Abstract
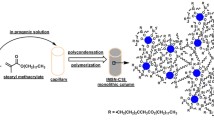
Innovative chiral capillary silica monoliths (CSMs) were developed based on DNA nanoflowers (DNFs). Baseline separation of enantiomers such as atenolol, tyrosine, histidine, and nefopam was achieved by using DNF-modified CSMs, and the obtained resolution value was higher than 1.78. To further explore the effect of DNFs on enantioseparation, different types of chiral columns including DNA strand containing the complementary sequence of the template (DCT)-modified CSMs, DNF2-modified CSMs, and DNF3-modified CSMs were prepared as well. It was observed that DNF-modified CSMs displayed better chiral separation ability compared with DCT-based columns. The intra-day and inter-day repeatability of model analytes’ retention time and resolution kept desirable relative standard deviation values of less than 8.28%. DNF2/DNF3-modified CSMs were able to achieve baseline separation of atenolol, propranolol, 2′-deoxyadenosine, and nefopam enantiomers. Molecular docking simulations were performed to investigate enantioselectivity mechanisms of DNA sequences for enantiomers. To indicate the successful construction of DNFs and DNF-modified CSMs, various charaterization approaches including scanning electron microscopy, agarose gel electrophoresis, dynamic light scattering analysis, electroosmotic flow, and Fourier-transform infrared spectroscopy were utilized. Moreover, the enantioseparation performance of DNF-modified CSMs was characterized in terms of sample volume, applied voltage, and buffer concentration. This work paves the way to applying DNF-based capillary electrochromatography microsystems for chiral separation.
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nanomaterials (NMs) occupy a crucial position in materials science due to their prominent advantages including large surface-to-volume ratios and multiform morphologies [1, 2]. There exist all sorts of pretty flowers in nature, whereas the life of flowers is ephemeral. The artistic structures of nanoflowers can sustain the beauty of natural flowers. Compared with traditional spherical NMs, nanoflowers possess higher surface area owing to their topographic properties of nanolayers. As innovative generation NMs, nanoflowers give rise to diversified applications in biocatalysis, biosensing, drug delivery, and cancer therapy [3,4,5,6]. DNA nanoflowers (DNFs) are a type of nanoflowers composing ultralong-strand DNA and inorganic metal-ion frameworks [7,8,9,10,11]. The DNFs are self-assembled by the liquid crystallization of replicated DNA building blocks without relying on Watson–Crick base-pairing. Depending on their programmability, high stability, and intrinsic functionalities, DNFs have attracted significant attention in biomedical field. However, the employment of DNFs for chiral separation is seldom reported.
Chirality is one of the predominant attributes of nature [12,13,14,15,16]. Efficient enantiomeric separation is of crucial significance. Endogenous biomolecules including DNA, protein, and polysaccharide can construct the basic molecular units of life. The inherent chirality of biomolecules enables them to be satisfactory chiral selectors [17]. For instance, polysaccharide-based chiral stationary phases were widely used in chromatography enantioseparation [18]. Stereoselective antibodies were able to be applied for detecting stereoisomers in immunoassays and sensors, and separating stereoisomers in chiral immunoaffinity chromatography [19]. The combination of biomolecules and NMs provides a marvellous opportunity for preparing various chiral materials [20,21,22]. A composite chiral interface was obtained based on bovine serum albumin (BSA), multi-walled carbon nanotubes (MWCNTs), and methylene blue [23]. Results indicated that the chiral identification efficiency was able to be enhanced by combining BSA and MWCNTs. In addition to proteins, a dipeptide was chosen as the chiral selector to develop dipeptide-functionalized graphene oxide (GO) membranes [24]. Compared with traditional chiral separation membranes, the resulting membranes presented improved separation factor. Although distinct strategies have been applied for stereorecognition, chiral separation still brings about great challenge due to the similar physicochemical behaviors of the enantiomers.
Among versatile approaches employed for chiral separation, capillary electrochromatography (CEC) is considered a desirable technique based on its high efficiency and specific selectivity [25,26,27,28]. We reviewed recent advances in fabricating CEC systems for chiral separation [29, 30]. Developing new chiral selectors has occupied a predominant position in chiral CEC. Investigating NM-based chiral stationary phases offers a prospective direction for the development of chiral CEC microsystems [31,32,33]. NMs with large surface area are beneficial to improve biomolecules’ immobilization efficiency and enantioseparation performance in terms of efficiency and selectivity. Wang et al. developed an innovative chiral CEC microsystem depending on zeolitic imidazolate framework-8 NMs utilizing l-histidine as the chiral carbon center [34]. The dual-chiral β-cyclodextrin covalent organic frameworks were applied as chiral stationary phase in capillary columns [35]. The resulting columns have been successfully employed to the separation of various enantiomers. DNA is a beautiful example of the natural self-assembling biomolecules. Diverse NMs have been applied for developing state-of-the-art chiral sensors by choosing DNA as a chiral donor [36,37,38,39,40]. It provides a predominant inspiration to integrate DNA and NMs in CEC chiral separation.
Herein, we developed innovative GO-modified capillary silica monoliths by choosing DNFs as the chiral selector. The DNFs were self-assembled by the liquid crystallization of replicated DNA building blocks. In addition to choosing appropriate chiral selectors, a desirable support for biomolecule immobilization should also be carefully considered. Therefore, capillary silica monoliths with high surface areas and desirable stability were chosen for DNF immobilization [41, 42]. The influence of DNFs on DNF-GO@CSM enantioseparation ability was explored to explain the chiral discrimination mechanisms from material perspective. To provide microscopic insight into the enantiorecognition mechanism between DNA and enantiomers, molecular docking (MD) simulations were performed by selecting histidine and nefopam as model analytes. To further evaluate the enantioseparation capability of DNFs, DNF2-GO@CSM and DNF3-GO@CSM were designed and applied for chiral separation. The optimized columns presented desirable chiral selectivity for various enantiomers including atenolol, tyrosine, histidine, nefopam, propranolol, and 2′-deoxyadenosine. This work indicated that DNF-based capillary silica monoliths have a significant advantage for chiral separation.
Materials and methods
Chemicals and materials
All DNA oligonucleotides were synthesized and HPLC purified by Sangon Biotechnology Co., Ltd. (Shanghai, China), and the sequences are shown in Table S1. DNFs were prepared according to the approach described in previous paper [43]. Tetramethoxysilane (TMOS), 3-aminopropyl trimethoxysilane (APTS), polyethylene glycol (PEG), disodium hydrogen phosphate, sodium dihydrogen phosphate, urea, 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (EDCI), agarose, atenolol, tyrosine, histidine, nefopam, propranolol, and 2′-deoxyadenosine enantiomers were purchased from Aladdin Co., Ltd. (Shanghai, China). T4 DNA ligase and Phi29 DNA polymerase were purchased from New England BioLabs (Beverly, MA, USA). Commercial GO dispersion was purchased from Xianfeng Nano-materials Technology Co., Ltd. (Nanjing, China). Acetic acid, phosphoric acid, hydrochloric acid, toluene, methanol, and sodium hydroxide were obtained from Nanjing, Chemical Reagent Co., Ltd. (Nanjing, China). Ultrapure water (18.2 MΩ·cm) purified by Milli-Q system (Millipore, Bedford, MA, USA) was used for the preparation of all solutions. The capillary silica monoliths were prepared by a sol–gel process according to the method described in previous paper [44, 45].
Sample and mobile phase preparation
The background electrolytes of phosphate buffer were obtained by dissolving an exact amount of disodium hydrogen phosphate in deionized water. Phosphoric acid solution was utilized to adjust the pH value of the buffer solution. The sample solutions (1 mg mL−1) of atenolol, tyrosine, histidine, nefopam, propranolol, and 2′-deoxyadenosine enantiomers were prepared by dissolving analytes in the buffer solution. DNA strands employed in this work were dissolved in deionized water. All above solutions were stored at 4 °C in a refrigerator and filtered through 0.22-μm pore size nylon membrane filters before use.
Preparation of DNA nanoflower-modified capillary silica monoliths
To maximize the number of silanol groups on monolith surface, 1 M HCl solution was pumped through capillary silica monoliths for 3 h. The columns were rinsed to neutral with water followed by methanol, and then dried by nitrogen. Afterwards, amino-functionalized capillary silica monoliths were obtained by introducing 10% (v/v) APTS toluene solution into the columns and reacting at 110 °C for 6 h. To eliminate the unreacted APTS, the columns were flushed with toluene and methanol, and dried by a nitrogen stream. A certain volume of aqueous GO solution containing potassium hydroxide (2 mg mL−1) was drawn into the columns and then reacted at 40 °C for 2 h. Subsequently, the GO-modified capillary silica monoliths were flushed with water and methanol and dried by nitrogen. Furthermore, 20 mM EDCI dissolved in 100 mM sodium phosphate solution (pH 5.0) was continued to flow through the GO-modified capillary silica monoliths for 1 h. Finally, the DNF solution was pumped through the EDCI-activated columns for 5 h. These proposed DNF-immobilized columns were called DNF-GO@CSMs. The preparation procedure for DNF-GO@CSMs is shown in Fig. 1.
To explore the effect of DNF on chiral separation, DNA strand containing the complementary sequence of the template was coated onto the EDCI-activated columns as well. These ssDNA-functionalized capillary silica monoliths were called DCT-GO@CSMs. The resulting columns were rinsed with sodium phosphate buffer solution (20 mM phosphate buffer + 100 mM NaCl, pH 7.4) to remove unbound DNA. Before installation to the CEC instrument, the obtained DNA-modified columns were conditioned with sodium phosphate running buffer solution for 1 h.
Characterization
The surface morphologies of DNFs and DNF-immobilized capillary silica monoliths were visualized by using SEM (Hitachi S-3400N, Tokyo, Japan). FT-IR spectra (4000–400 cm−1) was collected based on a Magna-560 spectrometer (Nicolet, Madison, WI, USA) in KBr plate. DLS was utilized to measure the hydrated radius of DNFs. The agarose gel electrophoresis analysis of DNF and DNA strands was implemented by employing a 2% agarose gel in Tris–acetate-EDTA buffer. The gel was run at a constant voltage of 110 V for 40 min.
Electrochromatography procedures
The CEC experiments were performed on a CL1040 high-performance capillary electrophoresis system (Huayang liming instrument Co., China). Analytes were electrokinetic injected at a voltage and were detected at 215 nm. During the running process, the applied voltage ranged from 5 to 15 kV was utilized. And the capillary temperature was kept at 25 °C. All solutions were degassed and filtered by utilizing 0.22-μm pore size filters before running.
Molecular simulations
The docking of histidine/nefopam and DCT was completed in the software AutoDock Vina 1.1.2. The 3D structure of DCT was predicted by utilizing 3dRNA/DNA server (http://biophy.hust.edu.cn/new/3dRNA/create). The 3D structures of histidine/nefopam were obtained from PUBCHEM database. Moreover, ADFRsuite 1.0 was used to convert the processed small molecules and DCT into the PDBQT format required for AutoDock Vina 1.1.2 docking. In the docking process, the highest rated docking conformation output was considered the best receptor-ligand complex. The PyMol 2.5.4 was utilized for graphical presentation.
Results and discussion
Characterization of DNA nanoflowers
A phosphorylated linear template was utilized to synthesize a circular template for rolling circle amplification (RCA). During the amplification process, tens to hundreds of tandem repeats could be introduced into a long ssDNA product. Agarose gel electrophoresis was employed to confirm the formation of DNF. A representative gel image is presented in Figure S1. Owing to their different lengths, the template and primer exhibited different migration velocities. Assembled DNFs possessed a migration velocity approximately to zero due to their significantly larger molecular weight. These electrophoresis results suggested that DNFs were successfully prepared. Furthermore, the formation of DNF flower-like spherical morphology was characterized by SEM. As seen in Fig. 2, the monodispersed DNFs possessed petal-like structures on their surface. DLS analysis indicated that the resulting monodisperse DNFs presented an average diameter of approximately 500 nm (Figure S2).
Characterization of DNA nanoflower-modified capillary silica monoliths
After each step of reaction, EOF was determined to evaluate the properties of the resulting capillary columns. The negative charge density decreased when APTS was introduced into monolithic matrix, inducing an increase of the retention time for thiourea. Attributing to the modification of GO onto capillary monolith, the negative charge density increased and there was an enhanced EOF mobility on the GO-functionalized monolith. As seen in Figure S3, EOF mobility on DNF@CSM further increased when compared with GO-based monolith. This phenomenon was owing to the introduction of DNF with negative charge, indicating the successful immobilization of DNF onto GO-modified monolith. Furthermore, desirable repeatability of retention time for thiourea was observed with good relative standard deviation (RSD) value of 2.02%. Additionally, EOF mobility of DCT-GO@CSM with 5.25 × 10−8 m2 V−1 s−1 was obtained. The morphology of the resulting capillary silica monoliths and DNF-immobilized capillary silica monoliths were characterized by SEM (Fig. 3). Compared with the bare monolith, flower-like DNFs were seen in DNF@CSM. This result suggested the successful immobilization of DNFs onto the pore surface of capillary silica monoliths. As shown in Figure S4, the FTIR spectra of silica monolithic material exhibited the characteristic bands of O–H vibration from Si–OH groups at 3420–3250 cm−1 and Si–O–Si stretching at 1000–1100 cm−1. The bands centered at 2940–2920 cm−1 and 1650–1580 cm−1 were ascribed to the C–H vibration of –CH2 and N–H vibration of –NH2, respectively. These results indicated that APTS was immobilized effectively onto silica monolith. It can be seen that the bands strength of amino vibration decreased in Figure S4 (c), which was attributed to the reaction between amine groups and GO.
Effect of capillary electrochromatography conditions on chiral separation
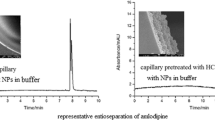
The resulting DNF-GO@CSM was able to baseline separate enantiomers including atenolol, tyrosine, histidine, and nefopam (Table S2). To investigate the effect of CEC conditions on chiral separation performance of DNF-GO@CSM, CEC conditions such as sample volume, applied voltage, and buffer concentration were optimized by choosing histidine and nefopam as the model analytes. As seen in Figure S5, the resolution of d,l-histidine increases with increasing injection volume from 10 kV, 1 s, to 10 kV, 3 s. Applied voltages of 5, 10, and 15 kV were explored under optimum injection volume 10 kV, 3 s (Figure S6). The influence of buffer concentration on d,l-histidine enantioseparation is presented in Figure S7. As seen in Figure S8, the chiral resolution of nefopam gets to a maximum value when electrokinetic injection is adopted at 10 kV for 3 s. Nevertheless, the resolution value displayed a decreased trend with larger injection volume. The optimum applied voltage of enantioseparating nefopam was 10 kV (Figure S9). The influence of buffer concentration on nefopam enantioseparation was also investigated, and 20 mM was selected as the optimal value (Figure S10).
Comparison with DCT-modified capillary silica monoliths
In addition to DNF-GO@CSM, two types of columns including DCT-GO@CSM and GO@CSM were prepared to explore the effect of DNF on chiral separation. Herein, chiral molecules such as histidine, atenolol, tyrosine, phenylalanine, and chlorpheniramine enantiomers were selected as the model analytes to evaluate the chiral recognition capability of DNF-GO@CSM under the optimized CEC conditions (Fig. 4). Results indicated that DNF-GO@CSM displayed better chiral separation performance compared to that of DCT-GO@CSM and GO@CSM, suggesting that the application of DNF was able to improve chiral separation ability of the column. As illustrated in Figure S11, no evidence of separation was observed for the analytes on GO@CSM. When DCT was utilized as the chiral selector, the enantiomers could be partially separated. The retention of enantiomers on DNF-GO@CSM increased relatively in comparison with DCT-GO@CSM. This phenomenon was attributed to the enhanced interaction between analytes and DNF-GO@CSM based on the high surface area of DNF. We suspected that the numerous tandem repeats in the elongated ssDNA were able to improve the interaction between DNF and analytes as well. Moreover, thiourea was chosen as EOF marker to determine EOF of these columns. The EOF increased accompanying with the increased negative charge density of DNA sequences. Considering the impact of EOF and chromatographic retention, the retention time of analytes increased in the order of GO@CSM < DCT-GO@CSM < DNF-GO@CSM. The stability of DNF-GO@CSM was evaluated by determining the repeatability of three successive enantioselectivity of d,l-histidine. The intraday repeatability of retention time kept desirable RSD values of less than 5.15%, along with good RSD values for resolution (Rs). Moreover, the interday repeatability was investigated by performing the chiral separation of d,l-histidine. RSD values of d,l-histidine retention time and Rs with 6.05% and 8.28% were obtained. Therefore, DNF-GO@CSM possessed satisfactory stability for chiral separation.
Enantioseparation of chiral compounds on DNA nanoflower-modified capillary silica monolith by capillary electrochromatography. A d,l-Histidine; B R,S-atenolol; C d,l-phenylalanine; D d,l-tyrosine; E R,S-chlorpheniramine. Conditions: injection, 10 kV, 3 s; running buffer, 20 mmol L−1 phosphate buffer (pH 7.4); applied voltage, 10 kV
Molecular simulations for investigating DNA sequences effect on chiral separation mechanism
To gain more insight into possible chiral separation mechanisms of DNA sequences for enantiomers, MD simulations were performed. Histidine and nefopam were selected as representative analytes to visualize the forces existing in the chiral recognition process. The docking energy were − 6.2 and − 6.5 kcal mol−1 for the d-histidine/DCT and l-histidine/DCT complexes, respectively, suggesting that l-histidine presented a greater stereoaffinity for DCT (Figure S12). As seen in Fig. 5, l-histidine molecules can interact with the pocket formed by deoxyribotide DG-90, DG-91, DA-67, and DC-66 in the DNA structure. The hydrogen-bonding interactions were generated between l-histidine and DG-90, DG-91, DA-67, and DC-66. All of these interactions further confirmed strong binding of l-histidine and DCT. Nevertheless, d-histidine interacted with the pocket formed by deoxyribotide DG-15, DC-24, DA-14, and DT-25. The docking energies were − 6.8 and − 6.7 kcal mol−1 for the R-nefopam/DCT and S-nefopam/DCT complexes, respectively, suggesting that R-nefopam presented a greater stereoaffinity for DCT (Figure S13). It was observed that R-nefopam molecules were able to interact with the pocket formed by deoxyribotide DA-30, DC-31, DA-68, DG-90, and DG-91 in the DNA structure (Figure S14). The hydrogen-bonding interactions and π-π conjugation effects were generated between R-nefopam and DG-90. Moreover, R-nefopam could interact with DC-31 to form a salt bridge force. All of these interactions further confirmed strong binding of R-nefopam and DCT. Nevertheless, S-nefopam only formed π-π conjugation effects with DG-90 and DG-91. Taken together, the MD simulation data indicated a greater stereoselective interaction of DCT to l-histidine/R-nefopam than to d-histidine/S-nefopam, ultimately leading to the chiral separation.
Predicting the interaction mechanism of the d/l-histidine and DNA complex. a Binding mode of d-histidine and DNA shown in cartoon; b binding mode of l-histidine and DNA shown in cartoon. The red and blue sticks respectively represent l-histidine and d-histidine, the yellow dotted line represents the H-bond interaction located in the d/l-histidine and DNA complex
Effect of different types of DNFs on chiral separation
To further explore the chiral separation ability of DNFs, template2/3 (template2 or template3) containing a distinct DNA sequence was respectively applied for developing DNF2/DNF3 (DNF2 or DNF3). It was observed that the proposed DNF2-GO@CSM presented satisfactory enantioseparation performance toward atenolol, propranolol, and 2′-deoxyadenosine (Fig. 6). Baseline separation of enantiomers including nefopam, propranolol, and 2′-deoxyadenosine was realized by utilizing DNF3-GO@CSM (Fig. 7). Results indicated that chiral CSMs based on DNFs provided an efficient chiral separation platform. A comparison of chiral materials, chromatographic parameters, and analytes for DNF-GO@CSM and other enantioseparation methods is summarized in Supplementary Table S3 [46,47,48]. Compared with traditional spherical nanomaterials, DNFs displayed higher surface area due to their topographic properties of nanolayers. Owing to their programmability, various types of DNF chiral selectors were able to be designed for separating a variety of enantiomers. DNF-based chiral selectors offered a reference for DNA enantiorecognition mechanism study.
Conclusion
Novel chiral capillary silica monoliths were successfully prepared by selecting DNFs as a chiral selector. Under the optimum conditions, baseline separation of enantiomers including atenolol, tyrosine, histidine, and nefopam was observed on DNF-GO@CSM. Moreover, DNF-GO@CSM presented satisfactory interday and intraday repeatability owing to DNF specific stability. Compared with DCT-GO@CSM and GO@CSM, DNF-GO@CSM displayed desirable chiral separation ability. We inferred that DNFs with high surface area and numerous tandem DCT repeats facilitated the improvement of enantioseparation performance. For exploring the interaction between DNA sequences and enantiomers, we further utilized MD simulations to investigate the enantioseparation mechanisms of DCT and model analytes. We deduced that the chiral separation capability of DCT was owing to the different interactions between enantiomers and DNA sequences. In addition to DNF-GO@CSM, DNF2-GO@CSM and DNF3-GO@CSM exhibited satisfactory chiral separation ability as well. Baseline separation of atenolol, propranolol, 2′-deoxyadenosine, and nefopam enantiomers was realized. In the following study, various types of DNF chiral selectors will be prepared due to their programmability. We anticipate that the developed DNF-GO@CSM will open up new prospects for developing bionanomaterial-based CEC enantioseparation systems. Since bionanomaterial chiral selectors can combine the merits of biomolecules and NMs, diverse bionanomaterials can be further investigated for chiral separation.
Data availability
Data will be made available on request.
References
Hefnawy M, El-Gendy M, Al-Salem H, Marenga H, El-Azab A, Abdel-Aziz A, Gamal AE, Alanazi M, Obaidullah A, Al-Hossaini A, Hefnawy A (2023) Trends in monoliths: packings, stationary phases and nanoparticles. J Chromatogr A 1691:463819
Wong XY, Sena-Torralba A, Álvarez-Diduk RÁ, Muthoosamy K, Merkoci A (2020) Nanomaterials for nanotheranostics: tuning their properties according to disease needs. ACS Nano 14:2585–2627
Bilal M, Asgher M, Shah SZH, Iqbal HMN (2019) Engineering enzyme-coupled hybrid nanoflowers: the quest for optimum performance to meet biocatalytic challenges and opportunities. Int J Biol Macromol 135:677–690
Zhao B, Liang J, Zou X, Zhang B, Zhang Y, Niu L (2023) Crystallization regulation engineering in the carbon nitride nanoflower for strong and stable electrochemiluminescence. ACS Appl Mater Interfaces 15:16723–16731
Perwez M, Lau SY, Hussain D, Anboo S, Arshad M, Thakur P (2023) Nanozymes and nanoflower: physiochemical properties, mechanism and biomedical applications. Colloid Surface B 225:113241
Saravanan N, Ganesh P, Pitchaimuthu S, Sundaramurthy A (2023) Nanozyme controlled photothermal heat generation on nanoceria decorated MoS2 nanoflowers for enhanced cytotoxicity in cancer chemo-photothermal therapy. Surf Interfaces 41:103225
Kim T, Nam K, Kim YM, Yang K, Roh YH (2021) DNA-assisted smart nanocarriers: progress, challenges, and opportunities. ACS Nano 15:1942–1951
Krissanaprasit A, Key CM, Pontula S, LaBean TH (2021) Self-assembling nucleic acid nanostructures functionalized with aptamers. Chem Rev 121:13797–13868
Tran TD, Nguyen PT, Le TN, Kim MI (2021) DNA-copper hybrid nanoflowers as efficient laccase mimics for colorimetric detection of phenolic compounds in paper microfluidic devices. Biosens Bioelectron 182:113187
Sheng J, Pi Y, Zhao S, Wang B, Chen M, Chang K (2023) Novel DNA nanoflower biosensing technologies towards next-generation molecular diagnostics. Trends Biotechnol 41:653–668
Zeng R, Wang J, Wang Q, Tang D, Lin Y (2021) Horseradish peroxidase-encapsulated DNA nanoflowers: an innovative signal-generation tag for colorimetric biosensor. Talanta 221:1–7
Cowan JA, Furnstahl RJ (2022) Origin of chirality in the molecules of life. ACS Earth Space Chem 6:2575–2581
Sanz-Hernández D, Hierro-Rodriguez A, Donnelly C, Pablo-Navarro J, Sorrentino A, Pereiro E, Magén C, McVitie S, María de Teresa J, Ferrer S, Fischer P, Fernández-Pacheco A (2020) Artificial double-helix for geometrical control of magnetic chirality. ACS Nano 14:8084–8092
Horrer A, Zhang Y, Gérard D, Béal J, Kociak M, Plain J, Bachelot R (2020) Local optical chirality induced by near-field mode interference in achiral plasmonic metamolecules. Nano Lett 20:509–516
Lee BH, Kotov NA, Arya G (2021) Reconfigurable chirality of DNA-bridged nanorod dimers. ACS Nano 15:13547–13558
Kang JS, Kim N, Kim T, Seo M, Kim BS (2022) Circularly polarized light-driven supramolecular chirality. Macromol Rapid Commun 43:2100649
Ma Y, Shi L, Yue H, Gao X (2020) Recognition at chiral interfaces: from molecules to cells. Colloid Surface B 195:1–15
De Gauquier P, Vanommeslaeghe K, Vander Heyden Y, Mangelings D (2022) Modelling approaches for chiral chromatography on polysaccharide based and macrocyclic antibiotic chiral selectors: a review. Anal Chim Acta 1198:1–16
Hofstetter H, Hofstetter O (2005) Antibodies as tailor-made chiral selectors for detection and separation of stereoisomers. TrAC Trends Anal Chem 24:869–879
Deng Y, Zhang Z, Pang Y, Zhou X, Wang Y, Zhang Y, Yuan Y (2022) Common materials, extraordinary behavior: an ultrasensitive and enantioselective strategy for D-Tryptophan recognition based on electrochemical Au@p-L-cysteine chiral interface. Anal Chim Acta 1227:340331
Upadhyay SS, Gadhari NS, Srivastava AK (2020) Biomimetic sensor for ethambutol employing β-cyclodextrin mediated chiral copper metal organic framework and carbon nanofibers modified glassy carbon electrode. Biosens Bioelectron 165:1–5
Zagitova L, Yarkaeva Y, Zagitov V, Nazyrov M, Gainanova S, Maistrenko V (2022) Voltammetric chiral recognition of naproxen enantiomers by N-tosylproline functionalized chitosan and reduced graphene oxide based sensor. J Electroanal Chem 922:1–11
Lu Q, Chen L, Meng Q, Jiang Y, Xie L (2021) A biomolecule chiral interface base on BSA for electrochemical recognition of amine enantiomers. Chirality 33:385–396
Gong H, Zhang S, Liu N, Zhang J, Chen Q, Liu H (2022) Retarded transport properties of graphene oxide based chiral separation membranes modified with dipeptide. Sep Purif Technol 288:1–10
Neequaye T, Rassi ZE (2023) Poly(carboxyethyl acrylate-co-ethylene glycol dimethacrylate) precursor monolith with bonded (S)-(-)-1-(2-naphthyl) ethylamine ligands for use in chiral and achiral separations by capillary electrochromatography. J Chromatogr A 1688:463713
Park JM, Park JH (2014) Enantiomer separations of basic chiral compounds by capillary electrochromatography on a phosphated-cyclodextrin-modified zirconia monolith. J Chromatogr A 1339:229–233
Qian H, Xu S, Yan X (2023) Recent advances in separation and analysis of chiral compounds. Anal Chem 95:304–318
Tran LN, Park JH (2015) Enantiomer separation of acidic chiral compounds on aquinine-silica/zirconia hybrid monolith by capillary electrochromatography. J Chromatogr A 1396:140–147
Hong T, Liu X, Zhou Q, Liu Y, Guo J, Zhou W, Tan S, Cai Z (2022) What the microscale systems ‘see’ in biological assemblies: cells and viruses? Anal Chem 94:59–74
Hong T, Zhou W, Tan S, Cai Z (2023) A cooperation tale of biomolecules and nanomaterials in nanoscale chiral sensing and separation. Nanoscale Horiz 8:1485–1508
Zhang M, Chen J, Xu G, Yu T, Du Y (2023) A chiral metal-organic framework synthesized by the mixture of chiral and non-chiral organic ligands for enantioseparation of drugs by open-tubular capillary electrochromatography. J Chromatogr A 1699:1–10
de Koster N, Clark CP, Kohler I (2021) Past, present, and future developments in enantioselective analysis using capillary electromigration techniques. Electrophoresis 42:38–57
Wang A, Liu K, Tian M, Yang L (2022) Open tubular capillary electrochromatography-mass spectrometry for analysis of underivatized amino acid enantiomers with a porous layer-gold nanoparticle-modified chiral column. Anal Chem 94:9252–9260
Wang T, Yang L, Cheng Y, Zhang Y, Ye J, Chu Q, Cheng G (2021) Evaluation of homochiral zeolitic imidazolate framework-8 supported open-tubular column by miniaturized capillary electrochromatography with amperometric detection. Microchim Acta 188:1–7
Wang X, Wu J, Liu X, Qiu X, Cao L, Ji Y (2022) Enhanced chiral recognition abilities of cyclodextrin covalent organic frameworks via chiral/achiral functional modification. ACS Appl Mater Interfaces 14:25928–25936
Ávalos-Ovando O, Besteiro LV, Movsesyan A, Markovich G, Liedl T, Martens K, Wang Z, Correa-Duarte MA, Govorov AO (2021) Chiral photomelting of DNA-nanocrystal assemblies utilizing plasmonic photoheating. Nano Lett 21:7298–7308
Kneer LM, Roller E, Besteiro LV, Schreiber R, Govorov AO, Liedl T (2018) Circular dichroism of chiral molecules in DNA assembled plasmonic hotspots. ACS Nano 12:9110–9115
Shen B, Linko V, Tapio K, Pikker S, Lemma T, Gopinath A, Gothelf KV, Kostiainen MA, Toppari JJ (2018) Plasmonic nanostructures through DNA-assisted lithography. Sci Adv 4:eaap8978
Tortora MMC, Mishra G, Prešern D, Doye JPK (2020) Chiral shape fluctuations and the origin of chirality in cholesteric phases of DNA origamis. Sci Adv 6:eaaw8331
Nguyen L, Dass M, Ober MF, Besteiro LV, Wang ZM, Nickel B, Govorov AO, Liedl T, Heuer-Jungemann A (2020) Chiral assembly of gold-silver core-shell plasmonic nanorods on DNA origami with strong optical activity. ACS Nano 14:7454–7461
Hlushkou D, Bruns S, Ltzel AH, Tallarek U (2010) From pore scale to column scale dispersion in capillary silica monoliths. Anal Chem 82:7150–7159
Yin J, Xu T, Zhang N, Wang H (2016) Three-enzyme cascade bioreactor for rapid digestion of genomic DNA into single nucleosides. Anal Chem 88:7730–7737
Jin Y, Li Z, Liu H, Chen S, Wang F, Wang L, Li N, Ge K, Yang X, Liang X, Zhang J (2017) Biodegradable, multifunctional DNAzyme nanoflowers for enhanced cancer therapy. NPG Asia Mater 9:1–10
Hong T, Liu X, Ji Y, Tan S, Cai Z (2023) Construction of chiral capillary electrochromatography microsystems based on Aspergillus sp. CM96. Microchimica Acta 90:357
Hong T, Chen X, Xu Y, Cui X, Bai R, Jin C, Li R, Ji Y (2016) Preparation of graphene oxide-modified affinity capillary monoliths based on three types of amino donor for chiral separation and proteolysis. J Chromatogr A 1456:249–256
Sun G, Tang W, Lu Y, Row KH (2022) Enantioseparation by simultaneous biphasic recognition using mobile phase additive and chiral stationary phase in capillary electrochromatography. J Chromatogr A 1666:462856
Sun X, Chen C, Li X, Du Y, Zhao S, Feng Z (2020) Gold nanoparticles coated with a tetramethylammonium lactobionate ionic liquid for enhanced chiral differentiation in open tubular capillary electrochromatography: application to enantioseparation of β-blockers. Microchim Acta 187:170
Li Y, Xu G, Chen J, Yu T, Miao P, Du Y (2023) One-step synthesis of chiral molecularly imprinted polymer TiO2 nanoparticles for enantioseparation of phenylalanine in coated capillary electrochromatography. Microchim Acta 190:279
Funding
This work was funded by the National Natural Science Foundation of China (Grant No. 81803495).
Author information
Authors and Affiliations
Contributions
Tingting Hong: investigation; conceptualization; methodology; writing—original draft; project administration; funding acquisition. Qi Zhou: investigation, methodology, validation. Yilian Liu: investigation, methodology, validation. Yibing Ji: conceptualization, methodology, project administration. Songwen Tan: conceptualization, writing—review and editing. Wenhu Zhou: conceptualization, writing—review and editing. Zhiqiang Cai: supervision, project administration, conceptualization, writing—review and editing. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethical approval
This research did not involve human or animal samples.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hong, T., Zhou, Q., Liu, Y. et al. Preparation of DNA nanoflower-modified capillary silica monoliths for chiral separation. Microchim Acta 191, 584 (2024). https://doi.org/10.1007/s00604-024-06663-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-024-06663-z