Abstract
A study was conducted to establish whether the wild thyme [Thymus polytrichus A. Kerner ex Borbás ssp. britannicus (Ronn.) Kerguelen (Lamiaceae)] growing in the metal-contaminated soils along the River South Tyne, United Kingdom, is colonised by arbuscular mycorrhizal (AM) fungi, and whether the degree of colonisation increases (perhaps suggesting increasing mycorrhizal dependence) or decreases (indicating possible inhibition of AM growth) with increasing degree of soil contamination. Seasonal changes in AM colonisation were also assessed. The AM fungal communities colonising T. polytrichus were also investigated, using the polymerase chain reaction with restriction fragment length polymorphism and sequencing of fungal DNA to establish whether AM species richness varied between sites, and whether fungal ecotypes specific to sites with different amounts of metal contamination could be identified. All plants examined were heavily colonised by AM fungi, and mean percentage root length colonised did not increase significantly with increasing soil metal contamination. However, AM vesicle abundance (percentage of mycorrhizal root length containing vesicles) at the most contaminated site was significantly greater than at the other sites. No significant seasonal variation in degree of colonisation or vesicle abundance was found. Glomus was the predominant AM genus detected at all sites. The number of AM genotypes colonising T. polytrichus roots was similar at all sites but, although some were common to all sites, certain strains appeared to be specific to either the most- or the least-contaminated site. This variation in species may account for the difference in vesicle abundance between sites. The consistently heavy AM colonisation of T. polytrichus found suggests that these fungi are not inhibited by soil heavy metals at these sites, and that the host derives some benefit from its AM symbiont.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
There is considerable evidence that colonisation by arbuscular mycorrhizal (AM) fungi can reduce plant uptake and/or phytotoxic effects of soil heavy metals (e.g. Gildon and Tinker 1983; Dehn and Schüepp 1989; Hetrick et al. 1994; Hildebrandt et al. 1999). However, mycorrhizal colonisation represents an energetic cost to the host in the form of carbon supplied to the mycobiont (Douds et al. 2000). It has therefore been suggested (e.g. Sanders and Fitter 1992a) that selective pressure would act to reduce colonisation unless the symbiosis provided some benefit to the host. Thus, in situations where mycorrhizal colonisation protects plants against heavy metal toxicity, colonisation may be positively correlated with plant-available metal concentrations.
There is also evidence, however, that heavy metals can inhibit mycorrhizal spore germination (Hepper and Smith 1976), growth of extraradical hyphae (del Val et al. 1999), and AM colonisation (Gildon and Tinker 1983), and that metal tolerance of mycorrhizal fungi differs between fungal species and ecotypes (del Val et al. 1999; Liao et al. 2003). As a consequence, both fungal spore density and species richness (Shetty et al. 1995) have been shown to vary with soil metal concentration, although most field studies of these phenomena have compared sites that also differ in other ways, for example, pH, organic matter content, and level of fertilisation—all factors that affect mycorrhizal growth and colonisation (Leyval et al. 1997).
The shingle banks along the Rivers Tyne and South Tyne in northern England tend to be heavily contaminated with Pb, Zn and other heavy metals, the result of centuries of mining of the local orefields (Raistrick and Jennings 1965), yet wild thyme [Thymus polytrichus A. Kerner ex Borbás ssp. britannicus (Ronn.) Kerguelen (Lamiaceae)] grows in abundance on these shingles, along with metallophytes such as Armeria maritima and Minuartia verna. A recent study (Whitfield 2002) demonstrated that these plants have a greater Zn tolerance than plants from populations growing at uncontaminated sites, and that offspring from the former grown in uncontaminated soil also had a high tolerance. High Zn tolerance in this subspecies does not, therefore, appear to be constitutive; rather, tolerant ecotypes have developed at these sites. However, although colonisation of T. polytrichus by AM fungi has been reported briefly (Grime et al. 1988), whether AM colonisation varies with soil metal concentrations, or reduces plant metal concentrations, in these populations is unknown.
The first objective of the present study, therefore, was to establish whether the T. polytrichus plants growing in the heavy-metal-contaminated soils along the River South Tyne are mycorrhizal, and whether the degree of AM colonisation increases (suggesting increasing mycorrhizal dependence) or decreases (indicating possible inhibition of AM fungi) with increasing soil contamination. The second objective was to assess the range of AM fungal species colonising T. polytrichus at each site, to establish whether AM species richness varies between sites, and whether fungal ecotypes specific to sites with different metal contamination levels could be identified.
Since it is not possible to identify AM fungi to species from their intraradical hyphal morphology (Gerdemann 1965), identification has traditionally been based on the morphology of the resting spores found in the rhizosphere. However, there is some evidence that fungal species diversity based on spore frequencies may not be well correlated with the species mix actively colonising plant roots (Clapp et al. 1995; Turnau et al. 2001); increasingly, therefore, molecular genetic methods are being used to characterise the AM fungal DNA within the roots. In this study a section of the 18S small subunit (SSU) ribosomal DNA (rDNA) was isolated from fungal DNA in root samples using specific primers, amplified using the polymerase chain reaction (PCR), and analysed using restriction fragment length polymorphism (RFLP) and DNA sequencing.
Materials and methods
Assessment of percentage root length colonised
Root and soil samples (15 cm ×15 cm ×10 cm deep) were collected from 15 mats of T. polytrichus at each of three sites along the River South Tyne (see Table 1 for details of sites and collection dates). The mats were at least 2 m apart and separated by areas with no visible thyme shoots, and were therefore assumed to be different clones or genets. Analysis of the soil samples showed that these sites had significantly different soil heavy metal concentrations but were otherwise very similar, being alluvial deposits with a near-neutral pH, and small concentrations of extractable P and organic matter (Table 1). Because some studies have shown mycorrhizal colonisation rates to fluctuate seasonally (Ietswaart et al. 1992; Sanders and Fitter 1992b), and because the above samples were collected at different times of year, root samples were collected from the same part of eight of the clones at two sites on 21 May, 26 July and 1 October 2000, and assessed for temporal changes in AM colonisation.
Samples of fine roots were washed and preserved by storing in 50% ethanol at 5°C. The roots were cleared with 5% KOH and hot-stained with acid fuchsin, then cut into 1 cm sections, and 20–30 randomly selected sections analysed by the line intersection method under ×200 magnification (McGonigle et al. 1990). Percentage root length colonised by AM hyphae (counting only those connected to an identifiable mycorrhizal structure), as well as percentage of mycorrhizal root length containing vesicles (vesicle abundance) and arbuscules, was assessed based on 150 intersections per sample.
Assessment of AM fungal species diversity
Fresh root samples were collected from three T. polytrichus clones at the most (site M4) and least contaminated (site M1) of the above sites on 2 June 2001, washed with tap water and distilled water, dried overnight at 70°C, stored in a desiccator and analysed within a few days. The other site (site M3) could not be accessed owing to foot and mouth disease restrictions, so two samples collected and dried the previous year had to be used for molecular analysis.
Subsamples of root were placed in a glass homogeniser with liquid nitrogen and ground, then ground with 500 μl hexadecyltrimethylammonium bromide (CTAB) buffer, and transferred to an Eppendorf tube, rinsing out the residue with a further 300 μl buffer. The mixture was incubated at 65°C for 1 h, and DNA extracted by addition of an equal volume of phenol/chloroform, followed by centrifugation, then addition of an equal volume of chloroform to the aqueous layer and further centrifugation. The DNA was precipitated with 1.5 volumes of isopropanol, incubated on ice for 15 min, centrifuged, and the dried pellet resuspended in 100 μl H2O, then purified using a BRL Life Technologies Concert purification column (Gibco-BRL, Paisley, UK).
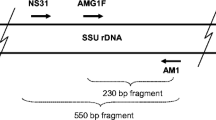
Primary PCR was performed using the universal eukaryotic primer NS31 (Simon et al. 1992) and the general fungal primer AM1 (Helgason et al. 1998) to isolate a section of the 18S SSU rDNA (ca. 550 bp) of any fungi present (Helgason et al. 1999), with amplification using Pfu DNA polymerase (Stratagene, La Jolla, Calif.). The reaction was performed with 0.2 mM dNTPs,10 pmol of each primer, and the manufacturer’s reaction buffer on a PTC-100 thermal cycler (M.J. Research, Boston, Mass.). The PCR was carried out for 29 cycles (9 cycles at 94°C for 1 min, 58°C for 1 min and 72°C for 1.5 min, 19 cycles at 94°C for 30 s, 60°C for 1 min and 72°C for 1.5 min, and 1 cycle at 94°C for 30 s, 58°C for 1 min, and 72°C for 10 min).
Following verification by gel electrophoresis, the PCR products were cloned by ligation with the pBluescript vector (Stratagene), and transformation into Escherichia coli competent cells. The transformed cells were spread onto LB agar plates containing ampicillin, X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), and IPTG (isopropyl β-d-thiogalactoside) and incubated overnight at 37°C. Eight randomly selected colonies per sample were replica-plated, and used as the basis of a second PCR with the T3 and T7 universal primers to isolate and amplify individual cloned 18S sequences.
The secondary PCR products were digested with the restriction enzymes HinfI and HSP92-II (Promega, Madison, Wis.) according to the manufacturer’s instructions, and grouped according to the resulting fragment sizes, measured by eye against known size standards on an electrophoretic gel. A selection of the T3/T7-amplified products, chosen to represent most of the RFLP types found, were purified using Qiaquick PCR purification spin columns (Qiagen, Seattle, Wash.) and sequenced on a Perkin-Elmer ABI 310 Genetic Analyser (Perkin-Elmer, Boston, Mass.) according to the manufacturer’s instructions, using both T3 and T7 primers. As the sequences obtained (NS31-AM1 sequence plus flanking vector sequences) were about 650 bp long, and clear sequences could generally be obtained for the first 400–450 bases with each primer, it was possible to verify most of each sequence by checking against its reverse complement.
The sequences were checked visually against their chromatograms using ProSequence release 2.9 (Filatov 2001) and accurate RFLP fragment lengths calculated for each restriction enzyme digestion. Closest matches to each sequence were found in the National Center for Biotechnology Information (NCBI) GenBank database (http://www.ncbi.nlm.nih.gov/Genbank), and ClustalX version 1.81 (Thompson et al. 1997) was used to align the sequences along with reference sequences from the database. Phylogenetic relationships between the sequences were examined with a neighbour-joining method (Saitou and Nei 1987) using ClustalX, and displayed using TreeView version Win32 (Page 1996). Confidence levels for the resulting sequence groupings were assessed by bootstrap analysis using ClustalX (1,000 bootstrap iterations).
Statistics
Percentage mycorrhizal colonisation data were arcsine-transformed where necessary to fit the data to a normal distribution (assessed by the Kolmogorov-Smirnoff test). Variations in colonisation between sites were examined using one-way analysis of variance (ANOVA) with least significant difference (LSD) tests, or by Student’s t-test if variances could not be homogenised. Seasonal colonisation data were analysed by repeated-measures ANOVA. Significance of correlations between soil metal concentrations and mycorrhizal colonisation were tested using linear regression analysis. All analyses were performed using the SPSS statistical package, release 10 (SPSS, Statsoft, Cary, N.C.).
Results
Mycorrhizal colonisation
All of the root samples examined were found to be colonised by AM fungi, and mycorrhizal vesicles were generally abundant, but although arbuscules were seen in most samples, the numbers counted were low and very variable (means for sites M1, M3, M4: 4.7%, 8.8%, 2.9% of colonised root length; not significant, ANOVA). This was felt to be because the roots were difficult to clear, and frequently coated with a brown fungus (probably Rhizoctonia spp.), so that although the hyphae and vesicles could be seen, the finer arbuscules were often obscured. Arbuscule numbers were therefore probably underestimated, and are not analysed in any more detail below.
There was no significant seasonal variation in mean percentage root length colonised with AM fungi at either site M1 or site M3 (Fig. 1). The mean vesicle abundance at site M3 showed a small decrease in October, but there was no significant seasonal variation at either site (Fig. 2). In addition, comparison of the root samples from site M4 collected on 16 August and 22 September 1999 showed no significant difference in vesicle abundance (means 52% and 59% respectively; P=0.32; t-test). To minimise any possible effect of seasonal variation, however, the samples collected from sites M1 and M3 in July 2000 were used for the main analysis (below).
Seasonal variation in percentage of Thymus polytrichus root length colonised by arbuscular mycorrhizal (AM) fungi at two South Tyne sites. a Site M1, b site M3 (mean±SE). Repeated-measures ANOVA showed no significant difference between sampling periods (site M1: F 2,12=0.32, P=0.73; site M3: F 2,14=0.55, P=0.59)
Seasonal variation in percentage of mycorrhizal root length containing vesicles in T. polytrichus roots from two South Tyne sites. a Site M1, b site M3 (mean±SE). Repeated-measures ANOVA showed no significant difference between sampling periods (site M1: F 2,12=0.21, P=0.82; site M3: F 2,14=3.11, P=0.08)
Mean percentage root length colonised did not differ significantly between sites (sites M1, M3, M4: 59.8%, 66.0%, 69.0%) (Fig. 3a). There was, however, significant variation in the mean vesicle abundance (sites M1, M3, M4: 44.8%, 43.3%, 56.8%; P<0.005, ANOVA), with no significant difference between sites M1 and M2, but a significantly greater vesicle abundance in roots from site M4 than in those from sites M1 (P<0.01) and M2 (P<0.005) (Fig. 3b).
a Percentage root length colonised by AM fungi in T. polytrichus samples from three South Tyne sites (mean±SE): there was no significant difference between sites (ANOVA: F 2,42=1.46, P=0.25). b Percentage of colonised root length containing vesicles (mean±SE), which varied significantly between sites (ANOVA: F 2,42=6.61, P<0.005). Significant between-site differences: M1 versus M4, P<0.01; M3 versus M4, P<0.005
Vesicle abundance showed positive relationships with extractable soil Cd and Zn concentrations, but only that with extractable Zn was significant (r 2=0.13; F 1,43=6.39; P=0.015).
Mycorrhizal DNA diversity
A total of 21 different RFLP types were found from visual inspection of the gel banding patterns from 60 cloned sequences (22 from site M1, 16 from site M2, and 22 from site M4). Full NS31-AM1 sequences (ca. 500 bp, excluding primers) representing different RFLP types were obtained for 16 samples (Table 2).
Accurate analysis of RFLP fragment sizes showed that the RFLP patterns originally typed as 2 and 10 were very similar, as were patterns for types 19 and 21, indicating that the differences in banding patterns on the electrophoresis gels were probably produced by variable flanking vector sequences, or insertion in the vector in opposite directions. Matching with sequences from GenBank confirmed that types 2 and 10 both seemed to be closely similar to Glomus sp. Glo4, and both clustered with this species on the neighbour-joining tree, although the sequences were not identical (Table 2; Fig. 4). However, although the type 19 and 21 sequences both clustered together with G. fasciculatum (Fig. 4), type 21 matched most closely with a different Glomus isolate (Glomus sp. Glo8; Table 2) according to the GenBank analysis.
Neighbour-joining tree showing relationships between NS31-AM1 rDNA sequences from T. polytrichus (see Table 2 for details of RFLP types and GenBank accession numbers) and reference sequences from GenBank, with Blastocladiella emersonii (ancestral to the Glomales) as the outgroup. All bootstrap values >70% are shown (1,000 replicates). Codes following reference isolates are GenBank accession numbers: see Table 2 for accession numbers of unlabelled isolates. Species in square brackets following reference isolates are the most closely related named species according to GenBank
Discussion
The present study appears to be the first to compare AM colonisation in a single wild host species, between sites with a long-term history of heavy metal contamination from the same source type, and with soils that differ in heavy metal content but are similar in origin (alluvial silts), pH, organic matter and available P concentration. The results bear out those of the above studies in that T. polytrichus at all three sites was well colonised by AM fungi, which would support the hypothesis that these fungi provide some benefit to their host, helping them to survive in these toxic soils.
The only significant between-site difference in colonisation was an apparent increase in vesicle abundance at site M4, the most contaminated site. This accords with the findings of Turnau et al. (1996), who noted greater AM vesicle numbers in the roots of Oxalis acetosella plants from woodland plots experimentally contaminated with Cd and Zn, compared with control plots. The vesicles produced by AM fungi are generally regarded as lipid storage structures (Gerdemann 1968; Mosse 1973), but their role—especially in relation to metal tolerance—has been little studied. One study investigating localisation of heavy metals and other elements in mycorrhizal tissues showed elevated concentrations of Zn associated with mycorrhizal hyphae, but the only elements found concentrated in vesicles were K, Fe and Ni (Kaldorf et al. 1999). However, in a microelement study of the grass Cynodon dactylon growing on gold and uranium mine tailings (Weiersbye et al. 1999), AM vesicles were found to accumulate Mn, Cu, Ni and U, leading the authors to suggest that sequestration of metals and radionuclides by the vesicles might reduce their toxicity to the host.
One other possibility that should be considered is that some of the “vesicles” counted in this study were intraradical spores, such as those produced by Glomus intraradices. These look much like, and may be formed from, vesicles (Gerdemann and Trappe 1974), and could be mistaken for these structures. Turnau (1998) reported large numbers of intraradical spores in E. cyparissias roots growing in Zn wastes, and suggested that since large metal concentrations are known to inhibit spore germination, this may be a fungal strategy to avoid exposure to toxic metals.
Whatever its functional significance, it is not known whether the greater vesicle concentration detected in roots from site M4 in the present study represented an increase in vesicle production by the same fungi, or an increased frequency of fungal species with a tendency to produce vesicles. However, the results of the DNA analyses, which show differences in the suites of fungal strains isolated from the most- and least-contaminated sites (M4 and M1), support the latter hypothesis. Moreover, in later experiments in which T. polytrichus plants were grown with AM inoculum from one of the field sites (Whitfield et al. 2003), vesicle abundance was just as great in mycorrhizas grown in uncontaminated soil as in those grown in Zn-spiked soil. This would suggest that the increased vesicle production seen in the field was related to differences in the AM species present, rather than a different response by the same fungi.
The results of the DNA analyses in the present study indicate that Glomus species predominate in the metal-contaminated alluvial sediments along the River South Tyne, but the suite of strains found in T. polytrichus roots differed between the most and the least contaminated site. RFLP types 1 and 5, which cluster most closely with the Glo1/G. mosseae/G. geosporum group, appeared to be the most common strains; these and types 2, 4 and 10 (Glo4 group) were found at all three sites, suggesting that these species can tolerate a range of heavy metal concentrations.
Some types, however, were found only at the least contaminated site (M1), possibly indicating more metal-sensitive strains: these too included types grouping with Glo1 (type 12) and Glo4 (types 3 and 6), but also types 7 and 11, which show the closest similarity to Glomus sp. Glo10. Some of the types only found at the more contaminated sites (M3 and M4) again clustered with Glo1 (types 17 and 18) and Glo4 (type 15), but others seem to be most similar to other species, including G. fasciculatum (types 19 and 21) and an Acaulospora isolate (type 16), perhaps indicating that these species are particularly able to tolerate large metal concentrations. The last group also included a sequence (type 20) matching most closely with a Glo8 isolate in the GenBank database, but clustering in the phylogenetic analysis more (but not very) closely with the Glo1/Glo4 groups.
Note, however, that it has recently been pointed out (Daniell et al. 2001) that, although the NS31-AM1 primer pair amplifies DNA sequences from the well established Glomalean families (Glomaceae, Acaulosporaceae and Gigasporaceae), it can amplify only a proportion of sequences from two newly proposed families with much more divergent SSU sequences, the Archaeosporaceae and the Paraglomaceae (Morton and Redecker 2001). It is possible, therefore, that other AM species than those detected are present in T. polytrichus at the South Tyne sites.
Daniell et al. (2001) found that Glomus species were predominant in roots at various arable sites, and suggested one reason for this may be the ability of the Glomaceae (unlike the Gigasporaceae) to recolonise roots from mycelial fragments, coupled with the ability of Glomus species, unlike Gigaspora or Scutellospora, readily to establish anastomoses between separate mycelia. These species may, therefore, be particularly well adapted to recolonise roots following disruption of the soil mycelium. The South Tyne sites are regularly affected by flooding (Macklin and Smith 1990), presumably causing major soil disturbance, and this could be another factor contributing to the predominance of Glomus species at these sites.
Daniell et al. (2001) also reported that AM species diversity at the arable sites investigated was very low, with just two Glomus species dominating colonisation in a range of crops. In contrast, a much greater rDNA sequence diversity was reported for AM fungi colonising bluebell roots in an undisturbed woodland (n=8; Helgason et al. 1999) and in a grassland ecosystem (n=24; Vandenkoornhuyse et al. 2002). A similar number of different sequence types to that found in the latter study was isolated from the sites with the greatest and smallest degrees of metal contamination in the present study, which would indicate a similar, and relatively large, AM species diversity at the two sites.
These results should, however, be treated with some caution, since there is increasing evidence that several different versions of the equivalent DNA sequence can occur within the same AM isolate, and even within a single spore (e.g. Antoniolli et al. 2000; Clapp et al. 2001; Rodriguez et al. 2001). Since AM mycelia and spores contain many nuclei, this has led to the intriguing suggestion (Clapp et al. 2001) that the phenotype exhibited by an AM fungus could be dictated by the relative proportions within it of nuclei containing different DNA sequences, with the possibility of ”morphological switching” mediated by exchange of nuclei between fungal mycelia via anastomoses. These authors point out that such a mechanism would explain the continuum in spore morphology exhibited between some Glomalean species.
In conclusion, this study has shown that T. polytrichus plants growing in soils containing phytotoxic concentrations of heavy metals are well colonised by AM fungi, suggesting that the host plant may derive some benefit from the symbiosis. The nature of this relationship is investigated in more detail in Whitfield et al. (2003). The greater vesicle abundance found at the most contaminated site probably reflects a difference in the fungal species mix colonising the roots—a theory borne out by the RFLP results—but whether vesicle production has any functional significance for the host or the mycobiont in relation to metal tolerance is uncertain. Glomus species predominated in the colonised roots at all sites, supporting findings from earlier studies that suggest that members of this genus are particularly tolerant of large concentrations of heavy metals. Identification and isolation of the most tolerant strains may have important implications for the future phytoremediation of heavy-metal-contaminated soils (Khan et al. 2000).
References
Antoniolli ZI, Schachtman DP, Ophel-Keller K, Smith SE (2000) Variation in rDNA ITS sequences in Glomus mosseae and Gigaspora margarita spores from a permanent pasture. Mycol Res 104:708–715
Clapp JP, Young JPW, Merryweather JW, Fitter AH (1995) Diversity of fungal symbionts in arbuscular mycorrhizas from a natural community. New Phytol 130:259–265
Clapp JP, Rodriguez A, Dodd JC (2001) Inter- and intra-isolate rRNA large subunit variation in Glomus coronatum spores. New Phytol 149:539–554
Daniell TJ, Husband R, Fitter AH, Young JPW (2001) Molecular diversity of arbuscular mycorrhizal fungi colonising arable crops. FEMS Microbiol Ecol 36:203–209
Dehn B, Schüepp H (1989) Influence of VA mycorrhizae on the uptake and distribution of heavy metals in plants. Agric Ecosyst Environ 29:79–83
Douds DD Jr, Pfeffer PE, Shachar-Hill Y (2000) Carbon partitioning, cost, and metabolism of arbuscular mycorrhizas. In: Kapulnik Y, Douds DD Jr (eds) Arbuscular mycorrhizas: physiology and function. Kluwer, Dordrecht, pp 107–129
Filatov D (2001) 2001 processor of sequences manual. University of Birmingham, http://www.biosciences.bham.ac.uk/labs/filatov/proseq.html
Gerdemann JW (1965) Vesicular-arbuscular mycorrhizae formed on maize and tuliptree by Endogone fasciculata. Mycologia 57:562–575
Gerdemann JW (1968) Vesicular-arbuscular mycorrhiza and plant growth. Annu Rev Phytopathol 6:397–418
Gerdemann JW, Trappe JM (1974) The Endogonaceae in the Pacific Northwest. Mycologia: Memoir no. 5
Gildon A, Tinker PB (1983) Interactions of vesicular-arbuscular mycorrhizal infection and heavy metals in plants. I. The effects of heavy metals on the development of vesicular-arbuscular mycorrhizas. New Phytol 95:247–261
Grime JP, Hodgson JG, Hunt R (1988) Comparative plant ecology: a functional approach to common British species. Unwin Hyman, London
Helgason T, Daniell TJ, Husband R, Fitter AH (1998) Ploughing up the wood-wide web? Nature 394:431
Helgason T, Fitter AH, Young JPW (1999) Molecular diversity of arbuscular mycorrhizal fungi colonising Hyacinthoides non-scripta (bluebell) in a seminatural woodland. Mol Ecol 8:659–666
Hepper CM, Smith GA (1976) Observation’s [sic] on the germination of Endogone spores. Trans Br Mycol Soc 66:189–194
Hetrick BAD, Wilson GWT, Figge DAH (1994) The influence of mycorrhizal symbiosis and fertilizer amendments on establishment of vegetation in heavy metal mine spoil. Environ Pollut 86:171–179
Hildebrandt U, Kaldorf M, Bothe H (1999) The zinc violet and its colonization by arbuscular mycorrhizal fungi. J Plant Physiol 154:709–717
Ietswaart JH, Griffioen WAJ, Ernst WHO (1992) Seasonality of VAM infection in three populations of Agrostis capillaris (Gramineae) on soil with or without heavy metal enrichment. Plant Soil 139:67–73
Kaldorf M, Kuhn AJ, Schroder WH, Hildebrandt U, Bothe H (1999) Selective element deposits in maize colonized by a heavy metal tolerance conferring arbuscular mycorrhizal fungus. J Plant Physiol 154:718–728
Khan AG, Kuek C, Chaudhry TM, Khoo CS, Hayes WJ (2000) Role of plants, mycorrhizae and phytochelators in heavy metal contaminated land remediation. Chemosphere 41:197–207
Leyval C, Turnau K, Haselwandter K (1997) Effect of heavy metal pollution on mycorrhizal colonization and function: physiological, ecological and applied aspects. Mycorrhiza 7:139–153
Liao JP, Lin XG, Cao ZH, Shi YQ, Wong MH (2003) Interactions between arbuscular mycorrhizae and heavy metals under sand culture experiment. Chemosphere 50:847–853
Macklin MG, Smith RS (1990) Historic riparian vegetation development and alluvial metallophyte plant communities in the Tyne basin, north-east England. In: Thornes JB (ed) Vegetation and erosion: processes and environment. Wiley, Chichester, UK, pp 239–256
McGonigle TP, Miller MH, Evans DG, Fairchild GL, Swan JA (1990) A method which gives an objective measure of colonization of roots by vesicular-arbuscular mycorrhizal fungi. New Phytol 115:495–501
Morton JB, Redecker D (2001) Two new families of Glomales, Archaeosporaceae and Paraglomaceae, with two new genera Archaeospora and Paraglomus, based on concordant molecular and morphological characters. Mycologia 93:181–195
Mosse (1973) Advances in the study of vesicular-arbuscular mycorrhiza. Annu Rev Phytopathol 11:171–196
Page RDM (1996) An application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358
Raistrick A, Jennings B (1965) A history of lead mining in the Pennines. Longman, London
Rodriguez A, Dougall T, Dodd JC, Clapp JP (2001) The large subunit ribosomal RNA genes of Entrophospora infrequens comprise sequences related to two different glomalean families. New Phytol 152:159–167
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sanders IR, Fitter AH (1992a) The ecology and functioning of vesicular-arbuscular mycorrhizas in coexisting grassland species. II. Nutrient uptake and growth of vesicular-arbuscular mycorrhizal plants in a semi-natural grassland. New Phytol 120:525–533
Sanders IR, Fitter AH (1992b) The ecology and functioning of vesicular-arbuscular mycorrhizas in coexisting grassland species. I. Seasonal patterns of mycorrhizal occurrence and morphology. New Phytol 120:517–524
Shetty KG, Hetrick BAD, Schwab AP (1995) Effects of mycorrhizae and fertilizer amendments on zinc tolerance of plants. Environ Pollut 88:307–314
Simon L, Lalonde M, Bruns TD (1992) Specific amplification of 18S fungal ribosomal genes from vesicular-arbuscular endomycorrhizal fungi colonizing roots. Appl Environ Microbiol 58:291–295
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Turnau K (1998) Heavy metal content and localization in mycorrhizal Euphorbia cyparissias from zinc wastes in southern Poland. Acta Soc Bot Pol 67:105–113
Turnau K, Miszalski Z, Trouvelot A, Bonfante P, Gianinazzi S (1996) Oxalis acetosella as a monitoring plant on highly polluted soils. In: Azcón-Aguilar C, Barea JM (eds) Mycorrhizas in integrated systems from genes to plant development. European Commission, Brussels, pp 483–486
Turnau K, Ryszka P, Gianinazzi-Pearson V, van Tuinen D (2001) Identification of arbuscular mycorrhizal fungi in soils and roots of plants colonizing zinc wastes in southern Poland. Mycorrhiza 10:169–174
Val C del, Barea JM, Azcón-Aguilar C (1999) Assessing the tolerance to heavy metals of arbuscular mycorrhizal fungi isolated from sewage sludge-contaminated soils. Appl Soil Ecol 11:261–269
Vandenkoornhuyse P, Husband R, Daniell TJ, Watson IJ, Duck JM, Fitter AH, Young JPW (2002) Arbuscular mycorrhizal community composition associated with two plant species in a grassland ecosystem. Mol Ecol 11:1555–1564
Weiersbye IM, Straker CJ, Przybylowicz WJ (1999) Micro-PIXE mapping of elemental distribution in arbuscular mycorrhizal roots of the grass, Cynodon dactylon, from gold and uranium mine tailings. Nucl Instrum Methods B 158:335–343
Whitfield L (2002). Heavy metal tolerance and mycorrhizal colonisation in Thymus polytrichus A. Kerner ex Borbás ssp. britannicus (Ronn.) Kerguelen (Lamiaceae). PhD thesis, University of Newcastle, UK
Whitfield L, Richards AJ, Rimmer DL (2003) Effects of mycorrhizal colonisation on Thymus polytrichus from heavy-metal-contaminated sites in northern England. Mycorrhiza DOI 10.1007/s00572-003-0269-y
Acknowledgements
Many thanks to Dr. Kirsten Wolff and Dr. Marie Hale (University of Newcastle) and Dr. Thorunn Helgason and Dr. Karyn Ridgway (University of York) for help with and advice on molecular techniques. Thanks also to Philip Green, Jackie Hodgson, Alan White, and the staff at Moor Bank Gardens and Close House Research Station for their valuable technical advice and support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Whitfield, L., Richards, A.J. & Rimmer, D.L. Relationships between soil heavy metal concentration and mycorrhizal colonisation in Thymus polytrichus in northern England. Mycorrhiza 14, 55–62 (2004). https://doi.org/10.1007/s00572-003-0268-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00572-003-0268-z