Abstract
Prostate diseases often occur in men. For further clinical treatment and diagnosis, we need to do accurate segmentation on prostate. There are already many methods that concentrate on solving the problem of automatic prostate MR image segmentation. However, the design of some hyperparameters of these methods is migrated from the models that are used for nature images which do not consider the difference between medical image and nature image. Besides, there is trend that researchers are likely to use deeper and more complicated networks to achieve high accuracy. The improvement is limited with surging parameters, computations, training time, and inference time. In this paper, we propose an efficient attention residual U-Net to segment the prostate MR image. We analyze the property of prostate MR image and fine-tune the architecture of U-Net. To accelerate the convergence of our method, residual connection and channel attention are added to our network. A set of experiments suggest our method can achieve a similar accuracy of state of the art with less parameters, less computations, shorter training time, and shorter inference time.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Prostate diseases (e.g., prostate cancer, prostatitis, and enlarged prostate) cause trouble for many men and usually can be judged by their magnetic resonance (MR) images. Therefore, segmenting the prostate from the MR image accurately is the key for further clinical treatment and diagnosis. In clinical practice, the prostate MR image composed of many slices represents a volume in physical space. Segmenting these slices by radiologist manually is quite time-consuming, cumbersome, costly, and subjective with limited reproducibility. In this connection, automatic prostate MR image segmentation is highly required in clinical practice.
Recently years, deep convolutional neural networks (CNNs) have achieved remarkable performance in many computer vision tasks [1,2,3]. He et al. [1] proposed the ResNet which has been the most popular network. Huang et al. [2] expanded the residual connection to densely connection, which connects each layer to every other layer in a feed-forward fashion. Hu et al. [4] proposed the squeeze-and-excitation (SE) block which further boosts the performances of networks. Benefited from the powerful feature extraction capabilities of CNNs, many researchers have employed CNNs in automated medical image segmentation [1, 5,6,7]. Most of them are based on U-Net [7, 8], which has a high performance in semantic segmentation. However, there is a trend that researchers are likely to use deeper and more complicated networks to achieve high accuracy. Yu et al. [5] used a U-Net with residual connection to segment the MR prostate image, where the model has 24 convolutional layers. He et al. [1] introduced residual connection and densely connection to the U-Net, leading the network has more than one hundred layers. Yang et al. [9] employed the adversarial training to segment the liver in a 3D manner. Except the segmentation network, they joined the discriminator network to learn a high consistency between the prediction and ground truth. So, the optimization objective for their network is trying to minimize a softmax cross-entropy loss together with an adversarial term that aims to distinguish between the ground truth and predicted segmentation map. We can see the capacities of segmentation networks have become larger and the architecture has become more complicated, which request more training time, larger computing power.
In fact, the semantic information of medical images is single. In most cases, the organs we are interested are just one or two. On the other hand, the nature images have more rich information and have hundreds of categories in them. Just migrating the architecture of networks that are originally designed for nature images does not make sense of medical images. For example, the ILSVRC classification competition has one thousand categories images, and the networks designed for them are big and the last convolutional layers of them output more than one thousand channels. For medical image, there are just two or three categories needed us to classify.
As a concrete example, the segmentation of prostate MR image is just a binary classification problem. Although there are big varieties in prostate, they are traceable. As shown in Fig. 1, according to the inherent property of prostate, the prostate can be divided into three approximately equal parts in the slice dimension: apex, middle, base, which has different appearance. And according to the scanning protocols, the prostate can be divided into with or without endorectal coil. These variations may challenge the performance of 2D segmentation network because it only sees one slice one time. But the appearance of 3D segmentation network has relieved the problem which consider the spatial contextual information. To explore the relationship between the performance and capacity of network in medical image dataset, we use U-Nets with different capacities to segment the prostate MR images. Figure 2 shows the curve of performance and its corresponding training time as the capacity changing. As the capacity of network increasing, the performance rising is rapidly then stable after the output channel of first convolutional layer is more than 16. Based on this fact, we think it is no need to design so big network to process the medical images.
In this paper, to reduce the time consumption and memory cost, we propose an efficient residual attention U-Net that can achieve the similar accuracy to state of the art, while the training time is decreased and the network parameter is less.
We introduce the residual connection to U-Net to segment the prostate MR image, where the residual connection is added to improve the training efficiency and accelerate the convergence speed.
We add the channel attention block to up-sampling path to improve the representational power of the segmentation network. Particularly, a channel attention block is added after the long connection in the U-Net, which is aimed to perform feature recalibration.
Finally, we fine-tuned the architecture of U-Net, which has less parameters and computation without accuracy losing.
2 Related works
2.1 Medical image segmentation
Medical image segmentation is a complex and critical step in medical image processing, and its purpose is to provide reliable basis for clinical diagnosis and treatment. Currently automatic medical image segmentation methods mainly include edge-based segmentation, region-based segmentation, and model-based segmentation [10]. For instance, Yuan et al. [11] proposed a contour evolution approach based on global optimization for the segmentation of prostate MR image. And Birkbeck et al. [12] leveraged the learning-based methods which used a statistical shape model to segment the prostate MR image. However, these methods have various shortcomings that limit the effectiveness in clinical practice, such as low accuracy, not robust enough, and sensitive to noise.
Recently, deep convolutional neural networks (CNNs) have achieved excellent performance in many tasks, which makes it promising to apply the medical image segmentation methods in clinical practice. For example, Ronneberger et al. [7] proposed the famous U-Net, the long connections added between encoder and decoder can recover the details lose during the down-sampling process. Milletari et al. [13] developed the U-Net to V-Net, which can make full use of the 3D spatial contextual information and make it works well in 3D space. Yu et al. [5] employed the residual connected mechanism for 3D prostate MR image segmentation and proposed a volumetric convnets with mixed residual connections, which won the champion in Promise12 [14] at 2017.
Meanwhile, the self-attention mechanism [15] has achieved promising progress in machine translation. In the field of video classification, Wang et al. [16] proposed the nonlocal block to capture the long-range dependencies. And Hu et al. [4] proposed the squeeze-and-excitation (SE) network which won the first place in the ILSVRC 2017 classification competition one the ImageNet dataset. The evaluation of SE blocks suggests the improvements induced by them can be applied in a wide range of architectures, not only deep networks (VGGNet [17], ResNet [1]), but also efficient networks (MobileNet [18], ShuffleNet [19]). This mechanism has also been used in medical image segmentation to force the network concentrate the organ we are interested. Roy et al. [20] modified the SE block, expanding it to three variants (cSE, sSE, scSE), which applied the self-attention in channel, spatial, and concurrent spatial and channel, respectively. Oktay et al. [21] proposed an attention gate (AG) model for medical imaging that can automatically learn to focus on target structures of varying shapes and sizes.
Furthermore, generative adversarial networks [22,23,24] have shown the potential ability in image-to-image translation. It consists of two modules, generator and discriminator, where the generator generates as realistic data as possible to cheat the discriminator, and the purpose of discriminator is to distinguish real data from the fake data generated by generator. The thought of adversarial training has been applied in many fields, such as domain adaption [25], knowledge distillation [26], and other tasks. In the field of medical image segmentation, Yang et al. [9] proposed a adversarial training approach to segment the liver CT image, which employed a deep convolutional network firstly to generate liver segmentation and then utilized a discriminator network to improve the shape consistency between prediction and ground truth. This method can overcome the limitation that softmax cross-entropy loss function cannot capture the relationship between pixels. [34] proposed SegAN, which also introduced the adversarial mechanism into the segmentation task, but it no longer used the concatenation operation when inputting the image and the segmentation map to the discriminator, instead of using the multiplication operation to fuse the information. Besides, they also designed a new L1 loss for the adversarial mechanism.
Generally, because of the 3D intrinsic properties of many medical images, 3D CNNs are more robust in medical image analysis tasks than 2D CNNs [8]. However, when the data are very nonuniform, a 2D CNNs may be a better choice. And if the data are very large, but the 3D CNNs cannot have an enough receptive field and capture sufficient contextual information limited by the GPU memory, then 2D CNNs may be better. On the other hand, compared with 2D CNNs, there are a much larger number of parameters in 3D CNNs, which makes them more difficult for optimization and more slower during the training phase.
3 Method
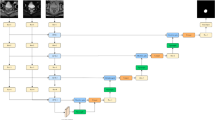
In this section, we describe the details of our proposed efficient residual attention U-Net from three aspects: the concepts of residual connection, channel attention to the architecture of our network, and the oversampling strategy for class imbalance. Figure 3 shows the overall network architecture of our proposed method.
3.1 Residual connection
Since ResNet [1] has a nice convergence behavior and can be easily combined with any existing architectures, it excels in many aspects. There have been many researches based on it [5, 27]. The main idea of ResNet is residual connection which is a kind of skip connection that represents the output as a linear superposition of the input and a nonlinear transformation of the input, and the ResNet explicitly reformulates the layers as learning residual functions with reference to the layer inputs. The original intention of these residual connections is to solve the problem of degradation, while adding more layers to network leads to higher training loss. The later research [cite] suggested that the residual connection keeps the gradient flow which is more resistant than plain network which makes the training easier. A residual block (RB) can be expressed as the following:
Here \( x_{l} \) and \( x_{l + 1} \) are the input and output vector of the l-th residual unit. The \( F\left( {x_{l} ,\left\{ {W_{l} } \right\}} \right) \) represents the residual function to be learned. Figure 4 shows the two structures of residual block used in our segmentation network.
3.2 Channel attention
Convolutional neural networks (CNNs) are mainly based on convolution operation, which can be thought as a filter fusing spatial and channel-wise information together to extract informative features. In the early stage, the number of filters is low and filters are mainly used to detect edges, corners, and contours. As the stage increases, the number of filters is going to high and filters are used to recognize the object. Therefore, filters are considered to extract edge feature at lower stages and semantic features at higher stages. However, not each filter can extract useful information, and some filter can only extract useless information. Hu et al. [4] proposed the squeeze-and-excitation (SE) block, in which the interdependencies between the features extracted by these filters are modeled specifically. In the SE block, the useful information can be selectively highlighted and the useless ones can be omitted by learning to use global information. We modified the SE block to make it available for segmentation task.
Specifically, we describe the feature map from the higher stage with \( X_{\text{high}} \) that has more semantic information and lower stage with \( X_{\text{low}} \) that has more edge information, where \( X_{\text{high}} \) is used to localize the object and \( X_{\text{low}} \) is used to recover the details. In original U-Net, as shown in Fig. 5, the authors do not use the attention mechanism to combine the \( X_{\text{high}} \) and \( X_{\text{low}} \). But, as mentioned above, not each feature map is useful, we should highlight the useful feature map and suppress the useless feature map. Inspired by the SE block, we propose the channel attention block (CAB) to recalibrate the feature map from lower stage and higher stage in up-sampling path (see Fig. 6). In details, firstly the CAB concatenates the two kinds of feature maps, \( X_{\text{high}} \) and \( X_{\text{low}} \), where \( X_{\text{high}} \) is resized to match the size of \( X_{\text{low}} \):
where \( \left[ \cdot \right] \) and \( F_{\text{upsample}} \) represent the concatenation operation and up-sampling operation followed by a Conv-BN layer, respectively.
Then we apply a global average pooling operation and two fully connection layers with activation function to capture channel-wise dependencies Z:
where \( F_{\text{pool}} \) denotes the global average pooling operation, W1 and W2 represent the fully connection layers, \( \sigma_{1} \) and \( \sigma_{2} \) denote the ReLU and sigmoid activation function, respectively.
Finally, multiply \( X_{\text{low}} \) by z to achieve the recalibration:
3.3 Network architecture
The task of segmentation network is to predict a category label to each pixel in the image from C categories. Inspired from [1, 4, 7], our segmentation network combines the U-Net with residual connection and channel attention, which takes an image I of size \( H \times W \) as input and outputs a probability map of size \( C \times H \times W \).
When designing the CNNs, a general practice is to double the number of filters as the number of stages increases. In [5], the numbers of output channel of convolutional layers are first 64 and then doubled after each down-sampling, which lead a big number of parameters, longer training time, and inference time. According to the fact Fig. 1 reveals, a larger number of output channel are not necessary. Therefore, we choose the 30 as the number of output channel of first convolutional layer for our network, which make a trade-off between efficiency and accuracy. On the other hand, inspired by [28], we increase the number of output channel gradually. Instead of doubling the output channel after each down-sampling, we just add a constant term. The feature map dimension in each stage is (30, 60, 90, 120, 150), while it is (64, 128, 256, 512, 1024) in [7]. Different from the original design of U-Net that each stage has the same number of convolutional layers, we fine-tuned the architecture. According to the design of residual block, we set different number of residual blocks in each stage. In the down-sampling path, which is an encoder, the number of residual blocks in each stage is (1, 1, 2, 3, 1). In the up-sampling path, which is a decoder, we set the number of convolutional layers in each stage to 1.
We add the residual connection to down-sampling path and channel attention block to up-sampling path. And after each convolution layer, batch normalization [29] is applied to stabilize the gradient flow. The segmentation network outputs a softmax result indicating the probabilities of each class. The loss function for this network is cross-entropy which can be formulated as
where \( c \) denotes the number of classes, \( y \) and \( \hat{y} \) denotes the ground truth and prediction result, respectively.
3.4 Oversampling for class imbalance
The class imbalance is a challenge for medical image segmentation, where the organs or lesions we are concerned just account for a small portion of the image. Unlike the method [30] that tried to design the loss function which is sensitive to the edge loss, we employ an oversample strategy to solve the problem at the source.
Explaining in detail, we calculate the bounding box for each prostate MR image, where the bounding box is the smallest rectangle that includes the prostate (see Fig. 7). In the training phase, we sample the training data that include the prostate with a certain probability p (0 ≤ p ≤ 1).
4 Experiment
4.1 Dataset and preprocessing
In this work, we evaluated our method on MICCAI Prostate MR Image Segmentation (PROMISE12) challenge dataset [12], its an ongoing benchmark for evaluating segmentation algorithms of the prostate from MR images. In total, 50 transversal T2-weighted MR images of the prostate are used for training and 30 MR images are used for testing. These data with differences in scanning protocol (e.g., differences in thickness, with/without endorectal coil) are come from multiple centers and vendors. We design 2D and 3D segmentation networks for this dataset. For 2D network, we only need to adjust the size of each slice in each MR image to the median size of the dataset which is 320 × 320 and then use zero mean and unit variance to normalize the intensities of each slice. For 3D network, each MR image is resized to have a same spacing \( 1.5 \times 0.625 \times 0.625 {\text{mm}} \) followed by a normalization which is done in the whole MR image.
4.2 Evaluation and comparison
We use the Dice coefficient (Dice)calculating in 3D to evaluate the performance of our proposed method. Furthermore, the number of parameters, complexity, training time, inference time of network are also considered to make a comprehensive comparison. We compared with U-Net [7], volumetric ConvNet [5], and U-Net with depth-wise separable convolution [18] (replace the standard convolution in down-sampling path with depth-wise separable convolution). Tables 1 and 2 show the quantitative results of 3D and 2D networks. Figure 8 shows the qualitative result of our method.
4.2.1 Comparison with U-Net
U-Net is a successful architecture for medical image segmentation which has an encoder and decoder. A long connection is used to connect the stage that has the same resolution. In this comparison, we modify the U-Net to fit our method. The changes include adding batch norm after convolution, removing the dropout layer, using padding to force the down-sampling path and up-sampling path have the same resolution, so we do not need to crop. And we replace the transposed convolution by linear interpolation. We design two kinds of U-Net with different capacities. One is the original size whose output channel of first convolutional layer is 64, while the other is 30. When using Dice as the comparison indicator, there is not much difference between U-Net and our method. When considering the number of parameters and complexity, there is a large margin between our method and U-Net. The parameter amount and complexity are reduced by 49–93% and 13–83% for different networks, respectively. And there is also a decline for training time and inference time except U-Net (c = 30). Although the parameter amount and complexity of our method are smaller than U-Net (c = 30), the training time and inference time are longer. We think the reason is that our method has more layers and GPU cannot advantage from serial task.
4.2.2 Comparison with volumetric ConvNet
Volumetric ConvNet is a 3D U-Net with mixed residual connections which won the first place in Promise12 challenge in 2017. It is worth noting that we report the original result (the number of parameters and complexity are not mentioned in its original paper) and our recurrent result of this network. Although the training time of original Volumetric ConvNet is much smaller than our recurrence, the dice coefficient is 2.5 percentage points lower than ours. It suggests that except the network architecture, other components (such as input size, optimizer, sampling strategy) in the medical image segmentation are also important.
4.2.3 Comparison with U-Net (DSC)
Inspired by MobileNets, we introduce the depth-wise separable convolution to U-Net. We replace the standard convolution by depth-wise separable convolution. From Tables 1 and 2, we can see the U-Net (DSC) has the smallest parameter size and complexity. However, what followed is the lowest Dice in all U-Nets. And we can see its training time and inference time are also longer than our method. The reason is that the depth-wise separable convolution split a standard convolution into two convolutions which has almost two times layers of original.
4.3 Ablation analysis
In this section, we would explore how the components we add influence the performance of our method. We remove the residual connection (RB) and channel attention block (CAB) in our network. Table 3 shows the comparison result. It is surprised that without the RB and CAB, the 2D network performs better, where 3D network performs worse. We think the reason may be the 2D network is easy to optimize so that the RB and CAB cannot boost the performance. And the 2D network also achieves the best result in validation set, which means the information in z-axis may be not important.
4.4 Implementation details
Our method uses Python and PyTorch to implement. All the training and experiments are carried out on a workstation with a TITAN XP GPU. In the training phase, all the date is preloaded to memory. The parameters and complexity are calculated by thop [31]. It is worth nothing that thop only calculates the convolutional layer, linear layer, batch norm layer, and ReLU layer, which means the up-sampling layer would not be calculated. (There are only 4 up-sampling layers, so it does not matter the result.) We employ the Adam optimizer [32] with a mini-batch size of 8 for all networks. The p in oversampling strategy is set to 1/3. The learning rate is set as 0.0003; the weights of all networks are initialized with xavier initialization [33]; all models are trained for 200 epochs, and each epoch we feed 2000 batches to networks. We utilized the data augmentation techniques to prevent overfitting, including elastic deformation, rotation with 90, 180, 270, and flip. The weights with best result on validation set will be saved and used for test set. In the inference phase of 3D networks, limited by the memory of GPU, we employ a sliding window strategy to predict the sub-volume in the prostate MR image. The stride for the sliding window is (16, 48, 48), and a Gaussian filter is used to weight the result.
5 Conclusion
In this paper, we propose an efficient residual attention U-Net for medical image segmentation. We analyze the property of prostate MR image and explore the relationship between the performance and the capacity of network. Based on it, we fine-tune the architecture of U-Net and add the residual connection and channel attention to it which balances trade-offs between accuracy and complexity. Furthermore, we propose an oversampling strategy to solve the class imbalance of medical image segmentation by calculating the bounding box for each class. The results suggest our method can achieve state of the art with less parameters, less computations, shorter training time, and shorter inference time.
References
He K, Zhang X, Ren S et al (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
Huang G, Liu Z, VanDerMaaten L et al (2017) Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 4700–4708
Long J, Shelhamer E, Darrell T (2015) Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE conference on computer vision and pattern recognition, 3431–3440
Hu J, Shen L, Sun G (2018) Squeeze-and-excitation networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 7132–7141
Yu L, Yang X, Chen H, et al (2017) Volumetric convnets with mixed residual connections for automated prostate segmentation from 3d mr images. In: Thirty-first AAAI conference on artificial intelligence
Chen H, Dou Q, Yu L et al (2018) VoxResNet: deep voxelwise residual networks for brain segmentation from 3D MR images. NeuroImage 170:446–455
Ronneberger O, Fischer P, Brox T (2015) U-net: Convolutional networks for biomedical image segmentation. In: International conference on medical image computing and computer-assisted intervention, Springer, Cham, pp 234–241
Isensee F, Petersen J, Kohl SAA et al (2019) nnU-Net: breaking the spell on successful medical image segmentation. arXiv preprint arXiv:1904.08128
Yang D, Xu D, Zhou SK et al (2017) Automatic liver segmentation using an adversarial image-to-image network. In: International conference on medical image computing and computer-assisted intervention, Springer, Cham, pp 507–515
Sharma N, Aggarwal LM (2010) Automated medical image segmentation techniques. J Med Phys Assoc Med Phys India 35(1):3
Yuan J, Qiu W, Ukwatta E et al (2012) An efficient convex optimization approach to 3D prostate MRI segmentation with generic star shape prior. Prostate MR Image Segment Chall MICCAI 7512:82–89
Birkbeck N, Zhang J, Requardt M, et al (2012) Region-specific hierarchical segmentation of MR prostate using discriminative learning. In: MICCAI grand challenge: prostate MR image segmentation
Milletari F, Navab N, Ahmadi SA (2016) V-net: fully convolutional neural networks for volumetric medical image segmentation. In: 2016 Fourth international conference on 3D vision (3DV), IEEE, pp 565–571
Litjens G, Toth R, van de Ven W et al (2014) Evaluation of prostate segmentation algorithms for MRI: the PROMISE12 challenge. Med Image Anal 18(2):359–373
Vaswani A, Shazeer N, Parmar N et al (2017) Attention is all you need. In: Advances in neural information processing systems, pp 5998–6008
Wang X, Girshick R, Gupta A et al (2018) Non-local neural networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 7794–7803
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556
Howard AG, Zhu M, Chen B et al (2017) Mobilenets: efficient convolutional neural networks for mobile vision applications. arXiv preprint arXiv:1704.04861
Zhang X, Zhou X, Lin M et al (2018) Shufflenet: an extremely efficient convolutional neural network for mobile devices. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 6848–6856
Roy AG, Navab N, Wachinger C (2018) Concurrent spatial and channel ‘squeeze and excitation’ in fully convolutional networks. In: International conference on medical image computing and computer-assisted intervention, Springer, Cham, pp 421–429
Oktay O, Schlemper J, Folgoc LL et al (2018) Attention u-net: learning where to look for the pancreas. arXiv preprint arXiv:1804.03999
Goodfellow I, Pouget-Abadie J, Mirza M et al (2014) Generative adversarial nets. In: Advances in neural information processing systems, pp 2672–2680
Isola P, Zhu J Y, Zhou T et al (2017) Image-to-image translation with conditional adversarial networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 1125–1134
Zhu JY, Park T, Isola P et al (2017) Unpaired image-to-image translation using cycle-consistent adversarial networks. In: Proceedings of the IEEE international conference on computer vision, pp 2223–2232
Dou Q, Ouyang C, Chen C et al (2019) PnP-AdaNet: plug-and-play adversarial domain adaptation network at unpaired cross-modality cardiac segmentation. IEEE Access 7:99065–99076
Liu Y, Chen K, Liu C et al (2019) Structured knowledge distillation for semantic segmentation. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 2604–2613
Drozdzal M, Vorontsov E, Chartrand G et al (2016) The importance of skip connections in biomedical image segmentation. In: Deep learning and data labeling for medical applications, Springer, Cham, pp 179–187
Han D, Kim J, Kim J (2017) Deep pyramidal residual networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 5927–5935
Ioffe S, Szegedy C (2015) Batch normalization: accelerating deep network training by reducing internal covariate shift. arXiv preprint arXiv:1502.03167
Kervadec H, Bouchtiba J, Desrosiers C et al (2019) Boundary loss for highly unbalanced segmentation. In: International conference on medical imaging with deep learning, pp 285–296
Kingma DP, Ba JA (2014) A method for stochastic optimization. arXiv preprint arXiv:1412.6980
Glorot X, Bengio Y (2010) Understanding the difficulty of training deep feedforward neural networks. In: Proceedings of the thirteenth international conference on artificial intelligence and statistics, pp 249–256
Xue Y, Xu T, Zhang H et al (2018) SegAN: adversarial network with multi-scale L1 loss for medical image segmentation. Neuroinformatics 16:383–392
Funding
This work is supported by the Science and Technology Major Project of Hubei Province (Next-Generation AI Technologies) under Grant No. 2019AEA170.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Mei, M., He, F. & Xue, S. Attention deep residual networks for MR image analysis. Neural Comput & Applic 35, 12957–12966 (2023). https://doi.org/10.1007/s00521-020-05083-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-020-05083-3