Abstract
Background
Deep-brain stimulation (DBS) of the internal globus pallidus (GPi) has shown remarkable therapeutic benefits for treatment-resistant neurological disorders including dystonia and Parkinson’s disease (PD). The success of the DBS is critically dependent on the reliable visualization of the GPi.
The aim of the study was to evaluate promising 3.0 Tesla magnetic resonance imaging (MRI) methods for pre-stereotactic visualization of the GPi using a standard installation protocol.
Methods
MRI at 3.0 T of nine healthy individuals and of one patient with PD was acquired (FLAIR, T1-MPRAGE, T2-SPACE, T2*-FLASH2D, susceptibility-weighted imaging mapping (SWI)). Image quality and visualization of the GPi for each sequence were assessed by two neuroradiologists independently using a 6-point scale. Axial, coronal, and sagittal planes of the T2*-FLASH2D images were compared. Inter-rater reliability, contrast-to-noise ratios (CNR) and signal-to-noise ratios (SNR) for the GPi were determined. For illustration, axial T2*-FLASH2D images were fused with a section schema of the Schaltenbrand-Wahren stereotactic atlas.
Results
The GPi was best and reliably visualized in axial and to a lesser degree on coronal T2*-FLASH2D images. No major artifacts in the GPi were observed in any of the sequences. SWI offered a significantly higher CNR for the GPi compared to standard T2-weighted imaging using the standard parameters. The fusion of the axial T2*-FLASH2D images and the atlas projected the GPi clearly in the boundaries of the section schema.
Conclusions
Using a standard installation protocol at 3.0 T T2*-FLASH2D imaging (particularly axial view) provides optimal and reliable delineation of the GPi.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Deep-brain stimulation (DBS) is a reversible stereotactic neurosurgical technique providing remarkable therapeutic benefits for otherwise treatment-resistant neurological and psychiatric disorders including dystonia, Parkinson’s disease (PD), and tremor [1, 58, 64, 76, 78]. The interventional procedure consists in the insertion of electrodes into specific target structures of the brain and subsequent electric stimulation by an implanted brain pacemaker [46]. Obviously, the surgical technique is critically dependent on the accurate placement of the DBS electrodes into the target site [61].
Various methods have been utilized for the target localization in functional stereotactic neurosurgery. The approaches can be classified as direct and indirect. The indirect methods are based on the identification of the anterior and posterior commissure using computed tomography (CT), magnetic resonance imaging (MRI) or ventriculography. Predefined calculated distances and coordinates from the intercommissural line are used to determine the target area [52]. Additionally, histologically defined atlas maps can be used to verify the target structures [36, 38]. In contrast, a direct targeting method, using stereotactic pre-operative MRI for visualization and targeting of the deep brain nuclei, is reasonably more appropriate for individual patients considering the given anatomical variability in position, functional segregation, and size [5, 78].
The internal globus pallidus (GPi) in particular represents a common target structure of the basal ganglia for DBS. It is surrounded by the optic tract ventrally, the external globus pallidus (GPe) laterally and dorsally and the internal capsule (Ci) posteromedially. A thin layer, the lamina pallidi medialis (LPm), separates the GPi from the GPe [29, 39].
The effectiveness of bilateral DBS of the GPi for the treatment of dystonia [12, 13, 41, 48], and PD [1, 4, 41, 47, 66] has recently been proven.
Nowadays, stereotactic imaging data, particularly for the GPi, are frequently calculated pre-operatively from T2-weighted fast spin echo [22, 24] or proton density weighted MRI [72]. In addition, various alternative MR imaging sequences and techniques, including quantitative T1 and T2 imaging [28, 30], T2* mapping [25], and susceptibility-weighted imaging (SWI) [32] have been proposed to improve the visibility of the DBS target structures.
However, the most promising new imaging techniques have not been directly compared.
Therefore, the aim of this study was to evaluate different promising MRI methods (sequence and orientation) for the visualization of the GPi at 3.0 T. In order to facilitate an easy implementation of the results for other neurosurgical centers we used commercially available sequences as provided by the vendor (standard installation protocol).
Material and methods
Participant characteristics
Nine healthy volunteers (five male, four female) with a mean age of twenty-five years (range 21 – 28 years) were recruited for this study.
Additionally, a 68-year-old male patient with longstanding PD was included in this study. The patient had been treated with antiparkinson drugs for the past seven years. In the last year prior to our study he began to experience intolerable motor dyskinesia predominantly of the hands despite adequate medication.
The study was approved by the local research ethics committee, and informed consent was obtained from all participants prior to their inclusion in the study. The approval also covers the analysis of other brain regions of the volunteers not addressed in this study.
Magnetic resonance imaging
For MR imaging of all subjects a 3.0 T system (Magnetom Trio, Siemens Healthcare, Erlangen, Germany) with a 32-channel receiver head coil was used. The following sequences were acquired: FLAIR (T2-weighted fluid attenuation inversion recovery), T1-MPRAGE (T1-weighted magnetisation-prepared rapid gradient-echo), T2-SPACE (T2-weighted sampling perfection with application of optimized contrasts using different flip angle evolutions), T2*-FLASH2D (T2*-weighted two-dimensional fast low angle shot magnetic resonance imaging with a standard bandwidth of 40 kHz), T2*-FLASH2D-HB (T2*-FLASH with a high bandwidth of 200 kHz), and SWI (susceptibility-weighted imaging). Slice thickness was manually adapted to completely cover the area of interest while maintaining an adequate acquisition time. Axial slices of the T1-MPRAGE images were reconstructed in-line with a slice thickness of 1 mm. The minimum intensity projections (MIP) of the SWI datasets were reconstructed in-line with a slice thickness of 9.6 mm. The specific imaging parameters are summarized in Table 1.
Qualitative evaluation of the data
Image viewing and analysis of the acquired sequences for all participants was performed using Osirix-software (OsiriX Imaging Software; Advanced Open-Source PACS Workstation DICOM viewer, http://www.osirix-viewer.com/index.html).
Two neuroradiologists graded the delineation of the GPi for each dataset independently. Raters were allowed to freely adjust window/level settings, but no automatic preprocessing was applied.
The criteria for the visualization of the GPi was the delineation between the GPi and the adjacent anatomical structure, i.e. the LPm, the margin to the GPe laterally and dorsally, and the Ci posteromedially, on the basis of each reader's own professional judgment. The visibility of the GPi was graded using a 6-point grading scale. The grading was as follows: 5 – excellent delineation; 4 – good delineation; 3 – moderate delineation; 2 – poor delineation; 1 – no delineation; 0 – no image / not evaluable.
The image quality of each sequence was evaluated by consensus regarding artifacts using a 6-point scale (5 - no artifacts; 4 - minimal artifacts; 3 - moderate artifacts; 2 - significant artifacts; 1 - massive artifacts; 0 - no image / not evaluable).
Quantitative analysis
The acquired sequences were assessed quantitatively using the Osirix-software.
Each dataset was carefully scrutinized to identify the sections with the structures of interest. The mean signal intensity (SI) was measured for all datasets by manually placing a region of interest (ROI) of approximately 0.2 cm2 within the GPi and the Ci considering intra-individual identical localization. The average standard deviation of noise was quantified by manually placing a ROI (approximately 2.0 cm2) outside the brain and away from phase-encoding artifacts. Identical ROI sizes were used for all corresponding images. ROI measurements were repeated three times and average values were taken.
After obtaining these measurements for each participant the signal-to-noise ratios (SNR) and the contrast-to-noise ratios (CNR) for the GPi were calculated for 18 cerebral hemispheres according to the equations:
where SIGPi represents the measured signal (mean) within the grey matter target structure (GPi), SICi the MRI signal value in the white matter tracts (internal capsule), and σ the average standard deviation of the noise.
To balance the differences of the sequences in slice thickness and pixel size the SNR and CNR values were additionally adjusted to a voxel of 1x1x1 mm3.
Statistical methods
Statistical calculations were performed using the Statistical Package for the Social Sciences software (SPSS 19, IBM Corporation, Somers, NY, USA). Inter-rater reliability for the delineation of the GPi was tested using Cohen’s kappa coefficient (κ) [19].
A p-value of 0.05 was used to indicate a statistical significance.
Differences in the delineation of the GPi between the sequences were statistically evaluated using a paired t-test.
The PD patient’s images were interpreted separately and were not included in the statistical analysis.
Fusion of the axial and coronal T2*-FLASH2D-imaging with the section schema of the Schaltenbrand and Wahren atlas
The axial T2*-FLASH2D imaging slice at the level of the GPi was superimposed on the corresponding schema of the Schaltenbrand and Wahren stereotactic atlas (plate 56) [69]. For the fusion of the MR image with the atlas data obviously delineated anatomic structures (i.e. pallidum, and lateral wall of the ventricle) in the MR image were used.
Results
Population
Imaging studies of nine healthy volunteers (five male, four female) with a mean age of twenty-five years (range 21 – 28 years) were included in the qualitative and quantitative analysis. Axial images of a representative healthy volunteer are shown in Fig. 1.
Representative axial images of a healthy volunteer at the level of the internal globus pallidus (GPi, top) and enlarged representation at the level of the GPi (bottom) for all sequences (left to right: a T1-MPRAGE transveral (tra); b FLAIR tra; c T2-SPACE tra; d T2*-FLASH2D tra; e T2*-FLASH2D-HB tra; f SWI tra; g SWI-MIP tra)
Qualitative results
Image quality was good to excellent in all acquired sequences and none of the images had to be excluded due to disturbing artifacts. No artifacts within the region of interest were visible in the T1-MPRAGE, minimal artifacts in FLAIR and T2*-FLASH2D imaging, and moderate artifacts in the T2*-FLASH2D-HB, T2-SPACE and susceptibility-weighted imaging (SWI-MIP and SWI) (Table 2).
Qualitative ratings for the visualization of the GPi are summarized in Fig. 2.
Delineation of the GPi vs. internal capsule (Ci) (a) and GPi vs. lamina pallidi medialis (LPm) (b) for all sequences independently analyzed by two radiologists for the healthy volunteers. The table (left) indicates the average delineation and the standard deviation of the mean for each sequence as well as the inter-rater reliability (κ) with the statistically significance (p-value) for each sequence. The diagram (right) demonstrates the average delineation for the GPi (error bars indicate the 95% confidence interval of the mean). Sequences with a statistically significant superior delineation compared to FLAIR imaging (paired t-test) are denoted (*).
Concerning inter-rater reliability, both readers graded the delineation of the GPi vs. Ci identical in FLAIR and T1-MPRAGE images. T2*-FLASH2D transversal (tra), T2*-FLASH2D coronal (cor), T2*-FLASH2D sagittal (sag), T2-SPACE, and SWI-imaging provided a substantial agreement, while T2*-FLASH2D-HB and SWI-MIP scans showed a moderate inter-rater reliability.
Inter-rater reliability for the grading of the demarcation between GPi and LPm demonstrated identical values for T1-MPRAGE, T2*-FLASH2D sag, T2*-FLASH2D-HB, and T2-SPACE imaging. FLAIR and coronal T2*-FLASH2D images provided at least a substantial agreement. The lowest inter-rater reliability was calculated for axial T2*-FLASH2D scans revealing a moderate inter-rater reliability for the delineation of both structures.
The T1-weighted images provided excellent gray-white matter contrast in the border regions of the brain, whereas only marginal contrast was visible within the basal ganglia in T1-MPRAGE imaging.
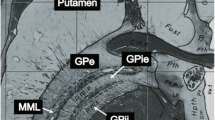
T2-weighted images demonstrated good contrast for white matter as well as for the basal ganglia. Still, we could not delineate the LPm, the anatomic boundary between the GPi and the GPe, in the axial T2*-FLASH2D-HB, T2-SPACE, FLAIR, and the SWI images. For this special region, the T2*-FLASH2D images provided a much better demarcation of the GPi. Moreover, particularly the T2*-FLASH2D images in axial orientation, and to a lesser degree in coronal orientation, clearly visualized the LPm, dividing the GPi from the GPe (Fig. 3).
Axial (a), coronal (b), and sagittal (c) views of the internal globus pallidus (GPi) (top) with magnified representation of the area of interest (bottom) in T2*-FLASH2D-sequences from a healthy volunteer. The GPi and the surrounding structures are indicated (GPi: internal globus pallidus; GPe: external globus pallidus; Put: putamen, LPm: lamina palladi medialis).
Furthermore, the T2*-FLASH2D in the axial and coronal view as well as the SWI sequences provided a statistically significant superior delineation of the GPi vs. the Ci compared to FLAIR images. The delineation of the GPi vs. the LPm was statistically superior particularly in the axial and to a lesser degree in the coronal T2*-FLASH2D sequences compared to the FLAIR images.
The LPm was discernible in the axial T2*-FLASH2D and coronal T2*-FLASH2D images in all 18 hemispheres.
To analyze the effect of different bandwidths on the delineation of the GPi we compared two T2*-FLASH2D sequences with bandwidths of 40 kHz (standard installation) and 200 kHz (HB). We found that the T2*-FLASH2D with 40 kHz was clearly superior to the high bandwidth of 200 kHz.
For the PD patient similar results were obtained. The GPi was optimally visualized on the T2*-FLASH2D images. Particularly, the axial and to a lesser degree the coronal T2*-FLASH2D-images demonstrated a clear demarcation of the GPi vs. the Ci and the LPm..
Quantitative results
To provide directly clinical relevant information, we analyzed the SNR and CNR of the sequences as provided by the manufacturer. Additionally, we adjusted the measurements for a comparison independent of the voxel volume.
SNR and CNR for the GPi are shown in Table 3.
In the non-adjusted measurements the SNR of the GPi in SWI, SWI-MIP, and T1-MPRAGE-images was substantially higher compared to the FLAIR, axial T2*-FLASH2D, T2*-FLASH2D-HB, and T2-SPACE.
The adjusted SNR values differ slightly: Besides the high SNR values for T1-MPRAGE and SWI, also T2-SPACE imaging provided a markedly higher SNR compared to FLAIR, T2*-FLASH2D, and T2*-FLASH2D-HB imaging. The lowest SNR was computed for the SWI-MIP.
After the SNR calculation and according to the Rose criterion all scans allowed the recognition of image features with a 100% certainty (all the values are higher than 5) [16].
The lowest non-adjusted CNR results were calculated for the T1-MPRAGE images. T2-weighted imaging provided good contrast for the structures of interest, with only minor variation between the mean CNR values for FLAIR, T2*-FLASH2D, T2*-FLASH2D-HB, and T2-SPACE images. The best CNR was present in the SWI-MIP and SWI scans.
The highest adjusted CNR values were computed for T2-SPACE and SWI tra. All other T2-weighted sequences exhibit largely similar, markedly lower CNR values. The lowest adjusted CNR values were computed for the T1-MPRAGE images.
The PD patient’s images showed varying results for the non-adjusted SNR-values in comparison with the images of the healthy volunteers. While T2-SPACE [32.56], and SWI-MIP [420.43] images provided a higher SNR for the PD patient, the SNR of T1-MPRAGE (186.19) and T2-weighted-imaging (FLAIR [40.78], T2*-FLASH2D tra [38.31], T2*-FLASH2D-HB [25.37] and SWI [83.83]) were slightly lower compared to the calculated SNR values for the healthy volunteers.
In contrast, the CNR of the patient’s images were promising for the future application in PD. Higher non-adjusted CNRs were observed for the PD patient compared to the healthy volunteers. Particularly, the susceptibility-weighted imaging (SWI-MIP [361.79] and SWI [68.37]) provided a considerably higher CNR. Also T2*-FLASH2D (T2*-FLASH2D tra [23.58]) as well as T2-weighted imaging (FLAIR [21.85], T2*-FLASH2D-HB [25.11], and T2-SPACE [24.09]) demonstrated a markedly higher CNR.
Fusion of the Schaltenbrand and Wahren atlas with T2*-FLASH2D-imaging
Figure 4 shows an example of an axial T2*-FLASH2D image at the level of the GPi fused with the commonly used Schaltenbrand and Wahren atlas for stereotaxy of the human brain (plate 56) [69]. With the help of the atlas the GPi and its adjacent structures (i.e. Ci and LPm) are easily identified in the corresponding localization of the section schema.
Fusion of the Schaltenbrand and Wahren atlas for stereotaxy of the human brain with the axial T2*-FLASH2D-acquisition of a healthy volunteer (a). Digitized coronal schema of the Schaltenbrand and Wahren atlas at the level of the internal globus pallidus (plate 56). Characterization of the main cerebral structures in the area of interest (b). The internal globus pallidus is clearly visible within the boundaries of the Schaltenbrand and Wahren atlas schema for the GPi.
Discussion
Deep brain stimulation of the GPi provides a therapeutic option for patients with otherwise treatment-resistant dystonia and PD [48, 83]. After the approval by the FDA and the European regulatory authority (CE-Mark) the technique got a considerably increase in interest among patients and physicians. Although the underlying principles and mechanisms of DBS are not fully understood [10], the precise and reliable identification of the anatomical borders of the target structure remains a critical, and essential step for clinical efficacy [46, 53, 56].
As already pointed out, the indirect targeting of the DBS target structure is inferior to direct methods given the inter-individual anatomical variability in position, size, and functional segregation of the target nuclei [5, 37]. Additionally, many neurosurgical centers try to confirm the exact localization of the target structure with intra-operative eletrophysiological mapping using simultaneous multitrack microelectrode recording [3, 15]. It is worth mentioning, that although microelectrode stimulation remains the gold standard for the intra-operative verification of DBS target, microelectrode mapping can prolong the intervention time and multiple electrode trajections to the brain can increase the risk of intracranial hemorrhage [8, 9, 62, 75, 85].
For several reasons DBS of the GPi in particular remains a great challenge if it is based on electrophysiological testing only. Firstly, intra-operative test stimulation for electrophysiological mapping is not always helpful in verifying the optimal site of electrode placement for the GPi in dystonia. This is on the one hand due to an often prolonged period until stimulation of the GPi in dystonia comes into effect [20, 44, 48, 84]. On the other hand microelectrode mappings are often inconclusive in dystonia due to similarities of the firing pattern of the GPi and GPe [74]. Secondly, the adjacent structures have to be respected. The GPi is surrounded by different vital structures such as the GPe laterally and dorsally, the optic tract ventrally, and the internal capsule with the pyramidal tract posteromedially. Adverse effects, including dysarthria or tetanic contractions due to stimulation of the contiguous pyramidal tract located in the posterior limb of the internal capsule must be avoided [63]. Thirdly, a further drawback of electrophysiological testing in dystonia is the necessity of general anesthesia complicating the evaluation of possible stimulation effects [63]. Fourthly, despite the relatively large target area and the known functional segregation of the GPi [39], the optimal target location of DBS electrodes for stimulation effects is still in discussion [11, 76–78]. Nevertheless, promising long-term benefits have been reported for subregions of the GPi, namely the most postero-ventral [12, 13, 37, 45, 54] and probably the medial [80] portion of the nucleus.
Hence, a direct DBS approach, using stereotactic pre-operative MRI for visualization and targeting of the GPi is reasonably more appropriate. Thereby both, inter-individual variations of the position [37] and size (especially in neurodegenerative disorders) are taken into account [40].
Prior studies using 1.5 Tesla MRI provided inconsistent results for the delineation of the GPi [18, 21, 81]. Deficits in contrast, signal, and resolution of 1.5 T MRI systems lead to an insufficient delineation of the borders of the target structures [18]. With the implementation of 3.0 T scanners in clinical practice [14], improvements in image resolution, CNR, and SNR were achieved [6]. Specialized neurosurgical facilities have already tried to integrate this 3.0 T MRI technology in stereotactic planning [5, 34, 72]. For PD patients initial results for pre-operative 3.0 T imaging have only been reported for the subthalamic nucleus, but to our knowledge not for the GPi [70, 79].
The aim of our study was to determine the currently optimal sequence and orientation for GPi targeting using a standard installation protocol at 3.0 T.
To our knowledge, a direct comparison of the most promising new sequences for the visualization of the GPi using 3.0 T MRI has not been published.
Our qualitative results show that T2*-FLASH2D imaging is superior to standard sequences (T1-MPRAGE, FLAIR), and promising new sequences (T2-SPACE, SWI imaging).
In contrast to T1-weighted imaging providing no obvious delineation of the GPi, axial standard T2-weighted imaging resulted in a good delineation between the GPi and the Ci. Still the LPm, segregating the GPi vs. the GPe, was not conclusively visualized in T2-weighted imaging. Susceptibility-weighted MR imaging (SWI and SWI-MIP) provided an excellent delineation between the GPi and the adjacent structures. Nevertheless, the demarcation of the LPm was restricted. As noted above, recent studies reported the importance of identifying the subregions of the GPi. An increase in efficiency and a decrease of adverse effects of DBS appears to be related to such subregions [13, 76, 78]. A major finding of the present study is that the GPi was optimal visualized on T2*-FLASH2D imaging with a clear delineation of the Ci and the LPm. This may for the first time facilitate the direct MRI-guided targeting of the GPi subregions.
Concerning the orientation especially the axial and to a lesser degree the coronal view of the T2*-FLASH2D images allowed a clear demarcation of the GPi and its neighboring structures, in particular the Ci and the LPm.
Coronal acquisition of the SWI-images may further optimize the delineation of the GPi.
For neurosurgical procedures based on imaging, a good inter-rater reliability of radiological assessment is essential. Up to now, inter-rater reliability at 3.0 T MRI has not been assessed for the visualization of the GPi. To proof the reliability of our results the inter-rater reliability of the delineation of the GPi vs. Ci and LPm for two readers were calculated using Cohen’s kappa (Fig. 2). In prior studies calculated values were characterized as < 0 as no agreement, 0 – 0.20 as slight, 0.21 – 0.40 as fair, 0.41 – 0.60 as moderate, 0.61 – 0.80 as substantial, and 0.81 – 1 as almost perfect agreement [50, 51]. In our study, a moderate agreement for the delineation of the GPi vs. the Ci for T2*-FLASH2D-HB and SWI-MIP sequences and for the GPi vs. the LPm in the axial T2*-FLASH2D images was seen. All further sequences provided an inter-rater reliability of at least substantial or above.
These values document, that using appropriate sequences at 3.0 T, the delineation of the globus pallidum and its subregions can be reliably performed by readers experienced in the imaging of the deep brain nuclei. However, technical progress has to be made to further ameliorate the inter-rater agreement.
To quantify the image quality of the acquired sequences SNR and CNR measurements were conducted.
In general, SNR and CNR values are linear proportional to slice thickness and pixel size [71]. Due to the use of standard parameters with an acquisition time appropriate for clinical conditions, slice thickness and pixel size differ between the sequences used. To obtain geometrically comparable values SNR and CNR values were adjusted to a voxel of 1x1x1 mm3. The geometric correction has been used previously to compare vendor optimized sequences with different voxel sizes [35].
Our quantitative analysis of the non-adjusted measurements provided the highest CNR values for the susceptibility-weighted sequences (SWI-MIP and SWI), while T1-MPRAGE demonstrate the lowest CNR results for the GPi. We assume that the high CNR of the SWI is secondary to a high iron content (see below).
For the adjusted measurements, the highest CNR results were calculated for SWI, and for T2-SPACE imaging. The CNR differences between non-adjusted and adjusted values can be ascribed to several factors.
Firstly, the CNR adjustments for the high resolution sequence T2-SPACE results in a high converting factor and consecutively in high CNR values.
Secondly, the SD of noise measured in the T2*-FLASH2D sequences was relatively high in comparison to the other sequences. This sequence characteristic also contributes to a low CNR for T2*-FLASH2D in comparison to T2-SPACE.
Thirdly, CNR changes because of intra-voxel signal decrease are not taken into account in the geometric voxel adjustment [17].
However, the high CNR of the GPi in T2-SPACE (relative to the internal capsule) did not entail a clear delineation of the GPi versus LPm/GPe.
In the PD patient the non-adjusted CNRs for the GPi were higher than in the group of healthy volunteers. A considerable increase of the CNR was particularly seen in susceptibility-weighted (SWI-MIP and SWI). These observations in a single individual correlate with previous studies resulting in a hypointense appearance of the basal ganglia in T2-weighted imaging due to a progressive rise in iron content with age [68], and in neurodegenerative diseases [27, 55].
To evaluate the influence of different bandwidths on the quality of GPi visualization, we performed an initial comparison of two T2*-FLASH2D sequences with different bandwidths. The rationale was that a higher bandwidth leads to a reduction of susceptibility artifacts [2] and therefore influences the appearance of iron containing structures [82]. Although, the two sequences did not demonstrate a major difference in quantitative values, we qualitatively did observe a much better delineation of the GPi using the low-bandwidth T2*-FLASH2D sequence. This finding is consistent with prior studies indicating higher susceptibility artifacts for lower bandwidths and therefore a better visualization of iron-rich structures [67]. Future studies are necessary to find the optimal bandwidth for FLASH sequences in GPi imaging.
The sensitivity of T2*-contrast images and SWI for the detection of iron rich structures like the GPi has previously been documented [43, 65].
For the imaging of age-related neurodegenerative processes iron plays a major role [73]. With advancing age by the third decade an increase of the nonheme iron concentration in the basal ganglia has been reported. Concerning the GPi a stabilization of the total amount of nonheme iron after the fourth decade was found [33, 42]. Therefore, T2-weighted sequences are considered the standard sequences for the identification of the basal ganglia [23, 24]. Beyond, iron still plays a major role in the research of neurodegenerative diseases [49, 57]. Several studies showed a progressive increase of iron concentrations also after the fourth decade in deep brain structures of PD patients [27, 86]. Although, the exact relationship between iron accumulation and PD remains still unclear, recent reports have suggested a non-specific effect of neuronal degeneration [22]. The remarkably high iron concentration of the basal ganglia in dystonia and PD corresponds to a hypointense signal of the GPi in T2-weighted sequences [24, 26]. With the development of gradient echo sequences with T2*-weighted images and later susceptibility weighted images [22, 60], the iron based imaging could further be optimized. SWI has lately been introduced to clinical MRI [32]. It provides a combination of gradient-echo magnitude and phase change images which are influences by the magnetic susceptibility of iron [31]. Due to the progressive iron accumulation in the DBS target structures in PD and dystonia, T2*, T2*-FLASH2D, and SWI imaging represent the currently optimal techniques for a reliable identification.
As an additional benefit we found that SWI provided an extraordinary visualization of deep cerebral veins and transparenchymal vessels. Pre-operatively planning the trajection line for DBS surgery, this anatomic detail is of special interest for the neurosurgeon. Hence, intracranial hemorrhages due to blood vessel injuries can possibly be avoided (Fig. 5) [62].
Susceptibility-weighted imaging indicating the main deep cerebral veins at the level of the internal globus pallidus (a-c). The deep cerebral veins (ICV: internal cerebral veins, TSV: thalamostriate veins, SV: septal veins) can be easily identified so that the trajectory can be readjusted to avoid hemorrhage.
Although the GPi is well visualized on T2*-FLASH2D images and the CNR and SNR are sufficient for the region of interest, the anatomic accuracy of the approach had to be confirmed. In a fusion of an axial T2*-FLASH2D image at the level of the GPi with the Schaltenbrand and Wahren atlas, the GPi obviously projects in the borders of the stereotactical schema. Even the LPm is accurately mapped in the images. This further confirms the correct identification and localization of the GPi in the acquired images. However, it has to be kept in mind that in general direct targeting based on pre-operative imaging has well-known limitations. In particular, distortions of MR images [7] and brain shift due to swelling or cerebrospinal fluid leakage and subsequent air invasion during surgery [59] may cause a discrepancy of the pre-operative images and the intra-operative anatomy.
Our study has a few potential limitations. First, we investigated a homogenous, young, and healthy volunteer collective without confounding factors that are typically present in a clinical population. This implies that in a clinical population slightly different results have to be expected. Especially the higher iron content in older patients and in patients with neurodegenerative diseases might influence the delineation of the basal ganglia. Second, the comparison of vendor optimized sequences was complicated by different voxel sizes which had to be adjusted. This geometric correction ignores intravoxel signal decay (see above). However, the primary use of sequences adjusted to equal voxel size resulting in impractical slice thicknesses and long scanning times was not within the scope of our study.
Nevertheless, despite the superior delineation of the GPi in T2*-FLASH2D imaging, the results can not be applied to clinical practice directly. Validation of our findings in a patient group (especially dystonia and PD) remains to be performed. Confirmation of the geometric accuracy for the direct targeting based on 3.0 T MR imaging should be proven in advance to clinical application. Furthermore, limitations of image quality have to be anticipated due to the use of head frames and motion artifacts of dystonia and PD patients.
Conclusion
Using a standard installation protocol at 3.0 T the GPi was reliably and optimally visualized in the T2*-FLASH2D images, particularly in the axial and to a lesser degree the coronal view. Due to the use of vendor optimized sequences our protocol can be easily implemented in clinical routine.
Consequently, for the clinical practice a combination of both techniques, T2*-FLASH2D and SWI, facilitates a more precise GPi targeting and possibly a reduction of intervention time and complicating hemorrhages in DBS surgery.
References
Deep-Brain Stimulation for Parkinson's Disease Study Group (2001) Deep-brain stimulation of the subthalamic nucleus or the pars interna of the globus pallidus in Parkinson's disease. N Engl J Med 345:956–963
Abosch A, Yacoub E, Ugurbil K, Harel N (2010) An assessment of current brain targets for deep brain stimulation surgery with susceptibility-weighted imaging at 7 tesla. Neurosurgery 67:1745–1756, discussion 1756
Amirnovin R, Williams ZM, Cosgrove GR, Eskandar EN (2006) Experience with microelectrode guided subthalamic nucleus deep brain stimulation. Neurosurgery 58:ONS96–102, discussion ONS196-102
Andrade P, Carrillo-Ruiz JD, Jimenez F (2009) A systematic review of the efficacy of globus pallidus stimulation in the treatment of Parkinson's disease. J Clin Neurosci 16:877–881
Ashkan K, Blomstedt P, Zrinzo L, Tisch S, Yousry T, Limousin-Dowsey P, Hariz MI (2007) Variability of the subthalamic nucleus: the case for direct MRI guided targeting. Br J Neurosurg 21:197–200
Bachmann R, Reilmann R, Schwindt W, Kugel H, Heindel W, Kramer S (2006) FLAIR imaging for multiple sclerosis: a comparative MR study at 1.5 and 3.0 Tesla. Eur Radiol 16:915–921
Balachandran R, Welch EB, Dawant BM, Fitzpatrick JM (2010) Effect of MR distortion on targeting for deep-brain stimulation. IEEE Trans Biomed Eng 57:1729–1735
Bejjani BP, Dormont D, Pidoux B, Yelnik J, Damier P, Arnulf I, Bonnet AM, Marsault C, Agid Y, Philippon J, Cornu P (2000) Bilateral subthalamic stimulation for Parkinson's disease by using three-dimensional stereotactic magnetic resonance imaging and electrophysiological guidance. J Neurosurg 92:615–625
Bejjani BP, Gervais D, Arnulf I, Papadopoulos S, Demeret S, Bonnet AM, Cornu P, Damier P, Agid Y (2000) Axial parkinsonian symptoms can be improved: the role of levodopa and bilateral subthalamic stimulation. J Neurol Neurosurg Psychiatry 68:595–600
Benazzouz A, Hallett M (2000) Mechanism of action of deep brain stimulation. Neurology 55:S13–16
Bergman H, Feingold A, Nini A, Raz A, Slovin H, Abeles M, Vaadia E (1998) Physiological aspects of information processing in the basal ganglia of normal and parkinsonian primates. Trends Neurosci 21:32–38
Bhidayasiri R (2006) Dystonia: genetics and treatment update. Neurologist 12:74–85
Bhidayasiri R, Tarsy D (2006) Treatment of dystonia. Expert Rev Neurother 6:863–886
Biswas J, Nelson CB, Runge VM, Wintersperger BJ, Baumann SS, Jackson CB, Patel T (2005) Brain tumor enhancement in magnetic resonance imaging: comparison of signal-to-noise ratio (SNR) and contrast-to-noise ratio (CNR) at 1.5 versus 3 tesla. Invest Radiol 40:792–797
Bour LJ, Contarino MF, Foncke EM, de Bie RM, van den Munckhof P, Speelman JD, Schuurman PR (2010) Long-term experience with intraoperative microrecording during DBS neurosurgery in STN and GPi. Acta Neurochir (Wien) 152:2069–2077
Bushberg JT, Seibert JT, Leidholdt EM Jr, Boone JM (2001) The Essential Physics of Medical Imaging. Lippincott Williams & Wilkins, Philadelphia
Chen N, Wyrwicz AM (1999) Removal of intravoxel dephasing artifact in gradient-echo images using a field-map based RF refocusing technique. Magn Reson Med 42:807–812
Cho ZH, Min HK, Oh SH, Han JY, Park CW, Chi JG, Kim YB, Paek SH, Lozano AM, Lee KH (2010) Direct visualization of deep brain stimulation targets in Parkinson disease with the use of 7-tesla magnetic resonance imaging. J Neurosurg 113:639–647
Cohen JA (1960) A coefficient of agreement for nominal scales. Educ Psychol Meas 20:37–46
Coubes P, Cif L, Azais M, Roubertie A, Hemm S, Diakonoya N, Vayssiere N, Monnier C, Hardouin E, Ganau A, Tuffery S, Claustre M, Echenne B (2002) Treatment of dystonia syndrome by chronic electric stimulation of the internal globus pallidus. Arch Pediatr 9(Suppl 2):84s–86s
Cuny E, Guehl D, Burbaud P, Gross C, Dousset V, Rougier A (2002) Lack of agreement between direct magnetic resonance imaging and statistical determination of a subthalamic target: the role of electrophysiological guidance. J Neurosurg 97:591–597
Dormont D, Ricciardi KG, Tande D, Parain K, Menuel C, Galanaud D, Navarro S, Cornu P, Agid Y, Yelnik J (2004) Is the subthalamic nucleus hypointense on T2-weighted images? A correlation study using MR imaging and stereotactic atlas data. AJNR Am J Neuroradiol 25:1516–1523
Drayer B, Burger P, Darwin R, Riederer S, Herfkens R, Johnson GA (1986) MRI of brain iron. AJR Am J Roentgenol 147:103–110
Drayer BP (1989) Basal ganglia: significance of signal hypointensity on T2-weighted MR images. Radiology 173:311–312
Duyn JH, van Gelderen P, Li TQ, de Zwart JA, Koretsky AP, Fukunaga M (2007) High-field MRI of brain cortical substructure based on signal phase. Proc Natl Acad Sci U S A 104:11796–11801
Gavarini S, Vayssiere N, Delort P, Cif L, Biolsi B, Tancu C, Vasques X, Plagnol S, Bonafe A, Coubes P (2008) Stereotactic MRI in DYT1 dystonia: focal signal abnormalities in the basal ganglia do not contraindicate deep brain stimulation. Stereotact Funct Neurosurg 86:245–252
Griffiths PD, Dobson BR, Jones GR, Clarke DT (1999) Iron in the basal ganglia in Parkinson's disease. An in vitro study using extended X-ray absorption fine structure and cryo-electron microscopy. Brain 122(Pt 4):667–673
Gringel T, Schulz-Schaeffer W, Elolf E, Frolich A, Dechent P, Helms G (2009) Optimized high-resolution mapping of magnetization transfer (MT) at 3 Tesla for direct visualization of substructures of the human thalamus in clinically feasible measurement time. J Magn Reson Imaging 29:1285–1292
Gross RE, Krack P, Rodriguez-Oroz MC, Rezai AR, Benabid AL (2006) Electrophysiological mapping for the implantation of deep brain stimulators for Parkinson's disease and tremor. Mov Disord 21(Suppl 14):S259–283
Guo T, Finnis KW, Deoni SC, Parrent AG, Peters TM (2006) Comparison of different targeting methods for subthalamic nucleus deep brain stimulation. Med Image Comput Comput Assist Interv 9:768–775
Haacke EM, Ayaz M, Khan A, Manova ES, Krishnamurthy B, Gollapalli L, Ciulla C, Kim I, Petersen F, Kirsch W (2007) Establishing a baseline phase behavior in magnetic resonance imaging to determine normal vs. abnormal iron content in the brain. J Magn Reson Imaging 26:256–264
Haacke EM, Xu Y, Cheng YC, Reichenbach JR (2004) Susceptibility weighted imaging (SWI). Magn Reson Med 52:612–618
Hallgren B, Sourander P (1958) The effect of age on the non-haemin iron in the human brain. J Neurochem 3:41–51
Hamani C, Richter EO, Andrade-Souza Y, Hutchison W, Saint-Cyr JA, Lozano AM (2005) Correspondence of microelectrode mapping with magnetic resonance imaging for subthalamic nucleus procedures. Surg Neurol 63:249–253, discussion 253
Haneder S, Attenberger UI, Biffar A, Dietrich O, Fink C, Schoenberg SO, Michaely HJ (2011) Gadofosveset: Parameter Optimization for Steady-State Imaging of the Thoracic and Abdominal Vasculature. Invest Radiol 46:678–685
Hariz MI, Bergenheim AT (1990) A comparative study on ventriculographic and computerized tomography-guided determinations of brain targets in functional stereotaxis. J Neurosurg 73:565–571
Hirabayashi H, Tengvar M, Hariz MI (2002) Stereotactic imaging of the pallidal target. Mov Disord 17(Suppl 3):S130–134
Holtzheimer PE 3rd, Roberts DW, Darcey TM (1999) Magnetic resonance imaging versus computed tomography for target localization in functional stereotactic neurosurgery. Neurosurgery 45:290–297, discussion 297–298
Hutchison WD, Lozano AM, Davis KD, Saint-Cyr JA, Lang AE, Dostrovsky JO (1994) Differential neuronal activity in segments of globus pallidus in Parkinson's disease patients. Neuroreport 5:1533–1537
Katayama S, Watanabe C, Khoriyama T, Oka M, Mao JJ, Yamamura Y, Tahara E, Nakamura S (1998) Slowly progressive L-DOPA nonresponsive pure akinesia due to nigropallidal degeneration: a clinicopathological case study. J Neurol Sci 161:169–172
Katsakiori PF, Kefalopoulou Z, Markaki E, Paschali A, Ellul J, Kagadis GC, Chroni E, Constantoyannis C (2009) Deep brain stimulation for secondary dystonia: results in 8 patients. Acta Neurochir (Wien) 151:473–478, discussion 478
Kitajima M, Korogi Y, Kakeda S, Moriya J, Ohnari N, Sato T, Hayashida Y, Hirai T, Okuda T, Yamashita Y (2008) Human subthalamic nucleus: evaluation with high-resolution MR imaging at 3.0 T. Neuroradiology 50:675–681
Kosta P, Argyropoulou MI, Markoula S, Konitsiotis S (2006) MRI evaluation of the basal ganglia size and iron content in patients with Parkinson's disease. J Neurol 253:26–32
Krauss JK, Yianni J, Loher TJ, Aziz TZ (2004) Deep brain stimulation for dystonia. J Clin Neurophysiol 21:18–30
Kringelbach ML, Jenkinson N, Green AL, Owen SL, Hansen PC, Cornelissen PL, Holliday IE, Stein J, Aziz TZ (2007) Deep brain stimulation for chronic pain investigated with magnetoencephalography. Neuroreport 18:223–228
Kringelbach ML, Jenkinson N, Owen SL, Aziz TZ (2007) Translational principles of deep brain stimulation. Nat Rev Neurosci 8:623–635
Kumar R, Lang AE, Rodriguez-Oroz MC, Lozano AM, Limousin P, Pollak P, Benabid AL, Guridi J, Ramos E, van der Linden C, Vandewalle A, Caemaert J, Lannoo E, van den Abbeele D, Vingerhoets G, Wolters M, Obeso JA (2000) Deep brain stimulation of the globus pallidus pars interna in advanced Parkinson's disease. Neurology 55:S34–39
Kupsch A, Benecke R, Muller J, Trottenberg T, Schneider GH, Poewe W, Eisner W, Wolters A, Muller JU, Deuschl G, Pinsker MO, Skogseid IM, Roeste GK, Vollmer-Haase J, Brentrup A, Krause M, Tronnier V, Schnitzler A, Voges J, Nikkhah G, Vesper J, Naumann M, Volkmann J (2006) Pallidal deep-brain stimulation in primary generalized or segmental dystonia. N Engl J Med 355:1978–1990
Kurian MA, McNeill A, Lin JP, Maher ER (2011) Childhood disorders of neurodegeneration with brain iron accumulation (NBIA). Dev Med Child Neurol 53:394–404
Landis JR, Koch GG (1977) An application of hierarchical kappa-type statistics in the assessment of majority agreement among multiple observers. Biometrics 33:363–374
Landis JR, Koch GG (1977) The measurement of observer agreement for categorical data. Biometrics 33:159–174
Lee JY, Kim JW, Lim YH, Kim C, Kim DG, Jeon BS, Paek SH (2010) Is MRI a reliable tool to locate the electrode after deep brain stimulation surgery? Comparison study of CT and MRI for the localization of electrodes after DBS. Acta Neurochir (Wien) 152:2029–2036
Lee KH, Blaha CD, Garris PA, Mohseni P, Horne AE, Bennet KE, Agnesi F, Bledsoe JM, Lester DB, Kimble C, Min HK, Kim YB, Cho ZH (2009) Evolution of Deep Brain Stimulation: Human Electrometer and Smart Devices Supporting the Next Generation of Therapy. Neuromodulation 12:85–103
Lozano A, Hutchison W, Kiss Z, Tasker R, Davis K, Dostrovsky J (1996) Methods for microelectrode-guided posteroventral pallidotomy. J Neurosurg 84:194–202
Manova ES, Habib CA, Boikov AS, Ayaz M, Khan A, Kirsch WM, Kido DK, Haacke EM (2009) Characterizing the mesencephalon using susceptibility-weighted imaging. AJNR Am J Neuroradiol 30:569–574
McIntyre CC, Savasta M, Kerkerian-Le Goff L, Vitek JL (2004) Uncovering the mechanism(s) of action of deep brain stimulation: activation, inhibition, or both. Clin Neurophysiol 115:1239–1248
McNeill A, Chinnery PF (2011) Neurodegeneration with brain iron accumulation. Handb Clin Neurol 100:161–172
Mehdorn HM, Goebel S, Falk D, Volkmann J, Leplow B, Pinsker MO (2008) Deep brain stimulation for movement disorders and its neuropsychological implications. Acta Neurochir Suppl 101:9–12
Miyagi Y, Shima F, Sasaki T (2007) Brain shift: an error factor during implantation of deep brain stimulation electrodes. J Neurosurg 107:989–997
Ordidge RJ, Gorell JM, Deniau JC, Knight RA, Helpern JA (1994) Assessment of relative brain iron concentrations using T2-weighted and T2*-weighted MRI at 3 Tesla. Magn Reson Med 32:335–341
Papavassiliou E, Rau G, Heath S, Abosch A, Barbaro NM, Larson PS, Lamborn K, Starr PA (2004) Thalamic deep brain stimulation for essential tremor: relation of lead location to outcome. Neurosurgery 54:1120–1129, discussion 1129–1130
Park JH, Chung SJ, Lee CS, Jeon SR (2011) Analysis of hemorrhagic risk factors during deep brain stimulation surgery for movement disorders: comparison of the circumferential paired and multiple electrode insertion methods. Acta Neurochir (Wien) 153:1573–1578
Pinsker MO, Volkmann J, Falk D, Herzog J, Steigerwald F, Deuschl G, Mehdorn HM (2009) Deep brain stimulation of the internal globus pallidus in dystonia: target localisation under general anaesthesia. Acta Neurochir (Wien) 151:751–758
Rehncrona S, Johnels B, Widner H, Tornqvist AL, Hariz M, Sydow O (2003) Long-term efficacy of thalamic deep brain stimulation for tremor: double-blind assessments. Mov Disord 18:163–170
Rossi M, Ruottinen H, Elovaara I, Ryymin P, Soimakallio S, Eskola H, Dastidar P (2010) Brain Iron Deposition and Sequence Characteristics in Parkinsonism: Comparison of SWI, T2* Maps, T2-Weighted-, and FLAIR-SPACE. Invest Radiol 45:795–802
Rouaud T, Dondaine T, Drapier S, Haegelen C, Lallement F, Peron J, Raoul S, Sauleau P, Verin M (2010) Pallidal stimulation in advanced Parkinson's patients with contraindications for subthalamic stimulation. Mov Disord 25:1839–1846
Runge VM, Wood ML, Kaufman DM, Traill MR, Nelson KL (1988) The straight and narrow path to good head and spine MRI. Radiographics 8:507–531
Rutledge JN, Hilal SK, Silver AJ, Defendini R, Fahn S (1987) Study of movement disorders and brain iron by MR. AJR Am J Roentgenol 149:365–379
Schaltenbrand G, Wahren, W. (1977) Atlas for Stereotaxy of the Human Brain. Thieme Stuttgart
Slavin KV, Thulborn KR, Wess C, Nersesyan H (2006) Direct visualization of the human subthalamic nucleus with 3 T MR imaging. AJNR Am J Neuroradiol 27:80–84
Stark DD, Bradley WG (1999) Magnetic resonance imaging. C.V. Mosby, St. Louis
Starr PA, Vitek JL, DeLong M, Bakay RA (1999) Magnetic resonance imaging-based stereotactic localization of the globus pallidus and subthalamic nucleus. Neurosurgery 44:303–313, discussion 313–304
Strassmann G (1949) Iron and calcium deposits in the brain; their pathologic significance. J Neuropathol Exp Neurol 8:428–435, illust
Tang JK, Moro E, Mahant N, Hutchison WD, Lang AE, Lozano AM, Dostrovsky JO (2007) Neuronal firing rates and patterns in the globus pallidus internus of patients with cervical dystonia differ from those with Parkinson's disease. J Neurophysiol 98:720–729
Terao T, Takahashi H, Yokochi F, Taniguchi M, Okiyama R, Hamada I (2003) Hemorrhagic complication of stereotactic surgery in patients with movement disorders. J Neurosurg 98:1241–1246
Tisch S, Rothwell JC, Limousin P, Hariz MI, Corcos DM (2007) The physiological effects of pallidal deep brain stimulation in dystonia. IEEE Trans Neural Syst Rehabil Eng 15:166–172
Tisch S, Silberstein P, Limousin-Dowsey P, Jahanshahi M (2004) The basal ganglia: anatomy, physiology, and pharmacology. Psychiatr Clin North Am 27:757–799
Tisch S, Zrinzo L, Limousin P, Bhatia KP, Quinn N, Ashkan K, Hariz M (2007) Effect of electrode contact location on clinical efficacy of pallidal deep brain stimulation in primary generalised dystonia. J Neurol Neurosurg Psychiatry 78:1314–1319
Toda H, Sawamoto N, Hanakawa T, Saiki H, Matsumoto S, Okumura R, Ishikawa M, Fukuyama H, Hashimoto N (2009) A novel composite targeting method using high-field magnetic resonance imaging for subthalamic nucleus deep brain stimulation. J Neurosurg 111:737–745
Trepanier LL, Kumar R, Lozano AM, Lang AE, Saint-Cyr JA (2000) Neuropsychological outcome of GPi pallidotomy and GPi or STN deep brain stimulation in Parkinson's disease. Brain Cogn 42:324–347
Vayssiere N, Hemm S, Cif L, Picot MC, Diakonova N, El Fertit H, Frerebeau P, Coubes P (2002) Comparison of atlas- and magnetic resonance imaging-based stereotactic targeting of the globus pallidus internus in the performance of deep brain stimulation for treatment of dystonia. J Neurosurg 96:673–679
Vertinsky AT, Coenen VA, Lang DJ, Kolind S, Honey CR, Li D, Rauscher A (2009) Localization of the subthalamic nucleus: optimization with susceptibility-weighted phase MR imaging. AJNR Am J Neuroradiol 30:1717–1724
Vidailhet M, Vercueil L, Houeto JL, Krystkowiak P, Benabid AL, Cornu P, Lagrange C, Tezenas du Montcel S, Dormont D, Grand S, Blond S, Detante O, Pillon B, Ardouin C, Agid Y, Destee A, Pollak P (2005) Bilateral deep-brain stimulation of the globus pallidus in primary generalized dystonia. N Engl J Med 352:459–467
Volkmann J, Benecke R (2002) Deep brain stimulation for dystonia: patient selection and evaluation. Mov Disord 17(Suppl 3):S112–115
Xiaowu H, Xiufeng J, Xiaoping Z, Bin H, Laixing W, Yiqun C, Jinchuan L, Aiguo J, Jianmin L (2010) Risks of intracranial hemorrhage in patients with Parkinson's disease receiving deep brain stimulation and ablation. Parkinsonism Relat Disord 16:96–100
Zhang W, Sun SG, Jiang YH, Qiao X, Sun X, Wu Y (2009) Determination of brain iron content in patients with Parkinson's disease using magnetic susceptibility imaging. Neurosci Bull 25:353–360
Conflicts of interest
None.
Author information
Authors and Affiliations
Corresponding author
Additional information
Ingo S. Nölte and Lars Gerigk contributed equally to this article.
Rights and permissions
About this article
Cite this article
Nölte, I.S., Gerigk, L., Al-Zghloul, M. et al. Visualization of the internal globus pallidus: sequence and orientation for deep brain stimulation using a standard installation protocol at 3.0 Tesla. Acta Neurochir 154, 481–494 (2012). https://doi.org/10.1007/s00701-011-1242-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00701-011-1242-8