Abstract
A novel nanoparticle-based fluorescence probe was developed for NF-κB transcription factor detection and in situ imaging via steric hindrance. The probe contains gold nanoparticles (AuNPs) to quench fluorescence, and nucleic acids immobilized on the surface of AuNPs to output fluorescence. In the basal state, Cy5 labeled DNA1 folds its long chain into a hairpin structure and quenches fluorescence by forcing the Cy5 fluorophore close to the surface of AuNPs. After the probe enters the cell, the NF-κB transcription factor can bind to the κB site in the DNA duplex of the nucleic acids. The steric hindrance caused by NF-κB leads to the extension of the long chain of DNA1 and the removal of the Cy5 fluorophore from the surface of AuNPs, thereby restoring the fluorescence of the probe. By measuring NF-κB in cell lysis in vitro, the probe obtains a detection limit of 0.38 nM and the linear range from 0.5 to 16 nM. Repeated measurements showed the recovery in the cell nuclear extract was between 93.38 and 109.32%, with relative standard deviation less than 5%. By monitoring the sub-localization of the Cy5 fluorophore in single cell, the probe system can effectively distinguish active NF-κB (nucleus) and inactive NF-κB (cytoplasm) through in situ imaging. The well-designed probe will make up for the shortcomings of the existing technology, and reveal the regulatory role of transcription factors in many disease processes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Transcription factors (TFs) are DNA-binding proteins that can bind to DNA enhancers or silencers sequence-specifically, and regulate gene transcription by localizing in the 5′-upstream region of the target genes [1, 2]. As key proteins for the decision of cell fates, TFs participate in cell differentiation, signaling or regulatory pathway, and are potent markers for diagnostic and treatment of diseases [3, 4]. Nuclear factor kappa B (NF-κB), named for its ability to bind to the κB site in the DNA duplex, is an enhancer of the κ light chain consisting of 10 nucleotides (5’-GGGACTTTCC-3′) [5]. The NF-κB family is composed of five members that form hetero- and homo-dimers, which are RelA (p65), RelB, c-Rel, NF-κB1 (p50), and NF-κB2 (p52). Typically, p50/p65 (NF-κB1/RelA) hetero-dimer is the main form of NF-κB activity, although other dimers such as p50/Rel or p52/p65 (NF-κB2/RelA) often coexist [6, 7]. Over the past decades, a large number of approaches for detecting NF-κB have been developed, including classic methods such as electrophoretic mobility shift assay (EMSA), Western blotting, immunohistochemistry, and newly developed methods such as enzyme-linked immune sorbent assay (ELISA). Although the sensitivity and detection limits of these methods are within an acceptable level, they are usually time-consuming and labor-intensive, and most importantly high cost. Furthermore, most of the approaches cannot distinguish between active NF-κB (nucleus) and inactive NF-κB (cytoplasm). Therefore, new strategies for detection of active NF-κB were desired to develop [8, 9]. In comparison with Western blotting and EMSA technologies, in situ imaging of fluorescence in single cell based on a nanoprobe could solve the problems to a certain extent.
Gold nanoparticles (AuNPs) have unique optical properties that make them appealing for ultrasensitive detection and imaging-based therapeutic techniques in medical applications [10, 11]. For example, the aggregation of solutions containing plasmonic nanoparticles in the presence of molecules induces the color to change from red to blue, which provides a useful tool for colorimetric inspection without the use of advanced techniques [12,13,14,15]. Besides, plasmonic nanoparticles selectively quench the fluorescence of organic dyes, and the property offers highly sensitive methods for detecting targets in biological media [16, 17]. The localized surface plasmon resonance (LSPR) of AuNPs produces fluorescence quenching and fluorescence enhancement effects on the signal strength of fluorescent molecules. These two completely opposite effects depend on the distance between the fluorophore and the AuNPs. If the fluorophore is less than 5 nm away from AuNPs, non-radiative fluorescence resonance energy transfer (FRET) will dominate and cause fluorescence quenching, because the energy of the excited fluorophore is transferred to the AuNPs in totality. If the distance between the fluorophore and AuNPs increases to 5–10 nm, the local field enhancement will elevate the excitation rate of the fluorophore, leading to the increase in fluorescence [18, 19]. The quenching and coming back of the fluorescence indicate the interaction among nanoparticles, fluorophores, and biomolecules, so that they can be used for diagnostics.
AuNPs can directly conjugate and interact with various molecules through their surface, including drugs, nucleic acids (DNA or RNA), proteins, antibodies, and fluorescent dyes, which allow AuNPs to be functionalized and play its roles in biomedicine [20,21,22]. Herein, an AuNP-based fluorescent probe composed of AuNP-modified oligonucleotide and fluorescent dye was developed. The oligonucleotide is specific to NF-κB through its κB site and performs as a linker to change its structure through the steric hindrance of the target binding to NF-κB. The spatial structure change further alters the distance between the fluorophore and AuNPs, thereby changing the fluorescence intensity [19]. The probe performs well in biological samples due to low fluorescence contamination in the background, and non-specific adsorption generally does not change its configuration [9]. In addition, 13-nm AuNPs were chosen for their easy preparation, little particle size deviation (±2 nm), and a sharp plasmon absorption band at 520 nm [15]. In this study, we proposed a strategy for in situ imaging of active NF-κB in single cell. After one-step incubation, it can be expected that the well-designed probe would monitor the subcellular localization of NF-κB and predict the amount of active NF-κB (in the nucleus) in vitro and in situ.
Experiment section
Apparatus and reagents
Tecnai G2 F20 (FEI, USA), a high-resolution field-emission transmission electron microscope (FE-TEM), was applied for imaging of nanoparticles. The particle size and size distribution of gold nanoparticles were measured using a Zetasizer Nano S90 (Malvern, UK). The UV-vis spectrum was scanned with a multi-mode microplate reader SpectraMax M5 (Molecular Devices, USA). An Advanced Live Cell Imaging System XCellence (Olympus, Japan) was used for in situ imaging of single cell.
Gold chloride trihydrate (HAuCl4·3H2O) and Tris(2-carboxyethyl)phosphine hydrochloride (TCEP) were purchased from Aladdin (Shanghai, China). Trisodium citrate dihydrate, DL-Dithiothreitol (DTT), DAPI, and ethidium bromide solution were bought from Sigma-Aldrich (St. Louis, MO, USA). Agarose H and DNA Marker A (25 ~ 500 bp) were obtained from Sangon Biotech. (Shanghai, China). The oligonucleotides were synthesized by Genscript Biotech. (Nanjing, China), and the sequences were shown in Table 1. Recombinant Human TNFα was acquired from PeproTech (Cranbury, NJ, USA), while NF-κB p65 (Rel A) peptide was provided by Rockland (PA, USA).
Cell lines and cell culture
Human breast cancer cell line resistant to doxorubicin (MCF-7/Adr) was purchased from Bogoo Biotech. (Shanghai, China). Human mammary carcinoma cell line MDA-MB-231 was obtained from the National Infrastructure of Cell Line Resource (Shanghai, China). Both of them were cultured in RPMI-1640 medium containing 10% fetal bovine serum, 100 U/mL penicillin, and 100 μg/mL streptomycin at 37 °C in a humidified 5% CO2 incubator. To maintain the drug resistance of MCF-7/Adr, doxorubicin (1 μg/mL) was supplied to cell culture at regular intervals.
Preparation of the functionalized DNA-AuNP probe
Gold nanoparticles (AuNPs, ~13 nm) were prepared by citrate reduction of HAuCl4 [23,24,25]. The surface plasmon resonance wavelength of AuNPs was measured using UV-vis spectroscopy. The size of the synthetic AuNPs was determined by dynamic light scattering (DLS) and imaged by transmission electron microcopy (TEM).
Fifty microliters of 5 mM TCEP was added to 5 nmoL DNA1 powder and incubated for 10 min to reduce the disulfide bond of DNA1 to monothiol. To obtain the DNA1/DNA2 duplex, 50 μL of DNA1 (100 μM) was mixed at the same molar rations with DNA2, then the mixture was heated at 90 °C for 10 min and naturally cooled to room temperature for 4 h; 1.5 mL of 13 nm AuNPs were centrifuged at 13,000g for 10 min and resuspended in 0.3 mL Milli-Q water to remove the excess reaction solution. The salt-aging method is used to functionalize AuNPs with thiolated DNA [9, 26]. In brief, 20 μL of the DNA1/DNA2 duplex was added to 1 mL of AuNPs, and the mixture was incubated at room temperature for 16 h. To increase DNA loading on the surface of AuNPs, we salted the mixture with 2 M NaCl in 0.01 M PBS. Specifically, 10 μL of 2 M NaCl was slowly added to 1 mL of the mixture and incubated for 30 min, and the concentration of NaCl in the probe solution was increased to 0.02 M. The step was repeated 10 times to increase the concentration of NaCl to 0.2 M. After the salting process, the mixture was incubated overnight at room temperature to stabilize the probe. To remove excess reagents, the solution was centrifuged at 13,000g for 15 min and redispersed in 1 mL of PBS.
Polyacrylamide gel electrophoresis (PAGE)
DNA primers (DNA1 and DNA2) and the DNA1/DNA2 duplex mixed with 6 × loading buffer (Takara, Japan) were loaded onto an 18% non-denaturing PAGE gel and the gel was electrophoresed in 0.5 × TBE buffer (45 mM Tris-Boric-acid, 1 mM EDTA, pH 8.3) at 100 V for 2 h. Then, the gel was stained with 0.5 μg/ml EB solution for 15 min and photographed under UV light by the Tanon 2500R Gel Imaging System (Shanghai, China).
Preparation of cell nuclear extracts
MCF-7/Adr cells were plated into 100-mm dishes at 1.0 × 106 cells/dish and grown overnight. The culture medium was removed and replaced with fresh RPMI-1640 with or without 30 ng/mL TNFα, and then the cells were incubated for another 24 h. Cells were harvested by scraping and washed once with PBS. Then, nuclear and cytoplasmic proteins in cell pellets were obtained by a Nuclear and Cytoplasmic Protein Extraction Kit (Beyotime Biotechnology, Shanghai, China). Cell nuclear extracts were diluted five times and used as sample solutions for subsequent reactions.
Sample assay procedure
To detect the concentration of NF-κB p65 in cell nuclear extracts, the functionalized DNA-AuNP probe prepared in the “Preparation of functionalized DNA-AuNP probe” section was twofold diluted to 15 nM as the reaction system. Cell nuclear extract (fivefold dilution) was spiked with different concentrations of the NF-κB p65 standard, and then 5 μL of sample solution was added to 50 μL of the reaction system and incubated at 37 °C for 30 min. After that, the fluorescence intensity of Cy5 was determined at Ex649nm/Em670nm using a multi-mode microplate reader.
In situ imaging of NF-κB p65
MCF-7/Adr cells and MDA-MB-231 cells were plated into a 24-well glass bottom plate at 5 × 104 cells/well and grown overnight. The medium was replaced with 300 μL of fresh RPMI-1640 containing 30 ng/mL TNFα or not, and the cells were incubated for 24 h. After that, 25 μL of the DNA-AuNP (15 nM) probe was added to each well and incubated for a further 8 h. Cells were washed twice with PBS, and fixed for 15 min with 4% paraformaldehyde solution. After washing, the cell nucleus was stained with 5 μg/mL DAPI for 5 min. Cell imaging for sub-localization of NF-κB p65 in MCF-7/Adr and MDA-MB-231 cells was performed by XCellence Imaging Stations.
Results and discussion
Principles of the nanoparticle-based fluorescence probe
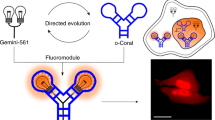
As shown in Scheme 1, the DNA1/DNA2 duplex containing the κB site that can bind to NF-κB p65 was modified to 13 nm AuNPs. The DNA1 is modified with a thiol group (-SH) at the 3′ end to make it immobilized on the surface of the AuNPs, while the 5′ end is labeled with a fluorophore (Cy5) for fluorescent analysis. DNA2 is the complementary sequence of the κB site of DNA1, so that the NF-κB p65 transcription factor can bind to the double-strand. The DNA1 has a hairpin structure that folds its long chains, bringing the Cy5 fluorophore close to the surface of AuNPs and fluorescence quenching happens. When NF-κB is present, it specifically binds to the κB site of the DNA1/DNA2 duplex. The steric hindrance caused by NF-κB extends the long chain of DNA1, and then the Cy5 fluorophore moves away from the AuNPs surface, and the fluorescent signal increases.
Characterization of the synthesized gold nanoparticles
The synthesized AuNPs were characterized by TEM and DLS. The TEM image showed that there is no aggregation in the synthesized AuNPs, and the particles have uniform size and good dispersion (Fig. 1A). The average diameter of AuNPs was 13.54 nm, which was quantified by the volume distribution of DLS (Fig. 1B). UV-vis spectra showed the surface plasmon resonance peak of AuNPs is 520 nm (Fig. 1C, curve a). Compared to AuNPs, a modest shift from 520 to 525 nm in the surface plasmon band was observed in DNA-modified AuNPs (Fig. 1C, curve b). Non-denaturing PAGE was applied to detect the DNA primers. In Fig. 1D, the band of DNA1(76 nt) with 5’-Cy5 fluorophores and 3’-SH was clearly displayed (lane 1), and DNA 2 (14 nt) was observed at a longer distance with lower brightness due to its short oligonucleotide chain (lane 2). Meanwhile, the DNA1/DNA2 duplex was obviously observed with a small shift compared to DNA1 alone, and the disappearance of DNA2 demonstrated the successful formation of the DNA duplex (lane 3).
Feasibility study
In this research, the DNA1/DNA2 duplex-modified AuNPs were used as a probe to detect the content of NF-κB p65. NF-κB can bind to the κB site of the GGG ACT TTC C sequence specifically. The binding of NF-κB p65 forced the hairpin structure of DNA1 to stretch by steric hindrance, which caused the Cy5 fluorophore to move away from the AuNPs and fluoresce (Scheme 1). The kinetics of the reaction between the probe and p65 revealed that 30 min was sufficient for structure respond of the DNA1/DNA2 duplex, and weaken the quenching of the Cy5 fluorophore by AuNPs. Therefore, a reaction time of 30 min was chosen in the subsequent experiments (Fig. S2).
The concentration of the functionalized nanoparticle probe exhibits a significant impact on the quenching effect of the Cy5 fluorophore. In the process of probe preparation, DNA and AuNPs were concentrated to enhance DNA loading on the surface of AuNPs. However, if the prepared probe was used for reaction directly, the excessively high density of AuNPs may quench the fluorescence of Cy5 in stretched DNA1 as well. Proper dilution of the probe can avoid this problem and obtain maximum fluorescence signal. As shown in Fig. S3, 15 nM of the probe was the optimal concentration and was selected for further experiments.
Sensitivity and specificity of the probe for detection of NF-κB p65
To verify the sensitivity of this method, the changes of Cy5 fluorescence intensity were monitored after the reaction of the probe (15 nM) with a series of concentrations of NF-κB p65. Figure 2 showed the correlation between the fluorescence intensity and NF-κB p65 concentration. Within the concentration range from 0.5 to16 nM, there is an excellent linear relationship between the fluorescence intensity and p65 concentration (Fig. 4B inset). The corresponding relationship is expressed by the formula Y = 39.44 + 50.38X (R2 = 0.9970), in which Y represents the fluorescence intensity of Cy5, and X represents the concentration of NF-κB p65. The limit of detection of the method is 0.38 nM, which was calculated by three times the standard deviation of the blank sample (3σ) divided by the slope of the regression line.
The specificity of the method was also investigated (Fig. 3). Other common proteins, including bovine serum albumin (BSA), hemoglobin (Hb), and tumor necrosis factor α (TNFα) were selected to confirm whether they could interfere with the reaction of NF-κB p65. In this assay, 300 nM of BSA, Hb, and TNFα were incubated with the DNA-AuNP probe (15 nM) for 30 min, and the fluorescence intensity of the reactions was compared with the fluorescence of p65 (16 nM). As shown in Fig. 3, the fluorescence signals of three common proteins were nearly equal to the blank signal, even if the concentration of them is 20 times higher than that of p65. The results indicated that other common proteins had no binding activity to the κB site of the DNA duplex, and NF-κB proteins were the only specific targets of the detection method.
Measurement of NF-κB p65 in the cell nuclear extract
To investigate whether the probe can be applied to quantify NF-κB p65 in biological samples, a fivefold diluted cell nuclear extract was used to determine p65 levels to evaluate the effect of the cell nuclear extract on actual detection conditions. Detected by the method, we found the baseline concentration of NF-κB p65 in control cell nuclear extracts was 2.78 nM, when the cells were induced by 30 ng/mL TNFα, the active NF-κB p65 increased sharply to 12.85 nM, which was in line with our expectation [27, 28]. Different concentrations of the p65 standard were spiked into the control cell nuclear extract, and we found the recovery in cell nuclear extract was between 93.38 and 109.32%, while the relative standard deviation (RSD) ranged from 2.02 to 4.96% (Table 2). The fluorescence signals of spiked NF-κB p65 in different concentrations under practical detection conditions were compared with the standard line measured in PBS. Under the influence of cell nuclear extract components, the background of the fluorescent signal increased. As shown in Fig. 4, the calibration plots measured in cell nuclear extracts (orange line, Y = 225.26 + 53.53X, R2 = 0.9969) and in PBS (black line, Y = 39.44 + 50.38X, R2 = 0.9970) were relatively parallel, suggesting that the interference of cell nuclear extract components has no significant impact on detecting NF-κB p65. Therefore, we concluded that this method is suitable for detection of NF-κB p65 in cell nuclear extraction.
In situ imaging of NF-κB p65
The kinetics of cellular uptake of DNA-AuNP was determined to optimize the probe incubation time and obtain better image performance. As shown in Fig. S4, 8 h is the optimal incubation time of the probe for cell imaging. MCF-7/Adr, a doxorubicin-resistant human breast cancer cell line, was first incubated with or without 30 ng/mL TNFα for 24 h to induce NF-κB activation. Then, 25 μL of the DNA-AuNP probe (15 nM) was added to the medium and incubated for 8 h. Fluorescence imaging showed that most of the NF-κB p65 protein (red) was located in the cytoplasm, while the position of the nucleus (blue) was almost empty in the basic expression of MCF-7/Adr cells. When cells were induced by TNFα, the fluorescence signals were enhanced and more active p65 entered the nucleus, indicating that TNFα had effectively induced the activity of NF-κB (Fig. 5).
To further prove the conclusion, another human mammary carcinoma cell line, MDA-MB-231, was applied to determine the sub-localization of NF-κB p65 in situ. As shown in Fig. 6, most of NF-κB p65 accumulated around the nucleus in cells with the basic expression of p65. After TNFα induction, NF-κB p65 was activated and entered the nucleus, which was indicated by the red fluorescence signals inside the nucleus. The results were very consistent with the results of MCF-7/Adr.
Conclusions
A novel nanoparticle-based fluorescence probe with target selective, specific self-delivered, and one-step operation properties was developed for NF-κB transcription factor detection and in situ imaging in single cell by steric hindrance. In this system, Cy5 labeled DNA1 has a hairpin structure to fold its long chain and quenches the Cy5 fluorescence by forcing the Cy5 fluorophore close to the surface of AuNPs. The binding of NF-κB leads to the extension of the long chain of DNA1 and the removal of the Cy5 fluorophore from the surface of AuNPs, thereby enhancing the fluorescence signal. By measuring NF-κB in cell lysis in vitro, the probe obtains a detection limit of 0.38 nM and the linear range from 0.5 to 16 nM. Compared with other reported methods [29,30,31,32,33], this method is not as sensitive as them (Table S1). However, the most advantage of this method is that the probe can be used for in situ imaging in single cell. By incubating single cells with the DNA-AuNP probe in one-step, the fluorescence signal reflects the expression level of NF-κB p65, and the sub-localization of the fluorophore indicates the activity of NF-κB p65. The method can effectively distinguish between active NF-κB (nucleus) and inactive NF-κB (cytoplasm) through in situ sub-localization. We expect the well-designed probe will replace the existing technologies that were time-consuming and labor-intensive, and an in-depth understanding of the function of NF-κB in cells will facilitate the study of transcription factors involved in many disease processes.
References
Adcock IM, Caramori G (2009) Transcription factors. In: Barnes PJ, Drazen JM, Rennard SI and Thomson NC (eds) Asthma and COPD (second edition), Chapter 31
Yusuf D, Butland SL, Swanson MI, Bolotin E, Ticoll A, Cheung WA, Zhang XYC (2012) The transcription factor encyclopedia. Genome Biol 13:R24
Fan Z, Wang J, Hao N, Li Y, Yin Y, Wang Z, Ding Y, Zhao J, Zhang K, Huang W (2019) Ultrasensitive detection of transcription factors with a highly-efficient diaminoterephthalate fluorophore via an electrogenerated chemiluminescence strategy. Chem Commun 55:11892–11895. https://doi.org/10.1039/C9CC05692K
Li B, Xia A, Zhang S, Suo T, Ma Y, Huang H, Zhang X, Chen Y, Zhou X (2021) A CRISPR-derived biosensor for the sensitive detection of transcription factors based on the target-induced inhibition of Cas12a activation. Biosens Bioelectron 173:112619. https://doi.org/10.1016/j.bios.2020.112619
Tong L, Yuan Y, Wu S (2015) Therapeutic microRNAs targeting the NF-kappa B signaling circuits of cancers. Adv Drug Del Rev 81:1–15. https://doi.org/10.1016/j.addr.2014.09.004
Guerrini L, Blasi F, Denis-Donini S (1995) Synaptic activation of NF-kappa B by glutamate in cerebellar granule neurons in vitro. Proc Natl Acad Sci U S A 92:9077–9081. https://doi.org/10.1073/pnas.92.20.9077
Lin YC, Brown K, Siebenlist U (1995) Activation of NF-kappa B requires proteolysis of the inhibitor I kappa B-alpha: signal-induced phosphorylation of I kappa B-alpha alone does not release active NF-kappa B. Proc Natl Acad Sci U S A 92:552–556. https://doi.org/10.1073/pnas.92.2.552
Li J-W, Zhang X-Y, Wu H, Ba Y-P (2020) Transcription factor engineering for high-throughput strain evolution and organic acid bioproduction: a review. Front Bioeng Biotech 8:98. https://doi.org/10.3389/fbioe.2020.00098
Zhang K, Fan Z, Huang Y, Xie M, Zhao J, Wang J (2020) A well-designed gold nanoparticle based fluorescence probe for assay Argonaute2 and Let-7a interaction in living cells. Sensor Actuat B: Chem 312:128000. https://doi.org/10.1016/j.snb.2020.128000
Mieszawska AJ, Mulder WJM, Fayad ZA, Cormode DP (2013) Multifunctional gold nanoparticles for diagnosis and therapy of disease. Mol Pharm 10:831–847. https://doi.org/10.1021/mp3005885
Singh P, Pandit S, Mokkapati VRSS, Garg A, Ravikumar V, Mijakovic I (2018) Gold nanoparticles in diagnostics and therapeutics for human cancer. Int J Mol Sci 19:1979. https://doi.org/10.3390/ijms19071979
Smith JE, Chávez JL, Hagen JA, Kelley-Loughnane N (2016) Design and development of aptamer–gold nanoparticle based colorimetric assays for in-the-field applications. J Vis Exp 112:54063. https://doi.org/10.3791/54063
Chen GH, Chen WY, Yen YC, Wang CW, Chang HT, Chen CF (2014) Detection of mercury(II) ions using colorimetric gold nanoparticles on paper-based analytical devices. Anal Chem 86:6843–6849. https://doi.org/10.1021/ac5008688
Chang CC, Chen CP, Wu TH, Yang CH, Lin CW, Chen CY (2019) Gold nanoparticle-based colorimetric strategies for chemical and biological sensing applications. Nanomaterials (Basel) 9. https://doi.org/10.3390/nano9060861
Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA (1997) Selective colorimetric detection of polynucleotides based on the distance-dependent optical properties of gold nanoparticles. Science 277:1078–1081. https://doi.org/10.1126/science.277.5329.1078
Iglesias MS, Grzelczak M (2020) Using gold nanoparticles to detect single-nucleotide polymorphisms: toward liquid biopsy. Beilstein J Nanotech 11:263–284. https://doi.org/10.3762/bjnano.11.20
Kohout C, Santi C, Polito L (2018) Anisotropic gold nanoparticles in biomedical applications. Int J Mol Sci 19:3385. https://doi.org/10.3390/ijms19113385
Saha A, Basiruddin SK, Sarkar R, Pradhan N, Jana NR (2009) Functionalized plasmonic−fluorescent nanoparticles for imaging and detection. J Phys Chem C 113:18492–18498. https://doi.org/10.1021/jp904791h
Bai X, Wang Y, Song Z, Feng Y, Chen Y, Zhang D, Feng L (2020) The basic properties of gold nanoparticles and their applications in tumor diagnosis and treatment. Int J Mol Sci 21:2480. https://doi.org/10.3390/ijms21072480
Hu X, Zhang Y, Ding T, Liu J, Zhao H (2020) Multifunctional gold nanoparticles: a novel nanomaterial for various medical applications and biological activities. Front Bioeng Biotech 8:990–990. https://doi.org/10.3389/fbioe.2020.00990
Dykman LA, Khlebtsov NG (2019) Gold nanoparticles in chemo-, immuno-, and combined therapy: review [invited]. Biomedical Optics Express 10:3152–3182. https://doi.org/10.1364/BOE.10.003152
Lévy R, Shaheen U, Cesbron Y, Sée V (2010) Gold nanoparticles delivery in mammalian live cells: a critical review. Nano Rev 1. https://doi.org/10.3402/nano.v1i0.4889
Frens G (1973) Controlled nucleation for the regulation of the particle size in monodisperse gold suspensions. Nat Phys Sci 241:20–22. https://doi.org/10.1038/physci241020a0
Hinman SS, McKeating KS, Cheng Q (2017) DNA linkers and diluents for ultrastable gold nanoparticle bioconjugates in multiplexed assay development. Anal Chem 89:4272–4279. https://doi.org/10.1021/acs.analchem.7b00341
Storhoff J, Elghanian R, Mucic R, Mirkin C, Letsinger R (1998) One-pot colorimetric differentiation of polynucleotides with single base imperfections using gold nanoparticle probes. J Am Chem Soc 120:1959–1964. https://doi.org/10.1021/ja972332i
Dam DHM, Lee RC, Odom TW (2014) Improved in vitro efficacy of gold nanoconstructs by increased loading of G-quadruplex aptamer. Nano Lett 14:2843–2848. https://doi.org/10.1021/nl500844m
Wu G-R, Mu T-C, Gao Z-X, Wang J, Sy M-S, Li C-Y (2017) Prion protein is required for tumor necrosis factor α (TNFα)-triggered nuclear factor κB (NF-κB) signaling and cytokine production. J Biol Chem 292:18747–18759. https://doi.org/10.1074/jbc.M117.787283
Hayden MS, Ghosh S (2014) Regulation of NF-κB by TNF family cytokines. Semin Immunol 26:253–266. https://doi.org/10.1016/j.smim.2014.05.004
Li X, Yang J, Yuan R, Xiang Y (2019) Programming cascaded recycling amplifications for highly sensitive and label-free electrochemical sensing of transcription factors in tumor cells. Biosens Bioelectron 142:111574. https://doi.org/10.1016/j.bios.2019.111574
Rasheed PA, Lee J-S (2018) Ultrasensitive colorimetric detection of NF-κB protein at picomolar levels using target-induced passivation of nanoparticles. Anal Bioanal Chem 410:1397–1403. https://doi.org/10.1007/s00216-017-0783-y
Zou M, Li X, Li D, Yuan R, Xiang Y (2019) Target-induced steric hindrance protection of DNAzyme junctions for completely enzyme-free and amplified sensing of transcription factors. Sensor Actuat B: Chem 298:126865. https://doi.org/10.1016/j.snb.2019.126865
Li B, Chen Y, Wang J, Lu Q, Zhu W, Luo J, Hong J, Zhou X (2019) Detecting transcription factors with allosteric DNA-silver nanocluster switches. Anal Chim Acta 1048:168–177. https://doi.org/10.1016/j.aca.2018.10.023
Li B, Xu L, Chen Y, Zhu W, Shen X, Zhu C, Luo J, Li X, Hong J, Zhou X (2017) Sensitive and label-free fluorescent detection of transcription factors based on DNA-Ag nanoclusters molecular beacons and exonuclease III-assisted signal amplification. Anal Chem 89:7316–7323. https://doi.org/10.1021/acs.analchem.7b00055
Funding
This work was supported by the National Natural Science Foundation of China (Grant Nos. 81702993 and 21705061), Jiangsu Provincial Key Medical Discipline (Laboratory) (ZDXKA2016017), and Innovation Capacity Development Plan of Jiangsu Province (BM2018023).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Nanomaterials for biomedical imaging and targeting
Supplementary Information
ESM 1
(DOCX 2738 kb)
Rights and permissions
About this article
Cite this article
Ding, Y., Fan, Z., Yao, B. et al. Nanoparticle-based fluorescence probe for detection of NF-κB transcription factor in single cell via steric hindrance. Microchim Acta 188, 226 (2021). https://doi.org/10.1007/s00604-021-04878-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-021-04878-y