine-escape mutation [3, 20, 46, 53] since it was first discovered in an infant infected with HBV who also had protective titers of anti-HBs [3]. The sG145R mutation has been shown to decrease virion and HBsAg detection significantly [14], and sG145R/A has been shown to exhibit various degrees of altered binding of HBsAg in different commercial assays [1, 46]. In addition, several studies of genotypes B and C strains have shown that sG145R/A was associated with the occurrence of OBI [20]. More importantly, sG145R was stable over time and could result in horizontal transmission [5, 20]. Thus, the sG145R/A mutation in the surface gene might pose a threat to the global vaccination program in the future, and it should be closely monitored, especially in genotype C strains.
The HBV population in the host consists of genetically heterologous variants that exist in the form of quasispecies [38]. A recent report from Indonesia confirmed the presence of quasispecies variants in the MHR of HBV strains using ultrahigh-throughput next-generation sequencing (NGS)[50]. Because the direct PCR sequencing method can only detect variants representing ≥20% of the total population [16, 25, 50], the prevalence of variants in coexistence with the wild-type strains is probably higher than that determined in the current study. Previous reports have demonstrated that the secretion of mutant virions could be rescued efficiently (i.e., sE2G, sC69*, sP127S and sG145R) [48, 49] or moderately efficiently (i.e., sW172*) by co-expression with the WT surface protein [48]. The coexistence of WT strains and defective mutants might be a general survival requirement for some variants, and the clinical implications of HBV quasispecies need to be studied further.
In conclusion, the prevalence of escape mutations within the MHR of hepatitis B virus in patients in Dongguan, China, was systematically studied. The prevalence of amino acid substitutions within the MHR varied according to the HBV genotype. Our results show that monitoring the mutations within MHR is essential, especially for hot-spot escape mutations related to detection failure, vaccination escape, or HBIG therapy breakthrough.
References
Alavian SM, Carman WF, Jazayeri SM (2013) HBsAg variants: diagnostic-escape and diagnostic dilemma. J Clin Virol 57:201–208
Bartholomeusz A, Schaefer S (2004) Hepatitis B virus genotypes: comparison of genotyping methods. Rev Med Virol 14:3–16
Carman WF, Zanetti AR, Karayiannis P, Waters J, Manzillo G, Tanzi E, Zuckerman AJ, Thomas HC (1990) Vaccine-induced escape mutant of hepatitis B virus. Lancet 336:325–329
Chen J, Liu Y, Zhao J, Xu Z, Chen R, Si L, Lu S, Li X, Wang S, Zhang K, Li J, Han J, Xu D (2016) Characterization of novel hepatitis B virus PreS/S-gene mutations in a patient with occult hepatitis B virus infection. PLoS One 11:e0155654
Coppola N, Onorato L, Minichini C, Di Caprio G, Starace M, Sagnelli C, Sagnelli E (2015) Clinical significance of hepatitis B surface antigen mutants. World J Hepatol 7:2729–2739
Coppola N, Onorato L, Iodice V, Starace M, Minichini C, Farella N, Liorre G, Filippini P, Sagnelli E, de Stefano G (2016) Occult HBV infection in HCC and cirrhotic tissue of HBsAg-negative patients: a virological and clinical study. Oncotarget 7:62706–62714
Ding F, Miao XL, Li YX, Dai JF, Yu HG (2016) Mutations in the S gene and in the overlapping reverse transcriptase region in chronic hepatitis B Chinese patients with coexistence of HBsAg and anti-HBs. Braz J Infect Dis 20:1–7
Echevarria JM, Avellon A (2006) Hepatitis B virus genetic diversity. J Med Virol 78(Suppl 1):S36–S42
Gerlich WH (2015) Prophylactic vaccination against hepatitis B: achievements, challenges and perspectives. Med Microbiol Immunol 204:39–55
Gish RG, Gutierrez JA, Navarro-Cazarez N, Giang K, Adler D, Tran B, Locarnini S, Hammond R, Bowden S (2014) A simple and inexpensive point-of-care test for hepatitis B surface antigen detection: serological and molecular evaluation. J Viral Hepat 21:905–908
Golsaz-Shirazi F, Shokri F (2016) Hepatitis B immunopathogenesis and immunotherapy. Immunotherapy 8:461–477
Grabarczyk P, Garmiri P, Liszewski G, Doucet D, Sulkowska E, Brojer E, Allain JP, Polish Blood Transfusion Centres Viral Study G (2010) Molecular and serological characterization of hepatitis B virus genotype A and D infected blood donors in Poland. J Viral Hepat 17:444–452
Hirzel C, Pfister S, Gorgievski-Hrisoho M, Wandeler G, Zuercher S (2015) Performance of HBsAg point-of-care tests for detection of diagnostic escape-variants in clinical samples. J Clin Virol 69:33–35
Huang CH, Yuan Q, Chen PJ, Zhang YL, Chen CR, Zheng QB, Yeh SH, Yu H, Xue Y, Chen YX, Liu PG, Ge SX, Zhang J, Xia NS (2012) Influence of mutations in hepatitis B virus surface protein on viral antigenicity and phenotype in occult HBV strains from blood donors. J Hepatol 57:720–729
Ito K, Qin Y, Guarnieri M, Garcia T, Kwei K, Mizokami M, Zhang J, Li J, Wands JR, Tong S (2010) Impairment of hepatitis B virus virion secretion by single-amino-acid substitutions in the small envelope protein and rescue by a novel glycosylation site. J Virol 84:12850–12861
Jones LR, Sede M, Manrique JM, Quarleri J (2016) Virus evolution during chronic hepatitis B virus infection as revealed by ultradeep sequencing data. J Gen Virol 97:435–444
Kitab B, El Feydi AE, Afifi R, Derdabi O, Cherradi Y, Benazzouz M, Rebbani K, Brahim I, Salih Alj H, Zoulim F, Trepo C, Chemin I, Ezzikouri S, Benjelloun S (2011) Hepatitis B genotypes/subgenotypes and MHR variants among Moroccan chronic carriers. J Infect 63:66–75
Lada O, Benhamou Y, Poynard T, Thibault V (2006) Coexistence of hepatitis B surface antigen (HBs Ag) and anti-HBs antibodies in chronic hepatitis B virus carriers: influence of “a” determinant variants. J Virol 80:2968–2975
Lazarevic I, Cupic M, Delic D, Svirtlih NS, Simonovic J, Jovanovic T (2010) Prevalence of hepatitis B virus MHR mutations and their correlation with genotypes and antiviral therapy in chronically infected patients in Serbia. J Med Virol 82:1160–1167
Lazarevic I (2014) Clinical implications of hepatitis B virus mutations: recent advances. World J Gastroenterol 20:7653–7664
Lee SA, Cho YK, Lee KH, Hwang ES, Kook YH, Kim BJ (2011) Gender disparity in distribution of the major hydrophilic region variants of hepatitis B virus genotype C according to hepatitis B e antigen serostatus. J Med Virol 83:405–411
Liao X, Liang Z (2015) Strategy vaccination against hepatitis B in China. Hum Vaccines Immunother 11:1534–1539
Liaw YF, Chu CM (2009) Hepatitis B virus infection. Lancet 373:582–592
Liu W, Hu T, Wang X, Chen Y, Huang M, Yuan C, Guan M (2012) Coexistence of hepatitis B surface antigen and anti-HBs in Chinese chronic hepatitis B virus patients relating to genotype C and mutations in the S and P gene reverse transcriptase region. Arch Virol 157:627–634
Lok AS, McMahon BJ (2007) Chronic hepatitis B. Hepatology 45:507–539
Luo Z, Li L, Ruan B (2012) Impact of the implementation of a vaccination strategy on hepatitis B virus infections in China over a 20-year period. Int J Infect Dis: IJID 16:e82–e88
Mello FC, Martel N, Gomes SA, Araujo NM (2011) Expression of hepatitis B virus surface antigen containing Y100C variant frequently detected in occult HBV infection. Hepat Res Treat 2011:695859
Mohebbi SR, Amini-Bavil-Olyaee S, Zali N, Noorinayer B, Derakhshan F, Chiani M, Rostami Nejad M, Antikchi MH, Sabahi F, Zali MR (2008) Molecular epidemiology of hepatitis B virus in Iran. Clin Microbiol Infect 14:858–866
Moradi A, Zhand S, Ghaemi A, Javid N, Tabarraei A (2012) Mutations in the S gene region of hepatitis B virus genotype D in Golestan Province-Iran. Virus Genes 44:382–387
Mortality GBD, Causes of Death C (2015) Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 385:117–171
Oluyinka OO, Tong HV, Bui Tien S, Fagbami AH, Adekanle O, Ojurongbe O, Bock CT, Kremsner PG, Velavan TP (2015) Occult hepatitis B virus infection in nigerian blood donors and hepatitis B virus transmission risks. PLoS One 10:e0131912
Pineiro YLFG, Pezzano SC, Torres C, Rodriguez CE, Eugenia Garay M, Fainboim HA, Remondegui C, Sorrentino AP, Mbayed VA, Campos RH (2008) Hepatitis B virus genetic diversity in Argentina: dissimilar genotype distribution in two different geographical regions; description of hepatitis B surface antigen variants. J Clin Virol 42:381–388
Pourkarim MR, Sharifi Z, Soleimani A, Amini-Bavil-Olyaee S, Elsadek Fakhr A, Sijmons S, Vercauteren J, Karimi G, Lemey P, Maes P, Alavian SM, Van Ranst M (2014) Evolutionary analysis of HBV “S” antigen genetic diversity in Iranian blood donors: a nationwide study. J Med Virol 86:144–155
Qiu S, Zhang J, Tian Y, Yang Y, Huang H, Yang D, Lu M, Xu Y (2008) Reduced antigenicity of naturally occurring hepatitis B surface antigen variants with substitutions at the amino acid residue 126. Intervirology 51:400–406
Raimondo G, Allain JP, Brunetto MR, Buendia MA, Chen DS, Colombo M, Craxi A, Donato F, Ferrari C, Gaeta GB, Gerlich WH, Levrero M, Locarnini S, Michalak T, Mondelli MU, Pawlotsky JM, Pollicino T, Prati D, Puoti M, Samuel D, Shouval D, Smedile A, Squadrito G, Trepo C, Villa E, Will H, Zanetti AR, Zoulim F (2008) Statements from the Taormina expert meeting on occult hepatitis B virus infection. J Hepatol 49:652–657
Raimondo G, Pollicino T, Romano L, Zanetti AR (2010) A 2010 update on occult hepatitis B infection. Pathologie-biologie 58:254–257
Roque-Afonso AM, Ferey MP, Ly TD, Graube A, Costa-Faria L, Samuel D, Dussaix E (2007) Viral and clinical factors associated with surface gene variants among hepatitis B virus carriers. Antiviral Ther 12:1255–1263
Seo SI, Choi HS, Choi BY, Kim HS, Kim HY, Jang MK (2014) Coexistence of hepatitis B surface antigen and antibody to hepatitis B surface may increase the risk of hepatocellular carcinoma in chronic hepatitis B virus infection: a retrospective cohort study. J Med Virol 86:124–130
Servant-Delmas A, Mercier-Darty M, Ly TD, Wind F, Alloui C, Sureau C, Laperche S (2012) Variable capacity of 13 hepatitis B virus surface antigen assays for the detection of HBsAg mutants in blood samples. J Clin Virol 53:338–345
Shi Y, Wei F, Hu D, Li Q, Smith D, Li N, Chen D (2012) Mutations in the major hydrophilic region (MHR) of hepatitis B virus genotype C in North China. J Med Virol 84:1901–1906
Song JE, Kim DY (2016) Diagnosis of hepatitis B. Ann Transl Med 4:338
Svicher V, Cento V, Bernassola M, Neumann-Fraune M, Van Hemert F, Chen M, Salpini R, Liu C, Longo R, Visca M, Romano S, Micheli V, Bertoli A, Gori C, Ceccherini-Silberstein F, Sarrecchia C, Andreoni M, Angelico M, Ursitti A, Spano A, Zhang JM, Verheyen J, Cappiello G, Perno CF (2012) Novel HBsAg markers tightly correlate with occult HBV infection and strongly affect HBsAg detection. Antiviral Res 93:86–93
Tajiri K, Shimizu Y (2015) Unsolved problems and future perspectives of hepatitis B virus vaccination. World J Gastroenterol 21:7074–7083
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729
Tian Y, Xu Y, Zhang Z, Meng Z, Qin L, Lu M, Yang D (2007) The amino Acid residues at positions 120 to 123 are crucial for the antigenicity of hepatitis B surface antigen. J Clin Microbiol 45:2971–2978
Tong S, Revill P (2016) Overview of hepatitis B viral replication and genetic variability. J Hepatol 64:S4–16
Wang Z, Zeng J, Li T, Zheng X, Xu X, Ye X, Lu L, Zhu W, Yang B, Allain JP, Li C (2016) Prevalence of hepatitis B surface antigen (HBsAg) in a blood donor population born prior to and after implementation of universal HBV vaccination in Shenzhen, China. BMC Infect Dis 16:498
Warner N, Locarnini S (2008) The antiviral drug selected hepatitis B virus rtA181T/sW172* mutant has a dominant negative secretion defect and alters the typical profile of viral rebound. Hepatology 48:88–98
Xiang KH, Michailidis E, Ding H, Peng YQ, Su MZ, Li Y, Liu XE, Dao Thi VL, Wu XF, Schneider WM, Rice CM, Zhuang H, Li T (2016) Effects of amino acid substitutions in hepatitis B virus surface protein on virion secretion, antigenicity, HBsAg and viral DNA. J Hepatol 66:288–296
Yamani LN, Yano Y, Utsumi T, Juniastuti Wandono H, Widjanarko D, Triantanoe A, Wasityastuti W, Liang Y, Okada R, Tanahashi T, Murakami Y, Azuma T, Soetjipto Lusida MI, Hayashi Y (2015) Ultradeep sequencing for detection of quasispecies variants in the major hydrophilic region of hepatitis B virus in indonesian patients. J Clin Microbiol 53:3165–3175
Yao QQ, Dong XL, Wang XC, Ge SX, Hu AQ, Liu HY, Wang YA, Yuan Q, Zheng YJ (2013) Hepatitis B virus surface antigen (HBsAg)-positive and HBsAg-negative hepatitis B virus infection among mother-teenager pairs 13 years after neonatal hepatitis B virus vaccination. Clin Vaccine Immunol: CVI 20:269–275
Yu DM, Li XH, Mom V, Lu ZH, Liao XW, Han Y, Pichoud C, Gong QM, Zhang DH, Zhang Y, Deny P, Zoulim F, Zhang XX (2014) N-glycosylation mutations within hepatitis B virus surface major hydrophilic region contribute mostly to immune escape. J Hepatol 60:515–522
Zhu HL, Li X, Li J, Zhang ZH (2016) Genetic variation of occult hepatitis B virus infection. World J Gastroenterol 22:3531–3546
Acknowledgements
This work was supported by Dongguan Bureau of Science and Technology for the City Key Program of Science and Technology (Project Number: 2013108101038). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Ethical standards
This study was approved by the Ethics Committee of Dongguan Eighth People’s Hospital. All participating subjects or their guardians gave signed informed consent before the study.
Conflict of interest
There are no conflicts of interest.
Rights and permissions
About this article
Cite this article
Li, S., Xie, M., Li, W. et al. Prevalence of S gene mutations within the major hydrophilic region of hepatitis B virus in patients in Dongguan, southern China. Arch Virol 162, 2949–2957 (2017). https://doi.org/10.1007/s00705-017-3437-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-017-3437-7