Abstract
MicroRNAs (miRNAs) play a crucial role in the pathogenesis of type 2 diabetes (T2D); they regulate several metabolic pathways including insulin secretion, glucose homeostasis, so their potential as biomarkers of diagnosis and prognosis has became increasingly appreciated. In this study, we explore serum miRNA profiles in T2D patients. A total of ten candidate miRNAs were identified by Solexa sequencing scanning and followed by a stem-loop quantitative reverse transcription PCR (qRT-PCR) to assess these candidate serum miRNAs. The results of qRT-PCR assessment revealed low serum levels of miR-23a, let-7i, miR-486, miR-96, miR-186, miR-191, miR-192, and miR-146a in T2D. Except for significantly lower in T2D and pre-diabetes patients compared with normal glucose tolerance (NGT) controls (P = 2.87E−05 and P = 3.75E−02), the levels of miR-23a demonstrated also significant decline in T2D patients compared with pre-diabetes patients (P = 1.06E−02). This marker yielded an AUC of 0.835 (95 % CI 0.717–0.954). At a cutoff value of 1.645, the sensitivity was 79.2 % and the specificity was 75.0 % in discriminating T2D patients from NGT normal controls. These results revealed that serum miR-23a was a valuable biomarker for early detection of T2D and pre-diabetes with NGT.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Type 2 diabetes mellitus (T2D) has reached epidemic levels worldwide; it is a metabolic disorder that is characterized by hyperglycemia in the context of reduced insulin sensitivity and insulin resistance. T2D accounts for about 90 % of all diabetes cases. It will be affecting 285 million adults in 2010 and will increase to 439 million adults by 2030, the global diabetes epidemic continues to grow [29]. Recently, several surveys data indicate that previous modeled estimates have generally been under-estimating the prevalence of diabetes, especially based solely on fasting blood glucose, many cases of diabetes easily be missed [33, 36]. Although the oral glucose tolerance test (OGTT) is the gold standard for the diagnosis of diabetes, because the detection process more complicated, some patients will not be adopted, which will cause a certain degree of misdiagnosis.
MicroRNAs (miRNAs) are small noncoding RNAs, approximately 21–23 nucleotides in length, that modulate gene expression by suppressing translation and/or reducing the stability of their target mRNAs [19, 21, 34]. By influencing protein translation, miRNAs have emerged as powerful regulators of crucial cell processes such as proliferation, differentiation, and apoptosis. miRNAs play a crucial role in the pathogenesis of many diseases, including a variety of cancers, cardiovascular disease, and other chronic diseases [6, 9, 30]. Furthermore, miRNAs have been reported to regulate several metabolic pathways including insulin secretion, glucose homeostasis, and carbohydrate and lipid metabolism [7, 24, 28]. As translational repressors, miRNAs enable us to better understanding of gene expression regulatory mechanisms during the pathogenesis and development of T2D; it will have potential applications in its prevention, early diagnosis and treatment.
Recently, it was found that miRNAs are also stable present in blood, where they detected in plasma, serum, platelets, erythrocytes, and nucleated cells [5, 18, 23]. Plasma or serum miRNAs were found to be remarkably stable even under harsh conditions, such as boiling, low or high pH, long-time storage at room temperature, and multiple freeze–thaw cycles [5, 23], which makes peripheral circulating miRNAs potentially novel sources of noninvasive biomarkers of cancers and other diseases [3, 6, 14, 15].
In this study, we employed high-throughput Solexa (Illumina) sequencing scanning and followed by a stem-loop quantitative reverse transcription PCR (qRT-PCR) to assay systematically and extensively assess the serum miRNAs expression profiles of T2D and pre-diabetes, then to access the potential of serum miRNAs as noninvasive biomarker for diagnosis and prognosis of T2D and pre-diabetes, and to related these finding to the pathogenesis of T2D.
Materials and methods
Research subjects and design
All research subjects were Han Chinese individuals who received OGTT and other medical health checkups in Diabetes Specialties centre, Department of Endocrinology, Zhejiang Provincial Hospital of TCM in Hangzhou, China. This study comprised of three groups (n = 20 in each category), grouping criteria were based on the plasma glucose criteria by the fasting plasma glucose test (FPG; blood samples were drawn after 12 h of fasting) and OGTT test.
Subjects who were confirmed by OGTT to have FPG ≥ 7.0 mmol/l (126 mg/dl) and/or 2 h plasma glucose value (2hPG) ≥ 11.1 mmol/l (200 mg/dl) based on WHO consulting group criteria [1] were diagnosed as T2D patients. Subjects with impaired fasting glucose (IFG) (FPG 5.6–6.9 mmol/l (100–125 mg/dl) and 2hPG < 7.8 mmol/l (140 mg/dl), and impaired glucose tolerance (IGT) [(FPG < 5.6 mmol/l (100 mg/dl) and 2hPG 7.8–11.0 mmol/l (140–199 mg/dl)], and IFG + IGT [FPG 5.6–6.9 mmol/l (100–125 mg/dl) and 2hPG 7.8–11.0 mmol/l (140–199 mg/dl)] were all considered as diagnosed as pre-diabetes patients. Subjects with FPG < 5.6 mmol/l (100 mg/dl) and 2-h post-glucose value <7.8 mmol/l (140 mg/dl) were considered as normal glucose tolerance (NGT) individuals.
Sample preparation
Peripheral blood was venous blood and then store at room temperature for 1 h. Following centrifugation 5 min 2,000×g, serum was collected and centrifuged again at 12,000g for 15 min to completely remove cell debris. The serum was then aliquoted and stored separately at −80 °C until used for analysis.
Five T2D patients and five NGT individuals serum samples were selected separately for Solexa sequencing Screening (Table 1). Twenty-four T2D patients, twenty pre-diabetes (IFG/IGT) patients, and twenty NGT individuals serum samples were recruited separately for quantitative real-time RT-PCR validation (Table 2).
Solexa deep sequencing
The T2D and NGT groups serum samples were melted and mixed together, respectively, before proceeding to RNA isolation. A small RNA library was generated from the serum samples using the Illumina Truseq Small RNA Preparation kit (Illumina, San Diego, USA) according to Illumina’s TruSeq Small RNA Sample Preparation Guide by LC Sciences (Hangzhou, China & Houston, USA, www.lcsciences.com).
The purified cDNA library was used for cluster generation on Illumina’s Cluster Station and then sequenced on Illumina GAIIx following vendor’s instruction for running the instrument. Raw sequencing reads were obtained using Illumina’s Sequencing Control Studio software version 2.8 (SCS v2.8) following real-time sequencing image analysis and base calling by Illumina’s real-time analysis version 1.8.70 (RTA v1.8.70). Then, a proprietary pipeline script, ACGT101-miR v4.2 (LC Sciences, USA), was used for sequencing data analysis. Library construction and sequencing were performed at LC Sciences (Hangzhou, China & Houston, USA, www.lcsciences.com).
Data processing
A proprietary pipeline script, ACGT101-miR v4.2 (LC Sciences, Houston, Texas, USA) [20, 32], was used for sequencing data analysis. Briefly, after the raw sequence reads or sequenced sequences (sequ seqs) were extracted from image data; a series of digital filters (LC Sciences) were employed to remove various un-mappable sequencing reads. Then, various “mappings” were performed on unique sequences against pre-miRNA (mir) and mature miRNA sequences listed in the miRBase 18.0 [11–13], or genome based on the public releases of appropriate species. Mappings were also done on mirs of interest against genome sequence.
Quantitative real-time RT-PCR
To validate the Solexa sequencing data, we used a previously described miRNA quantification method [4]. RNA samples used in qPCR validation experiments were isolated from 24 T2D patients, 20 pre-diabetes (IFG/IGT) patients, and 20 NGT individuals serum samples (Table S2). Primer Premier 5.0 and DNAStar. Lasergene. v7.1 software were used to designed the primer sequences (Table S3).
Extracted total RNA from 200 μl of serum using TRK-1001 (LC-Bio, USA) according to the protocol, then eluting with 50-μl elution solution. 5 μl of serum RNA containing miRNA was reverse transcribed to cDNA, reverse transcription solution system includes 5 μl total RNA, 1 μl 5 × M-MLV Buffer, 1 μl M-MLV, 0.5 μl dNTP, and 0.5 μl RRI (Reverse Transcriptase M-MLV (RNase H-), TAKARA, CHINA). Reverse transcription reaction in PCR instrument (MyGene L96G, LongGene, CHINA) 42 °C 1 h. These cDNAs were stored at −20 °C in refrigerator.
Quantitative real-time RT-PCR (qRT-PCR) solution system including 10 μl SYBR Green I, 8.2 μL DDH2O, 0.8 μL primer mix, and 1 μl RT product. The thermal cycle start with 2 min at 50 °C, 2 min template denaturation at 95 °C, 15 s denaturation at 95 °C, and 30 s elongation at 60 °C, 40 cycles in total. Real-time PCR was performed using Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen: 11733-038). RNU6B stable stay in serum and change less in different people was used as control gene according to the Applied Biosystems Application Note [35], and it has been widely used in different fields including diabetic research [8, 17, 22, 25, 27], so it was selected as an internal control gene in this work. And all reaction was run in triplicate.
Statistical analysis
The ΔΔC T method was used to determine the differences in expression between groups [31]. Relative expression of miRNA was calculated using the fold change of miRNA expression method. The fold change of miRNA expression in each DM serum sample [including T2D and pre-diabetes (IFG/IGT) patients] relative to the average expression in normal control (NGT) was calculated based on the threshold cycle (CT) value using the following formula: \( {\text{RQ}} = 2^{{ - \varDelta \varDelta C_{\text{T}} }} \), where, \( \varDelta \varDelta C_{T} = \left( {C_{{{\text{T}}_{\text{miRNA}} }} - C_{{{\text{T}}_{\text{RNU6B}} }} } \right){\text{DM}} - \left( {C_{{{\text{T}}_{\text{miRNA}} }} - C_{{{\text{T}}_{\text{RNU6B}} }} } \right){\text{Mean}}_{\text{NGT}} \). One-way ANOVA was also employed to compare the miRNA expression among the three groups of subjects. To carry out an agglomerative hierarchical cluster analysis of the subjects based on miRNAs expression, we used a between groups linkage algorithm and a squared Euclidean distance measure. For each miRNA, we constructed the receiver operating characteristic (ROC) curve and calculated the area under the ROC curve (AUC) to evaluate the predictive power for T2D and pre-diabetes.
Risk score analysis was performed to evaluate the associations between the concentrations of the serum miRNAs and T2D or pre-diabetes. The risk score of each miRNA, denoted as s, was set to 1 if the expression level was greater than the upper 95 % reference interval for the corresponding miRNA level in controls and to 0 otherwise.
Statistic analysis was performed by SPSS Statistics.v19.0.0 (SPSS, Inc., Chicago, USA). All statistical tests were two-sided. The statistical significance was defined as P < 0.05.
Results
Patient description
All five T2D patients and five NGT individuals enrolled for Solexa sequencing in this study were clinically and pathologically diagnosed with OGTT test, HbAlc, etc. Clinical information about this research subjects is presented in Table 1. There were no significant differences in the distribution of age (P = 0.317), TC (P = 0.728), TG (P = 0.101), LDL (P = 0.510) between T2D and the NGT controls, but HDL did show significantly lower stepwise trends from NGT to T2D (P = 0.021).
A total of 64 participants including 24 T2D patients, 20 pre-diabetes patients, and 20 NGT individuals were recruited into quantitative real-time RT-PCR validation. Patient characteristics and clinicopathological parameters were summarized in Table 2. There were no significant differences of age (P = 0.201 and 0.204), BMI (P = 0.374 and 0.330), TC (P = 0.603 and 0.309), TG (P = 0.733 and 0.530), HDL (P = 0.146 and 0.285), LDL (P = 0.295 and 0.865) among T2D, pre-diabetes, and NGT three subject groups. For OGTT test results, 2hPG showed significantly higher stepwise trends from NGT to pre-diabetes and then T2D TG (P < 0.0001). For FPG, no significant difference was found between NGT and pre-diabetes (P = 0.558), but showed significantly higher stepwise trends from NGT and pre-diabetes to T2D (P < 0.0001).
Overview for deep sequencing
In the initial screening phase by Solexa deep sequencing, serum samples were pooled from five T2D patients or five NGT healthy donors (approximately 1.0 ml each). As shown in online Supplementary Table S1, a genome-wide expression profiling of serum miRNAs obtained by Solexa sequencing showed that serum from T2D patients had 61 upregulated miRNAs and 92 downregulated miRNAs compared with serum from NGT controls. miRNAs were considered differential level only if Solexa sequencing detected at least 50 copies in the control group and showed at least a twofold change between the two pooled samples. Among the miRNAs detected, 50 miRNAs were found to be differential levels in T2D sera compared with normal sera, of which 36 were downregulated and 36 were upregulated.
Validation by qRT-PCR
Subsequently, the significantly different miRNAs were validated in individual serum samples using qRT-PCR. As summarized in Table 2, there was no significant difference between the T2D, pre-diabetes patients, and the control individuals in age distribution, BMI, TC, TG, HDL, and LDL. Only miRNAs with above twofold mean change were selected from the training set for further validation. miRNAs with a CT value >35 and a detection rate <75 % in the patient or control group were excluded from further analysis. Findings were consistent for expression levels standardized to RNU6b. The thermal cycle was set to 40 cycles; threshold cycle (Ct) values of all ten miRNAs and RNU6B were presented (Fig. 1); in some assays, miRNAs remained undetectable after 40 cycles, the Ct values of which were calculated as 40, data shown as mean ± SEM. In this study, the mean of Cq value of RNU6B was 32.84 ± 0.64, CV% = 1.95 % (Table 3).
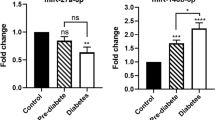
Using above criteria, Serum expression levels of the selected ten miRNAs were all downregulated to some extent among T2D and pre-diabetes patients. The levels of eight miRNAs, including miR-23a, let-7i, miR-486, miR-96, miR-186, miR-191, miR-192, and miR-146a were showed to be significantly decreased in T2D patients relative to normal NGT controls. And except for the levels of miR-23a, there were no other miRNA expressed significantly different between pre-diabetes patients to normal NGT controls (Fig. 2). The levels of miR-199a and miR-192 were observed at insignificantly decreased in T2D and pre-diabetes patients with normal NGT controls.
The levels of miR-23a were showed significantly down expression to some extent among T2D, pre-diabetes patients, and normal NGT controls (A). The levels of let-7i, miR-486, miR-96, miR-186, miR-191, miR-192, and miR-146a were significantly lower in T2D patients, but no significant association was found between pre-diabetes patients to normal NGT controls (B–I)
The levels of miR-23a were significantly lower in T2D and pre-diabetes patients compared with normal NGT controls (P = 2.87E−05 and P = 3.75E−02, Fig. 2a). Furthermore, the levels of miR-23a demonstrated significant decline in T2D patients with pre-diabetes patients (P = 1.06E−02, Fig. 2a).
Then, we used the ROC curve analysis to determine the usefulness of the selected miRNAs for detecting the T2D and pre-diabetes. In the analysis of T2D patients and NGT control groups, yielded the following AUCs results (Fig. 3a–c): the AUC was 0.835 for miR-23a [95 % confidence interval (CI) 0.717–0.954]; at the cutoff value of 1.645, the optimal sensitivity and specificity were 79.2 and 75.0 %, respectively, (Fig. 3a). The AUC was 0.771 for let-7i (95 % CI 0.629–0.913); at the cutoff value of 1.456, the optimal sensitivity and specificity were 75.0 and 70.0 %, respectively (Fig. 3b). The AUC was 0.698 for miR-486 (95 % CI 0.540–0.856); at the cutoff value of 0.920, the optimal sensitivity and specificity were 79.2 and 60.0 %, respectively (Fig. 3c). In the analysis of pre-diabetes and NGT control groups, the AUC was 0.690 for miR-23a (95 % CI 0.525–0.855); at the cutoff value of 1.320, the optimal sensitivity and specificity were 70.0 and 60.0 %, respectively (Fig. 3d). These results revealed that serum miR-23a was a valuable biomarker for differentiating T2D and pre-diabetes from NGT normal controls. For patients with pre-diabetes may be need further screening diagnosed in combination with other diagnostic methods.
Discussion
In recent years, serum or plasma miRNA has strong potential as a novel noninvasive biomarker for early detection of cancer, metabolic syndrome, cardiovascular diseases, and other chronic disease [3, 6, 10, 16, 18]. Results from recent studies revealed that circulating miRNAs are also potential diagnostic biomarkers and prognostic factors in diabetes [2, 17, 37]. For example, Zampetaki and colleagues explore plasma miRNA profiles in patients with DM; they revealed lower plasma levels of miR-20b, miR-21, miR-24, miR-15a, miR-126, miR-191, miR-197, miR-223, miR-320, and miR-486 in prevalent DM, but a modest increase of miR-28-3p [37]. Kone et al. explored the clinical significance of seven diabetes-related serum miRNAs during the pathogenesis of T2D; they found all 7 miRNAs which including miR-9, miR-29a, miR-30d, miR-34a, miR-124a, miR-146a, and miR-375 were significantly upregulated in n-T2D compared with s-NGT, and miR-9, miR-29a, miR-34a, miR-146a, and miR-375 were significantly upregulated compared with pre-diabetes, while miRNA expression was not significantly different between s-NGT and pre-diabetes [17]. Balasubramanyam et al. [2] revealed the miR-146a expression levels were significantly decreased in PBMCs from patients with T2D compared with control subjects, and they found miR-146a expression was negatively correlated with subclinical inflammation and insulin resistance.
By performing an Solexa sequencing scanning followed by individual qRT-PCR evaluations, we identified eight serum miRNAs, including miR-23a, let-7i, miR-486, miR-96, miR-186, miR-191, miR-192, and miR-146a, whose expression levels were significantly decreased in the serum of T2D patients compared with NGT normals controls. But as the results of T2D plasma miRNA profiles exploration by Zampetaki, except for miR-191 downregulated significantly, miR-186, miR-192, miR-96, and miR-146a expression were all no significantly different between T2D and s-NGT. Whether this results mean that the presence of fibrinogen or other substances in plasma will have some interference to the extraction and analysis of miRNA, further research maybe needs to confirmed it.
Several study related to T2D plasma or serum miRNA was all not related to miR-23a [2, 17, 37]. Karolina et al. [16] identified miR-23a as potential contributors of dyslipidemia in metabolic syndrome, but without further study of miR-23a between the T2D and pre-diabetes patients. Raychaudhuri et al. [26] revealed miR-23a targets SMAD4 indirectly that seem to be critical in regulating insulin-dependent glucose transport activity. In our study, we demonstrate that miR-23a was significantly lower in serum of T2D and pre-diabetes patients compared with NGT controls; serum miR-23a levels can also distinguish T2D from pre-diabetes.
In conclusion, miR-23a appears to be more potential novel biomarker for early diagnose of pre-diabetes and T2D in all the selected serum miRNAs. Serum miR-23 could be used as an objective complement to traditional diagnosis of pre-diabetes or T2D. In particular, it could be used to discriminate the undiagnosed cases of diabetes and pre-diabetes which easily be underestimated only by fasting glucose levels tests. The serum miRNA markers may improve the sensitivity and specificity of the assay for T2D and pre-diabetes screening. Maybe in the future preferable in large cases, prospective studies will be used to validate its clinical utility (individually or as part of a panel) as a novel noninvasive, economic screening tool for diabetes.
References
Alberti KGMM, Zimmet P (1998) Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus. Provisional report of a WHO consultation. Diabet Med 15:539–553
Balasubramanyam M, Aravind S, Gokulakrishnan K, Prabu P, Sathishkumar C, Ranjani H, Mohan V (2011) Impaired miR-146a expression links subclinical inflammation and insulin resistance in Type 2 diabetes. Mol Cell Biochem 351:197–205
Brase JC, Wuttig D, Kuner R, Sultmann H (2010) Serum microRNAs as non-invasive biomarkers for cancer. Mol Cancer 9:306
Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR (2005) Real-time quantification of microRNAs by stem–loop RT–PCR. Nucleic Acids Res 33:e179–e179
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X (2008) Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 18:997–1006
Creemers EE, Tijsen AJ, Pinto YM (2012) Circulating microRNAs novel biomarkers and extracellular communicators in cardiovascular disease? Circ Res 110:483–495
El Ouaamari A, Baroukh N, Martens GA, Lebrun P, Pipeleers D, Van Obberghen E (2008) miR-375 targets 3’-phosphoinositide-dependent protein kinase-1 and regulates glucose-induced biological responses in pancreatic β-cells. Diabetes 57:2708–2717
Feng B, Chen S, George B, Feng Q, Chakrabarti S (2010) miR133a regulates cardiomyocyte hypertrophy in diabetes. Diabetes/Metab Res Rev 26:40–49
Garzon R, Calin GA, Croce CM (2009) MicroRNAs in cancer. Annu Rev Med 60:167–179
Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N, Benjamin H, Kushnir M, Cholakh H, Melamed N (2008) Serum microRNAs are promising novel biomarkers. PLoS ONE 3:e3148
Griffiths-Jones S (2004) The microRNA registry. Nucleic Acids Res 32:D109–D111
Griffiths-Jones S, Grocock RJ, Van Dongen S, Bateman A, Enright AJ (2006) miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34:D140–D144
Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ (2008) miRBase: tools for microRNA genomics. Nucleic Acids Res 36:D154–D158
Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X (2010) Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer 127:118–126
Jackson DB (2009) Serum-based microRNAs: are we blinded by potential? PNAS 106:E5
Karolina DS, Tavintharan S, Armugam A, Sepramaniam S, Pek SLT, Wong MT, Lim SC, Sum CF, Jeyaseelan K (2012) Circulating miRNA profiles in patients with metabolic syndrome. J Clin Endocrinol Metab 97:E2271–E2276
Kong L, Zhu J, Han W, Jiang X, Xu M, Zhao Y, Dong Q, Pang Z, Guan Q, Gao L (2011) Significance of serum microRNAs in pre-diabetes and newly diagnosed type 2 diabetes: a clinical study. Acta Diabetol 48:61–69
Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, Banham AH, Pezzella F, Boultwood J, Wainscoat JS (2008) Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol 141:672–675
Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75:843–854
Li M, Xia Y, Gu Y, Zhang K, Lang Q, Chen L, Guan J, Luo Z, Chen H, Li Y (2010) MicroRNAome of porcine pre-and postnatal development. PLoS ONE 5:e11541
Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM (2005) Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433:769–773
Lovis P, Roggli E, Laybutt DR, Gattesco S, Yang J-Y, Widmann C, Abderrahmani A, Regazzi R (2008) Alterations in microRNA expression contribute to fatty acid–induced pancreatic β-cell dysfunction. Diabetes 57:2728–2736
Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A (2008) Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci 105:10513–10518
Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, MacDonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P (2004) A pancreatic islet-specific microRNA regulates insulin secretion. Nature 432:226–230
Poy MN, Hausser J, Trajkovski M, Braun M, Collins S, Rorsman P, Zavolan M, Stoffel M (2009) miR-375 maintains normal pancreatic α-and β-cell mass. Proc Natl Acad Sci 106:5813–5818
Raychaudhuri S (2012) MicroRNAs overexpressed in growth-restricted rat skeletal muscles regulate the glucose transport in cell culture targeting central TGF-β factor SMAD4. PLoS ONE 7:e34596
Roggli E, Britan A, Gattesco S, Lin-Marq N, Abderrahmani A, Meda P, Regazzi R (2010) Involvement of microRNAs in the cytotoxic effects exerted by proinflammatory cytokines on pancreatic β-cells. Diabetes 59:978–986
Rottiers V, Näär AM (2012) MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol 13:239–250
Shaw J, Sicree R, Zimmet P (2010) Global estimates of the prevalence of diabetes for 2010 and 2030. Diabetes Res Clin Pract 87:4–14
Ventura A, Jacks T (2009) MicroRNAs and cancer: short RNAs go a long way. Cell 136:586–591
Wang F, Zheng Z, Guo J, Ding X (2010) Correlation and quantitation of microRNA aberrant expression in tissues and sera from patients with breast tumor. Gynecol Oncol 119:586–593
Wei Z, Liu X, Feng T, Chang Y (2011) Novel and conserved micrornas in Dalian purple urchin (Strongylocentrotus nudus) identified by next generation sequencing. Int J Biol Sci 7:180
Whiting DR, Guariguata L, Weil C, Shaw J (2011) IDF diabetes atlas: global estimates of the prevalence of diabetes for 2011 and 2030. Diabetes Res Clin Pract 94:311–321
Wightman B, Ha I, Ruvkun G (1993) Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75:855–862
Wong L, Lee K, Russell I, Chen C (2007) Endogenous controls for real-time quantitation of miRNA using TaqMan® microRNA assays. Macmillan Publishers Limited, New York
Yang W, Lu J, Weng J, Jia W, Ji L, Xiao J, Shan Z, Liu J, Tian H, Ji Q (2010) Prevalence of diabetes among men and women in China. N Engl J Med 362:1090–1101
Zampetaki A, Kiechl S, Drozdov I, Willeit P, Mayr U, Prokopi M, Mayr A, Weger S, Oberhollenzer F, Bonora E (2010) Plasma MicroRNA profiling reveals loss of endothelial MiR-126 and other MicroRNAs in type 2 diabetes novelty and significance. Circ Res 107:810–817
Acknowledgments
This work was supported by Science and Technology Development Program of Hangzhou City (20110733Q21), Zhejiang Provincial Top Key Discipline of Biology, and Zhejiang Province Undergraduate Scientific and Technological Innovation Project (2011R406029). We also thanks to Dr. Ye Sende, Prevention and Health Care Outpatient Department of Hangzhou City for his help to collection of serum samples of T2D and normal subjects.
Conflict of interest
All the authors including Zhangping Yang, Haimin chen, Hongqiang si, Xuan Li, Xianfeng Ding, Qing Sheng, Ping Chen, and Hongqiang Zhang declare that they have no conflict of interest.
Ethical Standard
All human subjects used in the study “Serum miR-23a, a potential biomarker for diagnosis of pre-diabetes and type 2 diabetes” have been reviewed by the Research Ethics Committee, Hangzhou Center for Disease Control and Prevention, Hangzhou, Zhejiang Province, P. R. China and have been performed in accordance with the ethical standards laid down in an appropriate version of the 1964 Declaration of Helsinki. All samples were collected with informed consent of the patients. Ethics Committee specifically approved that not informed consent was required because data were going to be analyzed anonymously. Furthermore, there is no security and privacy violation to the patient's health in our study.
Human and Animal Rights
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Declaration of Helsinki 1975, as revised in 2008 (5).
Informed consent
Informed consent was obtained from all patients for being included in the study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Managed by Massimo Porta.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yang, Z., Chen, H., Si, H. et al. Serum miR-23a, a potential biomarker for diagnosis of pre-diabetes and type 2 diabetes. Acta Diabetol 51, 823–831 (2014). https://doi.org/10.1007/s00592-014-0617-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00592-014-0617-8