Abstract
Immunohistochemistry (IHC) and immunocytochemistry (ICC) are widely used to identify cancerous cells within tissues and cell cultures. Even though the optical microscopy evaluation is considered the gold standard, the limited range of useful labels and narrow multiplexing capabilities create an imminent need for alternative readout techniques. Laser-induced breakdown spectroscopy (LIBS) enables large-scale multi-elemental analysis of the surface of biological samples, e.g., thin section or cell pellet. It is, therefore, a potential alternative for IHC and ICC readout of various labels or tags (Tag-LIBS approach). Here, we introduce Tag-LIBS as a method for the specific determination of HER2 biomarker. The cell pellets were labeled with streptavidin-conjugated upconversion nanoparticles (UCNP) through a primary anti-HER2 antibody and a biotinylated secondary antibody. The LIBS scanning enabled detecting the characteristic elemental signature of yttrium as a principal constituent of UCNP, thus indirectly providing a reliable way to differentiate between HER2-positive BT-474 cells and HER2-negative MDA-MB-231 cells. The comparison of results with upconversion optical microscopy and luminescence intensity scanning confirmed that LIBS is a promising alternative for the IHC and ICC readout.
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The specific detection and imaging of biomarkers within cancerous tissues are crucial for early diagnosis and treatment. Traditionally, immunochemical methods combine the specificity of antibodies with various detection schemes [1]. In the most common approaches, antibodies immobilized in microtiter plates are used to capture the molecule of interest, followed by the detection step utilizing labels based on antibodies conjugated with either enzymes (enzyme-linked immunosorbent assay, ELISA) or fluorophores (fluorescence immunoassay, FIA) [2]. The same labels can also be employed for specific detection of target markers within tissue samples (immunohistochemistry, IHC) and cultured cells (immunocytochemistry, ICC) [3]. However, traditional labels suffer from several disadvantages; enzymes have high production costs, limited stability, and a time-consuming signal development [4], while fluorophores are prone to photobleaching and interference from autofluorescence [2].
In recent years, various kinds of nanomaterials have been used to overcome the properties of conventional labels in immunoassays and cell labeling, leading to enhanced detection capabilities [5, 6]. Typically, two basic schemes are implemented: either (i) catalytic nanoparticles (nanozymes), which allow a detection based on substrate conversion [4, 7], or (ii) luminescent nanoparticles, which allow a direct detection [8]. The most commonly used luminescent nanoparticle labels include quantum dots (QD) [9, 10]. However, other alternatives are being investigated, including carbon-based nanoparticles (CD) [11], or lanthanide-based photon-upconversion nanoparticles (UCNP) [12, 13]. We have recently investigated the feasibility of UCNP for the background-free visualization of the HER2 biomarker on the surface of breast cancer cells [14]. The luminescence readout provided a high sensitivity with an unsurpassed signal-to-background ratio (SBR). However, this technique allows detecting only the labels with luminescent properties, limiting the capabilities for multiplexing. Therefore, there is a need for novel readout techniques which would allow a universal detection of non-luminescent labels.
Laser-induced breakdown spectroscopy (LIBS) is an optical emission method complementary to conventionally utilized techniques in bioimaging applications [15]. It offers a fast analysis (reduction of turnaround time from minutes to microseconds per single spectrum acquisition), multi-elemental capability and selectivity, a possibility to detect halogens and light elements, and a reduction of operating and maintenance costs by order of magnitude [16, 17]. All these benefits are provided while satisfactorily conserving the sensitivity. The main advantage of LIBS is the ability of multi-elemental imaging with a fine resolution (units of micrometers) on a large scale (few centimeters) [18].
The analytical performance of LIBS has been beneficially used in many different biological applications. Recently, several reviews described the general scope of bioimaging [15, 19] or direct clinical usage [20]. NPs were used for LIBS signal enhancement of other analytes [21, 22]. For the characterization of NPs, optical catapulting-trapping LIBS was used [23]. Repeatedly, LIBS proved to be a useful readout method for various micro- and nanoparticle-based labels in the so-called tag-LIBS arrangement. Barcoding based on LIBS-encoded polystyrene microsphere was used as a coding method for suspension arrays [24]. Ovarian cancer biomarker CA-125 was detected via titanium dioxide and iron oxide microparticles-conjugated antibodies [25], metallothionein via Cd-based QDs [26], and human serum albumin via streptavidin-coated Ag NP labels [18]. The most recent work focused on the readout of lateral-flow immunoassays with Au NPs labeled Escherichia coli [27].
The LIBS technique proved to be a relevant alternative for the readout of immunoassays, appropriately applying all its advantages (multi-element capability and high-throughput). Recently, the emphasis has been given to the implementation of LIBS to immunoassays based on various supports (e.g., glass, plastics). Such experimental arrangement may be extended to the analysis of soft tissues or model cell pellets where proteins of interest are marked with various NPs. Selected NPs can be combined to create so-called barcode labels with a specific target conjugation as well as a characteristic emission response. With this approach, a great variety of micro- and nanoparticle labels is available to open new possibilities in barcoding via multi-elemental readout using LIBS. Recently, the need for new readout methods in bioanalytical chemistry (especially in immunoassay application) is increasing, and LIBS represents a suitable alternative. Several milestones were reached, from the identification of NP-labeled proteins [26] to applications in nanoparticle-based immunoassays [18], including lateral-flow assay [27].
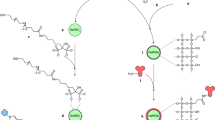
The novelty of this work lies in the specific immunochemical labeling of biomarkers on breast cancer tissues, followed by the LIBS readout. This approach pushes forward the LIBS technology in terms of the specific detection of cancer biomarkers up to the final goal—the development of routine cancer diagnostics for biochemical laboratories. Here, we pioneer the application of LIBS for the readout of nanoparticle-labeled immunocytochemistry sections. UCNP were conjugated with streptavidin (SA) and employed for the specific immunolabeling of the HER2 biomarker on the surface of breast cancer cell lines (Fig. 1a). The LIBS setup was optimized for imaging of the cells, and a data evaluation approach that enabled a reliable readout was introduced (Fig. 1b).
a Scheme of the ICC assay. The primary antibody is used to label the HER2 biomarker on the cell surface, followed by binding of the biotinylated secondary antibody and the UCNP-SA conjugate. b Schematic representation of LIBS readout, based on the ablation of material and the detection of Y II 437.49 nm signal
Experimental
Chemicals and reagents
Neridronate (Ner), nitrosyl tetrafluoroborate (NOBF4), copper (II) sulfate pentahydrate, L-ascorbic acid sodium salt, and Tween 20 were purchased from Merck (Germany). Dry dimethylformamide (DMF) and SuperBlock TBS (SB) were obtained from Thermo Fisher Scientific (USA). Streptavidin-azide was obtained from 7 Bioscience (Germany). α-N-hydroxysuccinimide-ω-alkyne polyethylene glycol (MW 3 kDa, Alkyne-PEG-NHS) was purchased from Iris Biotech (Germany). All other common chemicals were obtained in the highest quality available from Merck (Germany), Carl Roth (Germany), and Penta (Czech Republic).
Buffers included phosphate buffer (PB; 50 mM NaH2PO4/Na2HPO4, pH 7.4), phosphate-buffered saline (PBS; PB with 150 mM NaCl), Tris buffered saline (TBS; 50 mM Tris, 150 mM NaCl, pH 7.5), TBS-T buffer (TBS with 0.05% Tween 20), assay buffer (10% SuperBlock in TBS, 1 mM KF, 0.05% Tween 20, and 0.05% NaN3, pH 7.5), antibody dilution buffer (10% SuperBlock in PBS, 0.05% Tween 20, pH 7.5), 10 mM Tris-HCl (pH 7.5), and epitope retrieval buffer (10 mM Tris, 1 mM EDTA, 0.05% Tween 20, pH 9). The UCNP-SA labels were prepared according to our previous work [14].
Preparation of UCNP-based labels
UCNP-based labels were prepared according to our previous work [28]. The NaYF4:Yb3+, Tm3+-doped UCNP were synthesized by high-temperature co-precipitation [29].
To prepare the Alkyne-PEG-Ner linker, 30 mg of Ner was dissolved under sonication in 128 μL of 1 M NaOH, and 898 μL of PB (pH 7.6) was added. This was followed by the addition of 75 mg Alkyne-PEG-NHS in PB (pH 7.6) and incubation for 3.5 h at room temperature and at 4 °C overnight. The synthesized Alkyne-PEG-Ner linker was dialyzed in a Float-A-Lyzer G2 dialysis device (500–1000 Da MWCO, Fisher Scientific, Germany) against double-distilled water at 4 °C. The linker was then lyophilized using Alpha 1–2 freeze dryer (Christ, Germany) and stored at 4 °C [30].
Oleic acid-capped UCNP (10 mg) dispersed in cyclohexane were mixed with an equivalent volume of dimethylformamide (DMF), followed by the addition of 10 mg of NOBF4. The mixture was shaken at 30 °C for 20 min, during which the oleic acid was removed from the nanoparticle surface, and UCNP transferred from cyclohexane to DMF. The upper cyclohexane phase was discarded, followed by the addition of an excess of chloroform in order to precipitate the UCNP. The purification of UCNP was performed using centrifugation at 1000g for 15 min, followed by a redispersion in DMF, precipitation with chloroform, and another centrifugation step at 1000 g for 15 min. Then, 1 mL of double-distilled water with 2 mg of the Alkyne-PEG-Ner linker was used to redisperse the UCNP pellet, and the mixture was incubated at 4 °C overnight. The alkyne-PEG-Ner-UCNP conjugates were purified by dialysis in a Float-A-Lyzer G2 (100 kDa MWCO, Fisher Scientific) against double-distilled water with 1 mM KF at 4 °C.
For the click reaction, 10 mg of Alkyne-PEG-Ner-UCNPs in 700 μL water was first diluted with 100 μL of 325 mM Tris-HCl (pH 7.5), and 20 μL of 28.6 mg/mL of sodium L-ascorbate in double-distilled water was added. After purging the mixture for 40 min with argon, streptavidin-azide (100 μL of 1 mg/mL solution) was added, followed by purging for another 10 min. The start of the click reaction was catalyzed by the addition of 10 μL of 35.7 mM CuSO4 in double-distilled water, followed by purging with argon for 45 min and sonication for 10 min. The conjugates were purified by dialysis in Float-A-Lyzer G2 (100 kDa MWCO) against Tris-HCl (pH 7.5) with 1 mM KF and 0.05% NaN3 at 4 °C [12, 14].
Transmission electron microscopy (TEM) revealed a uniform spherical shape with an average diameter of 37 nm. The DLS number distribution of oleic acid-capped UCNP showed the size of 38.5 ± 1.8 nm, which increased to 63 ± 3 nm after the conjugation with streptavidin [28].
Immunochemical labeling of cells
The formalin-fixed paraffin-embedded (FFPE) slides with BT-474 (HER2 3+) and MDA-MB-231 (HER2 0–1+) cells [31] were obtained from AMS Biotechnology (UK). First, xylene (a mixture of isomers) was used to dewax the slides (3× 5 min), followed by the rehydration using 99% ethanol (2× 5 min), 90% ethanol (2× 5 min), and water (5 min). In the next step, the slides were placed in the epitope retrieval buffer (pre-heated to 60 °C) and heated for 20 min to 80 °C using a water bath to perform the heat-induced epitope retrieval. After 20 min of cooling down to room temperature, the slides were transferred to TBS for 2 min, taken out, and dried.
A PAP pen liquid blocker (Science Services, Germany) was used to encircle (~0.5 cm in diameter) the cell section, followed by a washing step in TBS for 1 min. The slide was gently dried; the pellet remained moist. The following steps were performed at room temperature. The solutions were added as 50-μL droplets, and before the incubation with a new reagent, the sample was pre-washed with the reagent applied. First, a blocking using concentrated SuperBlock with 0.05% Tween 20 was done for 60 min, followed by washing with TBS (3× 5 min). Second, the cells were incubated with a rabbit anti-HER2 antibody (ab134182, Abcam, UK) in a concentration of 1 μg/mL in antibody dilution buffer for 60 min. Third, the slides were washed with TBS (3× 5 min), followed by the incubation with 2 μg/mL of a biotinylated anti-rabbit antibody (111-065-144, Jackson ImmunoResearch, UK) in antibody dilution buffer for 60 min. Fourth, the cells were incubated for 60 min with the prepared nanoparticle conjugates (UCNP-SA in varying concentrations) in assay buffer. Finally, the slides were washed with TBS-T buffer (4× 5 min), TBS (5 min), 10 mM Tris (5 min), and dried by compressed air. The prepared samples were then analyzed with LIBS and the luminescence readout [14].
Laser-induced breakdown spectroscopy
The LIBS measurements were performed on a custom-made LIBS Discovery system (CEITEC, Czech Republic) consisting of an ablation chamber, a nanosecond laser (CFR Ultra 400, Quantel by Lumibird, France; Nd:YAG 532 nm, 20 Hz, 10 ns), and a Czerny-Turner spectrometer Shamrock (SR-500i-B2-R, Andor, UK) equipped with a detector iStar sCMOS (iSTAR-sCMOS-18F-E3, Andor, UK). The whole cell pellet was analyzed in a raster of 60 × 60 spots with a 100 μm step in both directions, one pulse per spot. The setting of the apparatus was as follows: gate width of 50 μs, gate delay of 500 ns, laser pulse energy of 10 mJ, with a purge of argon above the sample surface. The LIBS system was optimized a priori to achieve the best signal-to-noise ratio for the selected emission lines.
The emission line Y II 437.49 nm was chosen based on our previous experiments [32]. The LIBS data was processed using a custom-made C++ software and depicted in the ImageLab (National Institutes of Health, USA) and Origin 2019b (OriginLab, USA). First, each spectrum was internally standardized to the total emissivity to reduce pulse-to-pulse fluctuations. The emission line intensity was estimated as a sum of intensities over the selected wavelength range as done in our former work [18]. No background correction was utilized in the data processing. Consequently, the 2D maps of emission line intensities were created, representing the spatial distribution of Y in the cell pellet.
The data processing strategy was proposed to analytically compare the individual approaches in the sample preparation and intensity response of various label concentrations. LIBS analysis of one single pellet resulted in the map showing Y II 437.49 nm intensity distribution. Then, only spectra from the pellet region were considered; the intensity of Y was estimated for each spectrum (area under the peak) and summed up. Thus, the sample (cell pellet with varied UCNP concentration) was represented by the value of the average Y intensity from the pellet region and was considered as one point in the experiment design. In turn, the averaged Y II intensity value served as a figure of merit for comparing individual measurements. The signal-to-noise ratio (SNR) was estimated for each spectrum in the pilot sample and served for the optimization.
Upconversion microscopy imaging
The UCNP also provided the possibility to monitor their upconversion luminescence (emission of shorter wavelengths under near-IR excitation) as a reference readout technique. A modified wide-field epifluorescence inverted microscope (Eclipse Ti, Nikon, Japan) [33] was used for the visualization of the labeled cells. For the excitation of UCNP, a continuous-wave 980-nm laser diode (4 W; Wavespectrum, China) was coupled to the motorized TIRF illuminator unit (TI-TIRF-E, Nikon) via a multi-mode optical fiber (105 μm fiber core, NA = 0.22, Wavespectrum, China). The used filter set contained a long-pass excitation filter (cut-on 830 nm; Schott, Germany), a short-pass dichroic mirror (cut-on 875 nm; AHF Analysentechnik, Germany), and a NIR band-pass emission filter (809 ± 40 nm). The emission light was collected with a ×20 objective (NA 0.75) and recorded using a Neo 5.5 vacuum-cooled 5.5-megapixel sCMOS camera (Andor, UK), which provided a field of view of 832 × 702 μm. The software NIS Elements 4.5 Advanced Research (Nikon) was used to control the microscope. The images were evaluated by ImageJ [14].
Luminescence scanning
The upconversion luminescence was scanned using a Chameleon microplate reader (Hidex, Finland) modified for the measurement of upconversion. To obtain the images of the cell pellets, squares of 18 × 18 points with a step size of 500 μm were raster-scanned with an exposure time of 500 ms. The images were analyzed by ImageJ. The luminescence intensities of individual cell pellets were evaluated by placing circular regions of interest with an identical size over the pellet area, and the gray values of all pixels in the region of interest were averaged. After the quantitative evaluation, a bilinear interpolation was used to increase the resolution of images fivefold, and the resulting images were processed using Origin 2019b [34].
Results and discussion
Characteristic spectra
In conventional immunochemical methods, nanoparticles are used as specific labels to improve the detection performance. The application of nanoparticles also opens new perspectives in the readout strategy and implementation of novel techniques, such as LIBS. The presence of metallic nanoparticles within the sample is more favorable for the laser ablation and the consecutive laser-induced plasma formation resulting in a satisfactory analytical sensitivity. Thus, it is not necessary to target the investigated protein but indirectly evaluate its presence in terms of the characteristic NPs signal.
First, the LIBS experiment was optimized to achieve the highest SNR value for the selected analytical line. It was done in a pulse-to-pulse mode, where each spectrum was considered individually; gate delay and laser pulse energy were the only varied parameters. The optimal conditions were found to be 10 mJ per laser pulse and 500 ns gate delay; all settings are listed in Paragraph 2.5. Interestingly, the use of a 532 nm laser pulse led to a significant, undesirable damage to the glass substrate. Thus, lower pulse energy was chosen as a compromise between the glass damage and good analytical sensitivity. Further optimization is beyond the scope of this work when considering other options in the sample treatment (e.g., using other substrates - polystyrene microplate) and/or laser ablation conditions (266 nm laser pulse) and different experimental settings (e.g., ambient atmosphere and its pressure) [26].
The short-band spectra with characteristic spectral lines were detected for each laser pulse (Fig. 2). The spectral line of yttrium, Y II 437.49 nm, was selected based on our experience [32], and used throughout the experiment. The intensity profiles show distinct differences between positive and control samples (BT-474 or MDA-MB-231 cell pellet with 700 μg/mL of UCNP-SA). The intensity profile of a control spectrum reveals also the Y II 437.49 nm spectral line, which is at the detection limit. Its presence suggests a non-specific binding of UCNP for higher concentrations, which is also the case of the depicted spectra, and a further discussion follows in paragraph 3.2. In addition, the spectrum reveals the possibility of simultaneous detection of multiple elements along with yttrium, and the possibility of simultaneous detection of increased concentrations of other elements associated with cancer growth or with the development of other serious diseases [20]. However, this advantageous feature mainly suggests that LIBS has excellent potential in barcodes and multi-element label readout.
Elemental imaging
After a LIBS system optimization, the LIBS mapping raster was set to cover a square 60 × 60 points (step size 100 μm) with respect to the size of the cell pellets (ca 0.5 cm in diameter), and also to display the immediate surroundings. The center of the scanned area was always set to coincide with the center of a pellet. For the same conditions, the BT-474 pellet was mapped as a HER2-positive representative of breast cancer cell lines, and MDA-MB-231 was selected as a HER2-negative control.
Figure 3a shows the spatial distribution of yttrium-based UCNP in the cell pellets represented by the Y II 437.49 nm emission line intensity with varying UCNP-SA label concentrations. LIBS enabled to specifically map the presence of UCNP, showing an evident difference between the positive and negative samples. However, several undesirable effects were observed, as well. The isolated spikes on the maps, especially for higher label concentrations, probably indicate larger clusters of non-specifically bound nanoparticles [35]. Moreover, the signal tailing on the maps represented by high intensities appears at the top of the pellet map. It should be noted that the mapping was realized by consecutive laser pulses with a bottom-up direction. Lower intensity on the bottom of the pellet and its upward increase are most probably attributed to the redeposition of the ablated material. The redeposition phenomenon has already been observed [32] and is particularly limiting in terms of using the method for mapping the structures of cancerous tissues with an appropriate spatial resolution. As a zigzag-programmed sample motion was used, the signal shifting on every other line was also apparent.
a 2D LIBS maps of BT-474 and MDA-MB-231 cell pellets with HER2 marker labeled using the UCNP-SA conjugate with concentrations from 7 to 700 μg/mL; the raster size is 100 μm. b Dependence of Y II 437.49 nm signal on the UCNP concentration. The error bars correspond to the standard deviations of intensities within the cell pellet region. c Signal-to-background ratio evaluated by dividing the BT-474 and MDA-MB-231 cell pellet signals
The quantitative evaluation of the obtained intensity maps was based on the integration of yttrium intensities over the cell pellet region (Fig. 3b). This approach allows obtaining one representative value, which can be used for further processing. First of all, the area of the map should be carefully selected (in this case manually) because the pellet area and its position slightly change for each sample. For this purpose, not only maps themselves but also images of the sample before ablation were obtained.
The background signals in the case of the control sample can be considered as the sum of two contributions. The first one might be attributed to the continuum background of LIBS emission spectra which is always present. The background in each LIBS spectrum was not corrected during the evaluation; thus, Y II intensities were considered with a certain background. The second contribution might be attributed to the non-specific adsorption of UCNP on the sample controlled by an adsorption isotherm that could be derived from the slope of the control sample line. Furthermore, the dissolution of UCNP [36] can lead to a release of free Y3+ ions, which can non-specifically adsorb to the cells.
Thus, the diminishing of background signals is possible in two ways. Appropriate background subtraction and internal standardization to total emissivity can be used to minimize the LIBS optical background. The non-specific adsorption of the label can be reduced by optimizing the UCNP surface modification and modifying the labeling procedure to facilitate the removal of non-specifically bound UCNP. Both approaches deserve further investigation and will be the subject of our following experiments.
Overall, an increasing UCNP-SA concentration leads to the increasing signals for the HER2-positive cell line (BT-474) and only a small increase for the negative control (MDA-MB-231). The typical opening scissors shape graph for positive and control samples indicates a more sensitive detection for higher concentrations. The UCNP-SA concentration of 7 μg/mL is near the detection limit of the LIBS system, providing a barely visible signal change and a SBR (Fig. 3c) of 1.2. Increasing the label concentration up to 140 μg/mL led to the increase of the specific signal while keeping the low level of control, reaching a SBR of 3.3. However, further increasing the UCNP-SA concentration to 700 μg/mL was associated with an increase of the negative control signal, which led to a decrease of SBR to 2.7. This can be explained by the progressive saturation of the specific binding sites on the BT-474 cell line. On the other hand, the number of potential sites for non-specific binding is in excess, making the label concentration the most important factor affecting the SBR.
Finally, the four independent HER2-positive cell pellets were labeled using UCNP-SA conjugates in a concentration of 70 μg/mL to study the repeatability of the measurement (Fig. 4). All the samples provided similar results, showing the higher intensities on the edge of the pellet, which might be connected with the drying of the sample (“coffee ring” effect). Moreover, the rise in the intensity on the top of the map and beyond the pellet region may be attributed to the redeposition of the ablated matter. The variability of intensities within one spot was comparable among the samples; the overall relative standard deviation among the spots was 8% (Fig. 4b).
a 2D LIBS maps of BT-474 cell pellets labeled with UCNP-SA conjugate in a concentration of 70 μg/mL. b Average intensities within each of spots 1–4 (error bars correspond to the standard deviations of intensities within spots; marked in red) and the total average of the four mean values obtained from spots 1–4 (error bar corresponds to the standard deviation among the individual averages; marked in blue)
Comparison with reference techniques
To incorporate the LIBS method in the context of conventional techniques, the same ICC slides were analyzed by upconversion microscopy and luminescence scanning prior to the LIBS measurement. Optical microscopy is traditionally used as a gold standard for the evaluation of the ICC and IHC. We have recently introduced the application of UCNP-based labels in this field. The microscopy images after labeling the BT-474 and MDA-MB-231 cells using UCNP-SA conjugate in the concentration of 700 μg/mL are shown in Fig. 5. Specific labeling of HER2 on the cell membranes was visible on the positive sample of BT-474, whereas practically no signal was observed on the negative control of MDA-MB-231. The main advantage of microscopy compared to scanning-based approaches is the high resolution, allowing to follow the distribution of the target within the cellular structures. On the other hand, conventional optical microscopy is not optimal for the quantitative determination of the label amount within the whole spot of cells.
Next, LIBS was compared to upconversion luminescence scanning (Fig. 6). Both methods were successful in showing the difference between the HER2-positive and HER2-negative samples. LIBS provides a higher resolution (step size of 100 μm) compared to upconversion scanning (step size of 500 μm); however, it should be noted that both methods might be able to reach a rather high resolution after further optimizations. The LIBS provided a SBR of 5, whereas upconversion scanning reached a SBR of 159. Since the actual amount of UCNP label is the same in both scans, the lower SBR of LIBS is probably caused by a lower readout sensitivity. In comparison with luminescence-based methods, LIBS is not limited to labels with luminescent properties, which is an advantage that needs to be highlighted because it opens up possibilities for multiplexing.
Comparison of (a) LIBS and (b) upconversion scanning of BT-474 and MDA-MB-231 cells with HER2 biomarker labeled with UCNP-SA conjugate, using the same scanning area sizes. c The average intensities evaluated by the two methods. Error bars correspond to the standard deviation of intensities within the cell pellet
Laser ablation inductively coupled plasma mass spectrometry (LA-ICP-MS) method is another widely used laser ablation-based method for the imaging of biomolecules in tissues by using diverse labels. During the last decade, it became a recognizable tool in the identification and localization of various biomarkers by NP-labelling directly in tissue samples, in cultured cells, or even in single-cells [37]. LA-ICP-MS enables the elemental and isotopic determination of metals/metalloids in solid samples with a micrometer spatial resolution (down to 1 μm) at trace levels (achievable sensitivity down to picomoles) [38]. The widespread use of NP-based labels for specific-recognition reactions for the determination of biomolecules by LA-ICP-MS is summarized in the recent review [39]. Compared to LIBS, LA-ICP-MS is very instrumentation-demanding method with high operation and acquisition costs, without the possibility of construction of cheap, simple, and compact tabletop instruments. The technique also has other limitations, and its employing for straightforward, fast, or remote sensing is impossible.
Conclusions
LIBS was introduced as an alternative readout technique for ICC and IHC with upconversion nanoparticle labels. Cell pellets of BT-474 (HER2-positive) and MDA-MB-231 (HER2-negative) were labeled with streptavidin-conjugated UCNPs through primary anti-HER2 antibody and biotinylated secondary antibody.
LIBS enabled a 2D scanning of the sample surface (labeled cell pellets) through the detection of the characteristic signal of the Y II 437.49 nm emission line. This approach allowed indirect imaging of the cell pellets with a resolution of 100 μm. Other elements were detected along with the yttrium signal, which shows the multi-elemental feature of LIBS and outlines its further multiplexing and barcoding capabilities. We have shown that after an appropriate optimization, the LIBS analysis is stable and reproducible. Additionally, we have introduced a novel algorithm for LIBS data processing that provides a platform for the evaluation of 2D scans of biological samples. The results showed a desired LIBS sensitivity to the yttrium signal and specificity of UCNP-SA, clearly demonstrating the difference between the HER2-positive and negative cells in ICC readout. The increasing UCNP-SA concentration led to an increase in the specific binding with a small effect on the negative control, resulting in an optimal SBR at a concentration of 140 μg/mL. Finally, the LIBS results were supported by the upconversion optical microscopy and upconversion luminescence scanning.
Despite the success of our feasibility study, further improvements in terms of lateral resolution and analytical sensitivity are necessary in order to meet the needs of IHC and ICC readouts. In our future work, we want to focus on enhancing the scanning resolution while altering the LIBS experimental apparatus and applying different nanoparticle labels (including multi-metal alloys) to test the feasibility of multiplexing through multi-elemental detection.
References
Luppa PB, Sokoll LJ, Chan DW (2001) Immunosensors - principles and applications to clinical chemistry. Clin Chim Acta 314(1–2):1–26. https://doi.org/10.1016/s0009-8981(01)00629-5
Farka Z, Mickert MJ, Pastucha M, Mikusova Z, Skladal P, Gorris HH (2020) Advances in optical single-molecule detection: en route to supersensitive bioaffinity assays. Angew Chem Int Edit 59(27):10746–10773. https://doi.org/10.1002/anie.201913924
Susaki EA, Ueda HR (2016) Whole-body and whole-organ clearing and imaging techniques with single-cell resolution: toward organism-level systems biology in mammals. Cell Chem Biol 23(1):137–157. https://doi.org/10.1016/j.chembiol.2015.11.009
Wang QQ, Wei H, Zhang ZQ, Wang EK, Dong SJ (2018) Nanozyme: an emerging alternative to natural enzyme for biosensing and immunoassay. Trac Trends Anal Chem 105:218–224. https://doi.org/10.1016/j.trac.2018.05.012
Yezhelyev MV, Gao X, Xing Y, Al-Hajj A, Nie SM, O’Regan RM (2006) Emerging use of nanoparticles in diagnosis and treatment of breast cancer. Lancet Oncol 7(8):657–667. https://doi.org/10.1016/s1470-2045(06)70793-8
Farka Z, Juriik T, Kovaar D, Trnkova L, Sklaadal P (2017) Nanoparticle-based immunochemical biosensors and assays: recent advances and challenges. Chem Rev 117(15):9973–10042. https://doi.org/10.1021/acs.chemrev.7b00037
Dong JL, Song LN, Yin JJ, He WW, Wu YH, Gu N, Zhang Y (2014) Co3O4 nanoparticles with multi-enzyme activities and their application in immunohistochemical assay. ACS Appl Mater Interfaces 6(3):1959–1970. https://doi.org/10.1021/am405009f
Seydack M (2005) Nanoparticle labels in immunosensing using optical detection methods. Biosens Bioelectron 20(12):2454–2469. https://doi.org/10.1016/j.bios.2004.11.003
Zhou L, Yan J, Tong L, Han X, Wu X, Guo P (2016) Quantum dot-based immunohistochemistry for pathological applications. Cancer Transl Med 2(1):21–28. https://doi.org/10.4103/2395-3977.177562
Wu Y-T, Qiu X, Lindbo S, Susumu K, Medintz IL, Hober S, Hildebrandt N (2018) Quantum dot-based FRET immunoassay for HER2 using Ultrasmall affinity proteins. Small 14:1802266. https://doi.org/10.1002/smll.201802266
Tang DP, Lin YX, Zhou Q (2018) Carbon dots prepared from Litchi chinensis and modified with manganese dioxide nanosheets for use in a competitive fluorometric immunoassay for aflatoxin B-1. Microchim Acta 185(10):476. https://doi.org/10.1007/s00604-018-3012-2
Mickert MJ, Farka Z, Kostiv U, Hlavacek A, Horak D, Skladal P, Gorris HH (2019) Measurement of sub-femtomolar concentrations of prostate-specific antigen through single-molecule counting with an upconversion-linked immunosorbent assay. Anal Chem 91(15):9435–9441. https://doi.org/10.1021/acs.analchem.9b02872
He H, Howard CB, Chen YH, Wen SH, Lin GG, Zhou JJ, Thurecht KJ, Jin DY (2018) Bispecific antibody-functionalized upconversion nanoprobe. Anal Chem 90(5):3024–3029. https://doi.org/10.1021/acs.analchem.7b05341
Farka Z, Mickert MJ, Mikusova Z, Hlavacek A, Bouchalova P, Xu WS, Bouchal P, Skladal P, Gorris HH (2020) Surface design of photon-upconversion nanoparticles for high-contrast immunocytochemistry. Nanoscale 12(15):8303–8313. https://doi.org/10.1039/c9nr10568a
Modlitbova P, Porizka P, Kaiser J (2020) Laser-induced breakdown spectroscopy as a promising tool in the elemental bioimaging of plant tissues. Trac Trends Anal Chem 122:10. https://doi.org/10.1016/j.trac.2019.115729
Hahn DW, Omenetto N (2012) Laser-induced breakdown spectroscopy (LIBS), part II: review of instrumental and methodological approaches to material analysis and applications to different fields. Appl Spectrosc 66(4):347–419. https://doi.org/10.1366/11-06574
El Haddad J, Canioni L, Bousquet B (2014) Good practices in LIBS analysis: review and advices. Spectrochim Acta B At Spectrosc 101:171–182. https://doi.org/10.1016/j.sab.2014.08.039
Modlitbova P, Farka Z, Pastucha M, Porizka P, Novotny K, Skladal P, Kaiser J (2019) Laser-induced breakdown spectroscopy as a novel readout method for nanoparticle-based immunoassays. Microchim Acta 186(9):10. https://doi.org/10.1007/s00604-019-3742-9
Busser B, Moncayo S, Coll JL, Sancey L, Motto-Ros V (2018) Elemental imaging using laser-induced breakdown spectroscopy: a new and promising approach for biological and medical applications. Coord Chem Rev 358:70–79. https://doi.org/10.1016/j.ccr.2017.12.006
Gaudiuso R, Melikechi N, Abdel-Salam ZA, Harith MA, Palleschi V, Motto-Ros V, Busser B (2019) Laser-induced breakdown spectroscopy for human and animal health: a review. Spectrochim Acta B At Spectrosc 152:123–148. https://doi.org/10.1016/j.sab.2018.11.006
Dell’Aglio M, Alrifai R, Giacomo A (2018) Nanoparticle enhanced laser induced breakdown spectroscopy (NELIBS), a first review. Spectrochim Acta B At Spectrosc 148:105–112. https://doi.org/10.1016/j.sab.2018.06.008
Zhao X, Zhao C, Du X (2019) Detecting and mapping harmful chemicals in fruit and vegetables using nanoparticle-enhanced laser-induced breakdown spectroscopy. Sci Rep 9:906. https://doi.org/10.1038/s41598-018-37556-w
Fortes FJ, Fernandez-Bravo A, Laserna JJ (2014) Chemical characterization of single micro- and nano-particles by optical catapulting-optical trapping-laser-induced breakdown spectroscopy. Spectrochim Acta B At Spectrosc 100:78–85. https://doi.org/10.1016/j.sab.2014.08.023
He Q, Liu Y, He Y (2016) Digital barcodes of suspension array using laser induced breakdown spectroscopy. Sci Rep 6:36511. https://doi.org/10.1038/srep36511
Markushin Y, Sivakumar P, Connolly D, Melikechi N (2015) Tag-femtosecond laser-induced breakdown spectroscopy for the sensitive detection of cancer antigen 125 in blood plasma. Anal Bioanal Chem 407(7):1849–1855. https://doi.org/10.1007/s00216-014-8433-0
Konecna M, Novotny K, Krizkova S, Blazkova I, Kopel P, Kaiser J, Hodek P, Kizek R, Adam V (2014) Identification of quantum dots labeled metallothionein by fast scanning laser-induced breakdown spectroscopy. Spectrochim Acta B At Spectrosc 101:220–225. https://doi.org/10.1016/j.sab.2014.08.037
Gondhalekar C, Biela E, Rajwa B, Bae E, Patsekin V, Sturgis J, Reynolds C, Doh IJ, Diwakar P, Stanker L, Zorba V, Mao XL, Russo R, Robinson JP (2020) Detection of E. coli labeled with metal-conjugated antibodies using lateral-flow assay and laser-induced breakdown spectroscopy. Anal Bioanal Chem 412(6):1291–1301. https://doi.org/10.1007/s00216-019-02347-3
Peltomaa R, Farka Z, Mickert MJ, Brandmeier JC, Pastucha M, Hlaváček A, Martínez-Orts M, Canales Á, Skládal P, Benito-Peña E, Moreno-Bondi MC, Gorris HH (2020) Competitive upconversion-linked immunoassay using peptide mimetics for the detection of the mycotoxin zearalenone. Biosens Bioelectron 170:112683. https://doi.org/10.1016/j.bios.2020.112683
Wang F, Han Y, Lim CS, Lu YH, Wang J, Xu J, Chen HY, Zhang C, Hong MH, Liu XG (2010) Simultaneous phase and size control of upconversion nanocrystals through lanthanide doping. Nature 463(7284):1061–1065. https://doi.org/10.1038/nature08777
Kostiv U, Lobaz V, Kucka J, Svec P, Sedlacek O, Hruby M, Janouskova O, Francova P, Kolarova V, Sefc L, Horak D (2017) A simple neridronate-based surface coating strategy for upconversion nanoparticles: highly colloidally stable I-125-radiolabeled NaYF4:Yb3+/Er3+@PEG nanoparticles for multimodal in vivo tissue imaging. Nanoscale 9(43):16680–16688. https://doi.org/10.1039/c7nr05456d
Subik K, Lee J-F, Baxter L, Strzepek T, Costello D, Crowley P, Xing L, Hung M-C, Bonfiglio T, Hicks D, Ping T (2010) The expression patterns of ER, PR, HER2, CK5/6, EGFR, Ki-67 and AR by immunohistochemical analysis in breast cancer cell lines. Breast Cancer 4:35–41. https://doi.org/10.1177/117822341000400004
Modlitbova P, Hlavacek A, Svestkova T, Porizka P, Simonikova L, Novotny K, Kaiser J (2019) The effects of photon-upconversion nanoparticles on the growth of radish and duckweed: bioaccumulation, imaging, and spectroscopic studies. Chemosphere 225:723–734. https://doi.org/10.1016/j.chemosphere.2019.03.074
Farka Z, Mickert MJ, Hlavacek A, Skladal P, Gorris HH (2017) Single molecule upconversion-linked immunosorbent assay with extended dynamic range for the sensitive detection of diagnostic biomarkers. Anal Chem 89(21):11825–11830. https://doi.org/10.1021/acs.analchem.7b03542
Hlavacek A, Farka Z, Hubner M, Hornakova V, Nemecek D, Niessner R, Skladal P, Knopp D, Gorris HH (2016) Competitive upconversion-linked immunosorbent assay for the sensitive detection of diclofenac. Anal Chem 88(11):6011–6017. https://doi.org/10.1021/acs.analchem.6b01083
Skarkova P, Novotny K, Lubal P, Jebava A, Porizka P, Klus J, Farka Z, Hrdlicka A, Kaiser J (2017) 2d distribution mapping of quantum dots injected onto filtration paper by laser-induced breakdown spectroscopy. Spectrochim Acta B At Spectrosc 131:107–114. https://doi.org/10.1016/j.sab.2017.03.016
Dukhno O, Przybilla F, Muhr V, Buchner M, Hirsch T, Mely Y (2018) Time-dependent luminescence loss for individual upconversion nanoparticles upon dilution in aqueous solution. Nanoscale 10(34):15904–15910. https://doi.org/10.1039/c8nr03892a
Cid-Barrio L, Calderon-Celis F, Abasolo-Linares P, Fernandez-Sanchez ML, Costa-Fernandez JM, Encinar JR, Sanz-Meder A (2018) Advances in absolute protein quantification and quantitative protein mapping using ICP-MS. Trac Trends Anal Chem 104:148–159. https://doi.org/10.1016/j.trac.2017.09.024
Malile B, Brkic J, Bouzekri A, Wilson DJ, Ornatsky O, Peng C, Chen JIL (2019) DNA-conjugated gold nanoparticles as high-mass probes in imaging mass cytometry. ACS Appl Bio Mater 2(10):4316–4323. https://doi.org/10.1021/acsabm.9b00574
Lores-Padin A, Menero-Valdes P, Fernandez B, Pereiro R (2020) Nanoparticles as labels of specific-recognition reactions for the determination of biomolecules by inductively coupled plasma-mass spectrometry. Anal Chim Acta 1128:251–268. https://doi.org/10.1016/j.aca.2020.07.008
Acknowledgements
We thank Antonín Hlaváček for the synthesis of UCNPs and Matthias J. Mickert for the help with the optimization of the ICC experiments.
Funding
This work was financially supported by the Ministry of Education, Youth and Sports of the Czech Republic (MEYS CR) under the projects CEITEC 2020 (LQ1601), INTER-ACTION (LTAB19011), and by the CzechNanoLab Research Infrastructure supported by MEYS CR (LM2018110). We acknowledge CF Nanobiotechnology of CIISB, Instruct-CZ Centre, supported by MEYS CR (LM2018127). PP and PM gratefully acknowledge the financial support by the Czech Science Foundation within the project 20-19526Y. KV, IG, and KN acknowledge the support by the project 1390/2020 Analytical and physical chemistry in the research of biological, geological, and synthetic materials (BIOGEOSYNT). MP and ZF gratefully acknowledge the financial support by the Czech Science Foundation within the project GJ20-30004Y.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Pořízka, P., Vytisková, K., Obořilová, R. et al. Laser-induced breakdown spectroscopy as a readout method for immunocytochemistry with upconversion nanoparticles. Microchim Acta 188, 147 (2021). https://doi.org/10.1007/s00604-021-04816-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-021-04816-y