Abstract
Key message
Genome-wide characterization and development of first set of polymorphic Class I SSRs markers in pomegranate (Punica granatum L.) through in silico analysis using the draft genome sequence of pomegranate cv. Dabenzi (296 Mb) as reported by Qin et al. (2017).
Abstract
The availability of the draft genome sequence of pomegranate cv. Dabenzi presents unprecedented opportunities for the development of largescale genomic resources, such as DNA markers for genotyping applications. In this study, we identify a new set of highly polymorphic simple sequence repeat (SSR) markers by targeting the SSR motif lengths of ≥ 24 bp. A total of 1,73,633 SSRs were identified in the 296-Mb pomegranate genome assembly, reflecting an average density of 527.97 SSRs/Mb. Of these, 43,853 SSRs belong to Class I category (> 20 bp). Concerning the abundance of repeat types in the current dataset, dinucleotide (NN) repeats (31.19%) were the dominant class among all SSRs identified in the genome, followed by tetranucleotide (NNNN: 20.5%) and trinucleotide repeats (NNN: 16.8%). The top two SSR motifs in NN category were AT/AT (64.90%) and AG/CT (28.51%), whereas AAT/ATT (34.66%) and AAG/CTT (28.91%) were the most abundant among NNN repeats. Primer pairs were designed for a total of 2856 Class I SSRs and 110 primers were then assayed initially on eight pomegranate genotypes for polymorphism survey. Polymorphic fragments were obtained for 82 SSRs (77.36%), of which a subset of 13 informative SSRs was further employed to investigate genetic diversity among 46 pomegranate genotypes. Approaches, such as population structure, cluster and PCA elucidated genetic relationships among 46 diverse pomegranate genotypes. In summary, here we developed the first set of genome-wide SSRs in pomegranate that will serve as a powerful genomic tool for future genetic studies. These SSRs have widespread applications in QTL mapping and marker-assisted selection for breeding.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pomegranate (Punica granatum L.) is an economically important perennial crop, with high nutritional, medicinal and ornamental importance. It is believed to have originated from Iran and is widely cultivated in drier parts of Southeast Asia, Iran, China, Japan, the USA (California), West Indies, Tropical America and India (Holland and Bar-Yaakov 2014). Globally, India stands first in pomegranate cultivation with area 2.20 lakh ha, production and productivity of 27.95 lakh tonnes and 12 tonnes/ha, respectively (NHB 2018). Pomegranates are used as fresh fruit since long and continue to serve the food industry for production of juice, ready to serve beverages, alcoholic beverages and seed oil (Holland et al. 2009). The medicinal properties of pomegranate have been well documented. For instance, recent metabolomic analyses in pomegranate have revealed the presence of a wide range of phytochemicals, including gallotannins, ellagic acid, flavonoids, antioxidants, terpenoids and alkaloids in its different parts, such as arils, seed, rind, flower, bark and root (Caliskan and Bayazit 2013; Mayuoni-Kirshinbaum and Porat 2014; Ophir et al. 2014; Ahmed et al. 2014; Aslan et al. 2014; Bellesia et al. 2014). These bioactive compounds have been demonstrated to be beneficial in combating high blood pressure and other serious diseases, such as diabetes and various cancers (Shishodia et al. 2006). Given a growing body of literature reporting multiple health benefits of pomegranate, commercial cultivation has risen tremendously in the recent years. Importantly, pomegranate serves as a valued cash crop for small and marginal farmers owing to its inherent properties to grow with limited resources. Its cultivation remains highly remunerative in tropical and subtropical regions. Therefore, pomegranate cultivation is increasingly replacing subsistence farming to alleviate the livelihood of farmers (Priya et al. 2016).
The breeding efforts aimed at improving pomegranate has led to the development and release of improved varieties in India. These efforts mainly relied on conventional hybridization and selection from natural genetic variants (Jalikop et al. 2005). However, improving operational efficiency of pomegranate breeding programs demands integration of modern genomics tools. Molecular markers are the prerequisite to accelerate breeding program through genomics assisted breeding (GAB). The availability of appropriate DNA marker technology facilitates the identification of genetic determinants (genes/QTLs) underlying various traits of economic significance. Such developments pave the way for marker-assisted selection and help in designing future improvement strategies through guiding selection of parents with favourable alleles (Singh et al. 2015).
To date, a wide range of DNA marker systems like randomly amplified polymorphic DNA (RAPD) (Singh et al. 2013; Orhan et al. 2014), and inter simple sequence repeat (ISSR) (Narzary et al. 2010), amplified fragment length polymorphism (AFLP) (Jbir et al. 2008; Sarkhosh et al. 2011, 2012), sequence-related amplified polymorphism (SRAP) (Soleimani et al. 2012), simple sequence repeat (SSR) (Ravishankar et al. 2015) and single-nucleotide polymorphism (SNP) (Harel-Beja et al. 2015) have been deployed in pomegranate. Among these, SSRs are still preferred due to their genome abundance, high reproducibility, multi-allelic and co-dominant nature, which provides more information per unit assay as compared to other marker systems (Ebrahimi et al. 2017). In pomegranate, SSRs have been employed extensively to study genetic diversity and to understand population structure and association analysis (Curro et al. 2010; Pirseyedi et al. 2010; Singh et al. 2015). However, linkage mapping and QTL analysis based on SSR markers are currently lacking in pomegranate. A possible reason may be limited DNA polymorphism demonstrated in pomegranate by the currently available SSR markers. The length of the repeat motif is of paramount importance while surveying genetic polymorphisms with SSR markers. Temnykh et al. (2001) reported there are two major classes for SSR based on the length of repeat motifs (track length) i.e. SSRs with > 20 nucleotides and < 20 nucleotides are referred to as Class I and Class II, respectively. Further, the subsets, (i) Class I SSR with SSR lengths of > 20 bp and (ii) highly variable SSR with SSR lengths of > 50 bp, have shown significantly higher polymorphism in rice and pigeonpea (Temnykh et al. 2001; Singh et al. 2012; Bohra et al. 2017).
The widespread utility of such hyper or highly variable SSRs has been well established for genetic studies owing to ease of amplicon and scoring (Singh et al. 2012; Dutta et al. 2013; Bohra et al. 2017). Recent development of whole genome sequence of pomegranate by Qin et al. (2017) has offered unprecedented opportunities for genome-wide characterization of informative markers. Therefore, the present study was conducted with the aim to develop a comprehensive set of Class I SSRs (> 20 bp) through in silico analysis using the draft genome sequence of pomegranate cultivar Dabenzi. We then demonstrate the utility of these new SSR markers for genetic applications in pomegranate through analyzing genetic diversity among 46 pomegranate genotypes. The identified genome-wide Class I SSR markers will serve as an important resource for pomegranate genetics and genomics studies.
Materials and methods
Plant materials and DNA extraction
A total of 46 pomegranate genotypes, which are maintained at field gene bank of ICAR-National Research Centre on Pomegranate, India were collected. Details on passport data of genotypes are given in Table 1. Initially, a subset of eight pomegranate genotypes namely Bhagwa, Daru 17, G-137, Ganesh, Arakta, Dholka, Jodhpur Red and Solapur Lal, was used for experimental validation of the developed SSRs followed by diversity study of the 46 genotypes. Fresh leaf samples were collected from the genotypes and washed with sterile distilled water. Genomic DNA was extracted from the leaf samples following the modified CTAB method (Ravishankar et al. 2000). The quality and concentration of genomic DNA were determined on agarose gel electrophoresis (0.8%) by using uncut lambda DNA as standard. Final dilution of 10 ng/μl was made for the entire DNA samples for subsequent polymerase chain reactions (PCR).
In silico SSR mining and primer designing
The whole genome shotgun (WGS) sequences of Punica granatum cv. Dabenzi (296.38 Mb) was retrieved from the National Center for Biotechnology (NCBI) Genome Assembly database (GenBank accessions MTKT01000001 to MTKT01017405) (Qin et al. 2017). Total 17,405 shotgun sequences were surveyed for SSR motifs using MIcroSAtellite Identification (MISA) tool (https://pgrc.ipk-gatersleben.de/misa/). Keeping minimum repeat length of 12, the repeat unit was defined as 12, 6, 4, 3, 3 and 2 for mononucleotides, dinucleotides, trinucleotides, tetranucleotides, pentanucleotides and hexanucleotides, respectively. Compound SSRs were defined as two SSRs interrupted with 100 bases. The MISA statistics were further analyzed to draw frequency distribution graphs using Microsoft Excel.
The SSR loci identified from the genome assembly were recorded. Primer designing was performed using Batch Primer 3 v1.0 (https://wheat.pw.usda.Gov/demos/BatchPrimer3) with the criteria of minimum repeat length (≥ 24 bp). Primers were designed to generate amplicons of 100–400 bp in length with the following parameters: primer length (bp) 18–20, with 19 as the optimum; GC content (%) 40–60, with the optimum value being 50%; Tm (°C) 50–60, with 55 as the optimum. The other parameters used were as that of default program values.
SSR screening and genotyping
For experimental validation, a subset of 110 SSRs was selected for PCR amplification on eight pomegranate genotypes using Prime-96™ Thermal Cycler (Himedia, India) and resolved by gel electrophoresis. Subsequently, 13 SSRs were selected based on their clear amplification profile and screened on 46 pomegranate genotypes to evaluate genetic diversity. PCR amplification was carried out in 10 μl reaction volume containing 1.0 μl of 10X PCR buffer, 1 μl (1 mM dNTP mix), 0.5 μl each of forward and reverse primers (10 pmol), 0.2 μl of Taq DNA polymerase 5U/μl (Himedia, India) and 1 μl (10 ng) of template DNA. PCR condition was set as initial denaturation at 94 °C for 5 min, followed by 36 cycles of 94 °C for 1 min, 55 °C for 1 min, 72 °C for 2 min and a final extension at 72 °C for 7 min. PCR products were separated on 3% metaphor agarose gels containing 0.5 μg/ml ethidium bromide and 1X TBE running buffer at 130 V for 4 h, visualized and photographed in gel documentation system (Vilbert Dourmet, France).
Data collection and analysis
The PCR amplicons of each SSR marker obtained on all the samples were scored for allele size (bp). The genotype data were then used for estimating the following parameters using GenAlEx v. 6.5 (Peakall and Smouse 2012): the number of alleles (Na), the effective number of alleles (Ne), observed heterozygosity (Ho), expected heterozygosity (He) and polymorphic information content (PIC).
Population structure and Cluster analysis
A Bayesian clustering analysis implemented in Structure v. 2.3.3 (Pritchard 2000) was used to evaluate population genetic structure. An admixture model and correlated allele frequencies were applied to estimate the ancestry fractions of each cluster attributed to each genotype. For each value of K (range 1–10), five independent runs were performed with a burn-in period of 20,000 followed by 2,00,000 MCMC (Markov Chain Monte Carlo) repetitions. Parameters were set to the default values, and all genotypes were treated as having unknown origins. The delta K method (Evanno et al. 2005) implemented in Structure Harvester (Earl 2012) was used to determine the most probable value of K. UPGMA (Unweighted pair group method with an arithmetic mean), based neighbour-joining tree was constructed using DARwin v. 6.0.13 (Perrier and Jacquemoud-Collet 2006). A principal coordinate analysis (PCoA), based on the standardized covariance of genetic distances was performed using GenAlEx v. 6.5.
Results
SSR identification in the pomegranate genome
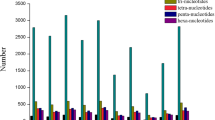
The SSR survey of the 17,405 pomegranate genome sequences resulted in the identification of a total of 1,73,633 SSRs, which translates to an average marker density of 527.97 SSRs/Mb (excluding mononucleotide SSRs 25,114). The general information on SSR containing sequences is summarized in Table 2. The total number of contigs with SSRs was 5,524; of which 2,377 contained more than one SSR and 15,483 were present in compound form. A total of 1,58,150 perfect SSRs (excluding compound SSRs 15,483) were identified in the genome. Further, the overall frequency distribution for SSR repeat units di, tri, tetra, penta and hexanucleotides revealed dominance of dinucleotides repeats (31.19%), followed by tetra (20.5%), tri (16.80%) and mono (14.46%) nucleotides (Fig. 1a, b). Interestingly, we found penta- and hexanucleotides in nearly equal proportions, i.e. 8.16% and 8.87%. Based on the hypervariable criteria (> 20 bp, Class I-SSRs), a total of 43,853 (29.53%) motifs were detected in the genome. Dinucleotide repeats (55.84%) were found dominated in this class followed by hexa- (16.87%), tri- (11.8%), tetra- (7.84%) and penta- (7.65%) nucleotides (Fig. 1a).
Frequency distribution graphs for dinucleotide and trinucleotide repeats were examined (Fig. 1c, d). In dinucleotides, maximum frequency of 35,149 (64.90%) was observed for classified motifs AT/AT, followed by AG/CT (28.51%), AC/GT (6.3%) and CG/CG (0.29%). The graph suggested a decline in SSR frequency with rise in the number of repeat units, and the trend was most pronounced for AT/AT followed by AG/CT, AC/GT and CG/CG repeat motifs. Similarly, for trinucleotides maximum frequency was observed for AAT/ATT (34.66%) and AAG/CTT (28.91%), followed by AGG/CCT (10.45%) and ATC/TAG (9%). The graph also outlined a gradual decrease in SSR frequency as the number of repeat units increased.
Development and validation of SSR markers
We designed primers for 2,856 Class I SSRs following survey of the whole genome sequence of pomegranate (Supl Table 2). Three hundred fifty contig sequences (2.01%) were selected randomly for primer designing, with SSRs distributed across the pomegranate genome. The majority of these primers were specific to dinucleotide motifs (primers 2429, 85.05%), followed by trinucleotide repeats (228, 7.98%). Further, we randomly selected a subset of 110 primers, referred to as NRCP_SSRs, for experimental validation on eight genotypes (Supl Table 1). As a result, 24 primers yielded monomorphic profiles while four markers did not show any amplification. Eighty-two primers (77.36%) yielded scorable amplicons with polymorphism (Fig. 2a). The number for alleles in the panel ranged from 1 to 3, with an average of 1.81. Similarly, PIC values ranged from 0 to 0.63, with an average of 0.28 (Supl Table 1). Of the total 82 polymorphic markers, 46 (56.10%) had PIC values between 0.4 and 0.6, whole remaining 36 primers showed PIC values ranging from 0.1 to 0.3.
Gel images showing allelic variations as revealed by SSR markers: a assaying on eight pomegranate genotypes using NRCP_SSR 18, 19, 23 and 24 and b assaying NRCP_SSR 92 and 93 on 46 pomegranate genotypes (where L-50 bp DNA ladder for Lane 1–8 eight set of pomegranate genotypes as mentioned in material and methods, L-100 bp DNA ladder for Lane 1–46 pomegranate genotypes as listed in Table 1, genotypes in lane 47 and 48 were excluded from analysis)
Assaying informative SSRs on a broad diversified panel
To further demonstrate the utility of newly developed SSR markers, we studied genetic diversity in a broader set of 46 pomegranate genotypes with 13 highly polymorphic SSRs. The SSR analysis generated 30 alleles across 46 genotypes, ranging from 2 to 4 alleles (at SSR 62 and 92), with an average of 2.30 alleles per locus (Table 3). The Ne values ranged from 1.55 alleles at locus SSR 97 to 3.32 at SSR 74, with a mean of 1.92 alleles per locus. The allele sizes varied from 160 bp at locus SSR 91 to 420 bp at locus SSR 74. The maximum allele frequency (0.77) was observed for allele 300 bp at SSR 97. In addition, the mean Ho and He values per locus ranged from 0 at SSR 24 to 0.57 at SSR 74 and from 0.36 at SSR 97 to 0.70 at SSR 74. The average PIC value of 0.46 suggested moderate diversity levels among the genotypes considered here. Four highly polymorphic SSR loci (PIC > 0.5) had PIC values of 0.51 (SSR 24, 92 and 93) and 0.71 (SSR 74). The remaining nine loci exhibited moderate polymorphic trends (0.3 < PIC < 0.5), ranging from 0.36 (SSR 97) to 0.48 (SSR 18) (Table 3). The average value of Shannon information index was 0.70 for the entire population. Representative gel images illustrating the SSR fingerprints of the 46 pomegranate genotypes are shown in Fig. 2a, b.
Population structure and principal coordinate analysis
Population structure analysis of 46 pomegranate genotypes by adopting non-stratified strategy showed clear peak for ΔK at K = 4, where entire genotypes were divided into four major groups, with few admixture among the groups (Fig. 3a). Group 3 contained the highest number of genotypes (16), followed by group 2 (11), group 4 (10) and group 1 (9). Statistical analysis indicated that the percentage of genotypes with a membership coefficient ≥ 90% was 54.34%, total of 23.91% of genotypes exhibited a membership coefficient ≥ 60% and only 10% of the genotypes exhibited a membership coefficient of 5% or less. A principal coordinate analysis (PCoA) also divided 46 genotypes into four clusters (Fig. 3b).
Principal coordinates (PCos) 1 and 2 explained 33.31% and 24.29% of the variance in the genotype data, respectively. More than 30% of the accessions were assigned to cluster 3, whose accessions were much more scattered than those in clusters 4, 2 and 1. The dendrogram demonstrated clustering of 46 genotypes into four groups (Fig. 3c).
Discussion
Identification of SSRs in the pomegranate genome
With the rapid developments in next-generation sequencing (NGS) technologies, WGS sequencing of plant genomes is gaining widespread attention. These sequences serve as valuable resources for genome-wide mining and development of genetic markers, such as SSR and SNPs (Bohra and Singh 2015; Simsek et al. 2018). Whole-genome assemblies have enabled the development of genome-wide SSR markers in many plant species, such as cotton (Gossypium sp) (Wang et al. 2015), tea plant (Camellia sinensis) (Liu et al. 2018) and pear (Pyrus spp) (Xue et al. 2018). Although reports on largescale development of EST–SSR markers from transcriptome data using NGS technologies have been reported in pomegranate (Ono et al. 2011; Simsek et al. 2018). However, largescale development of SSR markers from draft genome sequences of pomegranate has not been reported so far in pomegranate. The genomic Class I SSRs have tremendous utility for diversity analysis, varietal fingerprinting and molecular breeding applications in addition to enable efficient management of germplasm resources (Dutta et al. 2013). Therefore, we report development of the first set of genome-wide Class I SSRs in pomegranate and validate them on a set of diverse genotypes to show their immediate utility for genetic analysis. The relevance and importance of Class I SSRs to plant breeding is well described in various crops including rice and pigeonpea (Singh et al. 2010; Narshimulu et al. 2011; Bohra et al. 2017). The availability of such SSR markers spanning entire genome would facilitate genetic studies in pomegranate.
In the present study, we identified SSRs in the pomegranate genome with an average density of 527.97 SSRs/Mb, which is comparable to SSR density reported in other plant species like cucumber (552 SSR/Mb) and Arabidopsis (371 SSR/Mb) (Cavagnaro et al. 2010). Although analyzing the pomegranate WGS data, Ravishankar et al. (2015) have estimated the density of SSRs to be one SSR for every 5.56 Kbp based on the partial pomegranate genome sequence; however, here we found one SSR/1.90 Kbp.
We also analyzed the distribution and frequency of SSR motifs of 2–6 bp in the genome. We observed that the SSR frequency decreased with increase in the number of repeat units, and the change was more conspicuous in case of dinucleotides than the longer repeat motif types as observed in other studies (Liu et al. 2018). Following the definition of Class I SSRs (> 20 bp), we found 43,853 (29.53%) Class I SSRs in the pomegranate genome. Simsek et al. (2018) also reported largescale identification of 1900 SSR motifs in pomegranate genome through RNA seq using NGS technology. In our study, dinucleotide repeats (31.19%) were most abundant followed by tetra- (20.5%), tri- (16.8%) and mono- (14.46%) nucleotide repeats. Among the di-nucleotide repeats, AT/AT motifs were the most abundant (64.90%) followed by AG/CT repeat motifs (28.51%), while motifs AAT/ATT (34.66%) and AAG/CTT (28.91%) were the most frequent among tri-nucleotides. These observations were in close agreement with earlier findings that dinucleotides (73.33%) are the most abundant class of repeats in the pomegranate genome sequences (Ravishankar et al. 2015). Previously, authors have confirmed the abundance of AT/TA, TTA/TAT/ATT types in the pomegranate genome. However, the frequency of a given motif is also known to vary depending on sequence data sets; for instance, genome vs. transcriptome (Bohra et al. 2011; Varshney et al. 2005).
Marker validation and polymorphism survey
We successfully validated a set of randomly selected 110 SSR markers, of which 82 primers (77.36%) could reveal polymorphism among eight pomegranate genotypes with 2 to 3 alleles. The allele sizes (bp) obtained across pomegranate genotypes were similar to the expected sizes of the products for each locus. The PIC values for these markers ranged from 0.12 to 0.63, with a mean value of 0.36. Basaki et al. (2013) studied genetic diversity in 202 pomegranate genotypes belonging to 22 different provinces of Iran using polymorphic SSR as reported by Pirseyedi et al. (2010). The authors observed PIC values for these SSRs in the range of 0.01–0.56 on simple agarose gel detection system. The possible reason for lesser number of alleles and PIC values for SSR markers is limited resolution for agarose and metaphor gels as compared to automated capillary-based systems. Therefore, the SSRs developed here could show a higher level of polymorphism when assayed on polyacrylamide gel and capillary systems. Following the criterion laid by Botstein et al. (1980), 46 primers (56.10%) showed PIC values (> 0.4) in the current study, of which 15 SSRs met the criteria of highly polymorphic nature (PIC of 0.5). The most informative was the locus NRCP_SSR 2 with an average PIC value of 0.63, whereas the lowest average PIC value (0.12) was recorded for three loci-NRCP_SSR38, NRCP_SSR46 and NRCP_SSR 103 (Supl Table 1). Assaying 106 primers on eight genotypes produced 192 alleles with an average of 1.81 alleles per locus, which is comparable to 1.95 alleles per locus for 11 pomegranate genotypes as reported by Soriano et al. (2011).
In order to demonstrate the utility of these new SSR markers, 13 SSRs were selected based on their clear amplification profile with varying polymorphism rates to study the diversity in a broader panel of 46 genotypes. The allelic patterns suggested occurrence of a total of 30 alleles with an average of 2.31 alleles per locus. The PIC values ranged from 0.36 to 0.71 with a mean PIC value of 0.47. Similarly, Jian et al. (2012) reported 2–5 alleles with mean allele of 2.80 and PIC values ranged from 0.091 to 0.656 based on the analysis of 42 pomegranate accessions with 15 SSRs. Similar range of PIC values was obtained with SSR markers in pomegranate earlier by various research groups (Soriano et al. 2011; Zarei and Sahraroo 2018; Basaki et al. 2013). However, Ravishankar et al. (2015) reported a very high polymorphism (97.6%) for SSRs primers having allele range 1–14 with an average PIC value of 0.54 in pomegranate using high-throughput capillary-automated DNA Sequencer. We anticipate that the SSRs developed in this study might be highly informative when assayed on polyacrylamide and capillary-based system. To the best of our knowledge, this study reports for the first-time genome-wide SSR mining and designing of 2,856 primers with the SSR track length of ≥ 24 bp. For instance, a higher level of polymorphism has been revealed by SSRs (track length > 50 bp) on simple gels in different crops (Dutta et al. 2013; Bohra et al. 2015, 2017).
Out of 13 SSRs, four SSR loci were found to be highly polymorphic (PIC > 0.5) on 46 pomegranate genotypes, with PIC values ranging from 0.51 (SSR 24, SSR 92 and SSR 93) to 0.71 (SSR 74). Most of the selected primers confirm their polymorphism efficiency in larger set as per the initial screening results on eight pomegranate genotypes. Shannon information index for thirteen SSR loci ranged from 0.54 to 1.29 with an average value of 0.70, revealing higher genetic diversity among 46 genotypes. Similar observations were reported earlier in pomegranate using SSR markers (Pirseyedi et al. 2010; Raina et al. 2013). Further, relatively higher values of diversity index for these loci indicated the potential usefulness of these new SSR markers to analyze genetic diversity in pomegranate. In this study, we report successful development and validation of genome-wide SSR markers from draft genome sequence of pomegranate variety Dabenzi for genetic applications in pomegranate.
Genetic diversity
Examination of genetic variation in germplasm is key to accelerating genetic improvement of plants. To this end, molecular marker technologies including SSRs have emerged as a promising tool to uncover genetic polymorphism in a given set of genotypes/germplasms. In this context, structure and cluster analyses are effective means for studying genetic relationships related to germplasm resources (Goossens et al. 2002). Structure analysis showed that the grouping was largely consistent with the UPGMA clustering (Fig. 3a, c). The structure analysis revealed that K = 4 was the best value for classification of the 46 pomegranate genotypes, which remains in concordance with other studies. Singh et al. (2015) used 44 publicly available SSRs to study population structure among 88 pomegranate accessions and classified these into four populations. Considering the higher genetic diversity levels, cluster 4 constituted most diverse genotypes like introduced exotic pomegranate lines Tabesta and Alah (Iran), Shrin Anar, GR Pink and Gulesha Red (Russia), Spendander (Exotic cultivars), selections P-26, P-13, P-23 and P-16 of exotic line Muscat made at MPKV Rahuri, Maharashtra, India and few wild accessions IC1205, IC1182 and IC 1198 (Uttaranchal, India), which contributed for making this cluster most diverse. In this group, we find large fruits with either hard or soft-seeded arils.
Cluster 2 exclusively contained wild indigenous collection of Daru types of India. ‘Daru’ pomegranates are mainly grown in the forests on Himalayan slopes as wild pomegranates (Jalikop et al. 2005), and characterized by thorny bushes having small fruits with a sour and hard-seeded arils (Jalikop 2007). Cluster 3 covered highly domesticated pomegranate commercial varieties and cultivars, which are grown in India. This cluster included few introduced exotic breeding lines, such as Kandhari and Kabuli Yellow (Afghanistan) and commercial varieties, such as Nimali and Kalpitiya (Srilanka). The genotypes of this cluster are mainly characterized by large fruits with sweet and soft-seeded arils. Similarly, Singh et al. (2015) also found distinct grouping of cultivated pomegranate types, including commercial varieties, local types and introduced varieties, from wild accessions by screening 44 SSRs markers on 88 accessions. Cluster 1 constituted few cultivars of Tamil Nadu (Yercaud local, Yercaud HRS and Co-white) and few introduced exotic lines, i.e. Bedana Sedana and Kabuli Canoor (Afghanistan) and Spin Saccharin (Exotic Cultivar). The fruits are characterized by either sweet to acidic with hard-seeded arils.
PCoA plot explained 57.6% of the total variation, which is in close agreement with the results as observed for PCA explaining 60% variation among 88 pomegranate genotypes using 44 SSR markers (Singh et al. 2015). In PCoA plot, Axes 1 explained higher proportion of the variance (33.31%) compared with Axes 2 (24.29%). Probability it may be due to Axes 1 that separated CL 2 and CL4, which are representing highly diverse wild indigenous and introduced exotic lines. However, Axes 2 clearly separated CL1 from CL3, which represented commercial pomegranate lines of India and few introduced exotic lines with limited diversity.
The results of cluster analysis revealed grouping patterns of both geographical distributions and pedigree relationships. For instance, all Daru type wild accessions grouped together belong to Himachal regions of North India. The genotypes Yercaud HRS and Yercaud Local and Co-White grouped together are found in Tamil Nadu region of South India. Similarly, Jyoti and KRS belong to Karnataka regions. Proximity of Nimali and Kalpitiya may be due to their cultivation in Srilanka. Similarly, Tabesta and Alah grouped closer are from Iran. Remaining exotic lines dispersed across the clusters. Similarly, Jian et al. (2012) suggested geographical basis of clustering for 42 pomegranate accessions based on the expressed sequenced tag (EST)-SSR markers. With respect to pedigree, G-137, Arakta and Ganesh resided in the same cluster. Similarly, the genotypes P-26, P-13, P-23 and P-16, selected from Muscat, grouped together within the same cluster. The genotypes Yercaud HRS and Yercaud Local are also grouped together. Raina et al. (2013) observed grouping of some pomegranate genotypes on the basis of their parentage and pedigree.
Conclusion
The present work examines the SSR markers (Class I and II) in the draft genome sequence of pomegranate plant cv. Dabenzi providing a snapshot of the differential coverage and density of different SSR repeats in this species. The dinucleotides were most pronounced repeat types, accounting for up to 64.90% (AT/AT) of all the identified SSRs. The AAT/ATT (34.66%) and AAG/CTT (28.91%) were the abundant classes among trinucleotide repeats. A total of 43,853 motifs belonging to Class I SSRs (> 20 bp) were identified, and we designed primer pairs for 2856 SSRs targeting motifs ≥ 24 bp. Forty-six of the 82 primers (56.10%) showed polymorphic patterns, and a subset of 13 informative SSRs revealed genetic relationships among 46 pomegranate genotypes. Thus, the genome-wide development of such SSR markers could be very useful for various research areas in pomegranate, such as identification of the economically important pomegranate cultivars, study genetic diversity and evolutionary origin analysis, molecular fingerprinting, genetic linkage map construction, QTL/gene mapping and marker-assisted selection for breeding.
Author contribution statement
PGP, NVS, SP and JS designed the experiments. PGP, AB and RS performed the experiments and analyzed the data. NG, DM VS, SJ, AI, DB and NVS contributed to the collection of test materials. PGP and AB wrote the original draft.
Abbreviations
- QTL:
-
Quantitative trait loci
- CTAB:
-
Cetyl-trimethyl-ammonium-bromide
- NCBI:
-
National Centre for Biotechnology Information
- UPGMA:
-
Unweighted pair group method of arithmetic averages
- PIC:
-
Polymorphic information content
References
Ahmed MM, Samir ES, El-Shehawi AM, Alkafafy ME (2014) Antiobesity effects of Taif and Egyptian pomegranates: molecular study. Biosci Biotechnol Biochem 24:1–12. https://doi.org/10.1080/09168451.2014.982505
Aslan A, Can MI, Boydak D (2014) Anti-oxidant effects of pomegranate juice on Saccharomyces cerevisiae cell growth. Afr J Tradit Complement Altern Med 11:14–18
Basaki T, Nejat MA, Nejad RJ, Faraji S, Keykhaei F (2013) Identification of simple sequences repeat (SSR) informative markers associated with important traits in pomegranate (Punica granatum L.). Intl J Agron Plant Prod 4:575–583
Bellesia A, Verzelloni E, Tagliazucchi D (2014) Pomegranate ellagitannins inhibit alpha-glucosidase activity in vitro and reduce starch digestibility under simulated gastro-intestinal conditions. Int J Food Sci Nutr 18:1–8. https://doi.org/10.3109/09637486(953455)
Bohra A, Dubey A, Saxena RK, Penmetsa RV, Poornima KN, Kumar N, Farmer AD, Srivani G, Upadhyaya HD, Gothalwal R, Ramesh R, Singh D, Saxena KB, Kavi Kishor PB, Singh NK, Town CD, May GD, Cook DR, Varshney RK (2011) Analysis of BAC-end sequences (BESs) and development of BES-SSR markers for genetic mapping and hybrid purity assessment in pigeonpea. BMC Plant Biol 11:56
Bohra A, Jha R, Pandey G, Patil PG, Saxena RK, Singh IP, Singh D, Mishra RK, Mishra A, Singh F, Varshney RK, Singh NP (2017) New hypervariable SSR markers for diversity analysis, hybrid purity testing and trait mapping in pigeonpea [Cajanus cajan (L.) Millspaugh]. Front Plant Sci 8:377. https://doi.org/10.3389/fpls.2017.00377
Bohra A, Singh NP (2015) Whole genome sequences in pulse crops: a global community resource to expedite translational genomics and knowledge-based crop improvement. Biotechnol Lett 37:1529–1539. https://doi.org/10.1007/s10529-015-1836-y
Bohra A, Singh IP, Yadav AK, Pathak A, Soren KR, Chaturvedi SK, Nadarajan N (2015) Utility of informative SSR markers in the molecular characterization of cytoplasmic genetic male sterility-based hybrid and its parents in pigeonpea. Natl Acad Sci Lett 38:13–19
Caliskan O, Bayazit S (2013) Morpho-pomological and chemical diversity of pomegranate accessions grown in Eastern mediterranean region of Turkey. J Agric Sci Technol 15:1449–1460
Cavagnaro PF, Senalik DA, Yang L, Simon PW, Harkins TT, Kodira CD, Huang S, Weng Y (2010) Genome-wide characterization of simple sequence repeats in cucumber (Cucumis sativus L.). BMC Genomics 11:569. https://doi.org/10.1186/1471-2164-11-569
Curro S, Caruso M, Distefano G, Gentile A, La Malfa S (2010) New microsatellite loci for pomegranate, Punica granatum (Lythraceae). Am J Bot 97:e58–e60. https://doi.org/10.3732/ajb.1000143
Dutta S, Mahato AK, Sharma P, Ranjeet SR, Sharma TR, Singh NK (2013) Highly variable Arhar simple sequence repeat markers for molecular diversity and phylogenetic studies in pigeonpea (Cajanus cajan (L.) Millisp.). Plant Breed 132:1439–1523. https://doi.org/10.1111/pbr.12014
Earl DA (2012) Structure Harvester: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Ebrahimi A, Zarei A, Zamani FM, Lawson S (2017) Evaluation of genetic variability among “Early Mature” Juglans regia using microsatellite markers and morphological traits. Peer J 5:e3834. https://doi.org/10.7717/peerj.3834
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Goossens B, Funk S, Vidal C, Latour S, Jamart A, Ancrenaz M, Wicking E, Tutin C, Bruford M (2002) Measuring genetic diversity in translocation programmes: principles and application to a chimpanzee release project. Anim Conserv 5:225–236
Harel-Beja R, Sherman A, Rubinstein M, Eshed R, Bar-Ya’akov I, Trainin T, Ophir R, Holland D (2015) A novel genetic map of pomegranate based on transcript markers enriched with QTLs for fruit quality traits. Tree Genet Genomes 11:109. https://doi.org/10.1007/s11295-015-0936-0
Holland D, Bar-Ya’ako I (2014) Pomegranate: aspects concerning dynamics of health beneficial phytochemicals and therapeutic properties with respect to the tree cultivar and the environment. In: Yaniv Z, Dudai N (eds) Medicinal and aromatic plants of the Middle-East. Springer, Netherlands, pp 225–239
Holland D, Hatib K, Bar-Yaakov I (2009) Pomegranate: Botany, horticulture, breeding. Hortic Rev 35:127–191
Jalikop SH (2007) Linked dominant alleles or inter-locus interaction results in a major shift in pomegranate fruit acidity of ‘Ganesh’ 6 ‘Kabul Yellow’. Euphytica 158:201–207
Jalikop S, Rawal R, Kumar R (2005) Exploitation of sub-temperate pomegranate Daru in breeding tropical varieties. Acta Hortic 696:107–112
Jbir R, Hasanaoui N, Mars M, Marrakchi M, Trifi M (2008) Characterization of Tunisian pomegranate (Punica granatum L.) cultivars using amplified fragment length polymorphism analysis. Sci Hort 115:231–237. https://doi.org/10.1016/j.scienta.2007.09.002
Jian ZH, Liu XS, Hu JB, Chen YH, Feng JC (2012) Mining microsatellite markers from public expressed sequence tag sequences for genetic diversity analysis in pomegranate. J Genet 91:353–358
Liu S, An Y, Li F, Li S, Liu L, Zhou Q, Zhao S, Wei C (2018) Genome-wide identification of simple sequence repeats and development of polymorphic SSR markers for genetic studies in tea plant (Camellia sinensis). Mol Breed 38:59. https://doi.org/10.1007/s11032-018-0824-z
Mayuoni-Kirshinbaum L, Porat R (2014) The flavor of pomegranate fruit: a review. J Sci Food Agric 94:21–27. https://doi.org/10.1002/jsfa.6311
Narshimulu G, Jamaloddin M, Vemireddy LR, Anuradha G, Siddiq E (2011) Potentiality of evenly distributed hyper-variable microsatellite markers in marker-assisted breeding of rice. Plant Breed 130:314–320. https://doi.org/10.1111/j.1439-0523.2010.01834.x
Narzary D, Rana T, Ranade S (2010) Genetic diversity in inter-simple sequence repeat profiles across natural populations of Indian pomegranate (Punica granatum L.). Plant Biol 12:806–813. https://doi.org/10.1111/j.1438-8677.2009.00273.x
NHB (2018) National Horticultural Board, Gurgaon, India, URL: http://nhb.gov.in/PDFViwer.aspx?Enc=3ZOO8K5CzcdC/Yq6HcdIxC0U1kZZenFuNVXacDLxz28. Accessed 20 Aug 2019
Ono NN, Britton MT, Fass JN, Nicolet CM, Lin D, Tian L (2011) Exploring the transcriptome landscape of pomegranate fruit peel for natural product biosynthetic gene and SSR marker discovery. J Integr Plant Biol 53:800–813. https://doi.org/10.1111/j.1744-7909.2011.01073.x
Ophir R, Sherman A, Rubinstein M, Eshed R, Schwager SM, Harel-Beja R, Bar-Ya’akov I, Holland D (2014) Single-nucleotide polymorphism markers from de-novo assembly of the pomegranate transcriptome reveal germplasm genetic diversity. PLoS ONE 9:e88998. https://doi.org/10.1371/journal.pone.0088998
Orhan E, Ercisli S, Esitken A, Sengul M (2014) Molecular and morphological characterization of pomegranate (Punica granatum L.) genotypes sampled from Coruh Valley in Turkey. Genet Mol Res 13:6375–6382. https://doi.org/10.4238/2014
Peakall R, Smouse PE (2012) GenAlEx 6.5 genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539. https://doi.org/10.1093/bioinformatics/bts460
Perrier X, Jacquemoud-Collet JP (2006) DARwin software. http://darwin.cirad.fr/darwin
Pirseyedi SM, Valizadehghan S, Mardi M, Ghaffari MR, Mahmoodi P, Zahravi M, Zeinalabedini M, Khayam NSM (2010) Isolation and characterization of novel microsatellite markers in pomegranate (Punica granatum L.). Int J Mol Sci 11:2010–2016. https://doi.org/10.3390/ijms11052010
Pritchard JK, Stephens M, Donneily P (2000) Inference of population structure using multi-locus genotype data. Genetics 155:945–959
Priya BT, Murthy BNS, Gopalakrishnan C, Artal RB, Jagannath S (2016) Identification of new resistant sources for bacterial blight in pomegranate. Eur J Plant Pathol. https://doi.org/10.1007/s10658-016-0947-1
Qin G, Xu C, Ming R, Tang H, Guyot R, Kramer EM, Hu Y, Yi X, Qi Y, Xu X, Gao Z, Pan H, Jian J, Tian Y, Yue Z, Xu Y (2017) The pomegranate (Punica granatum L.) genome and the genomics of punicalagin biosynthesis. Plant J 91:1108–1128. https://doi.org/10.1111/tpj.13625
Raina D, Dhillon WS, Gill PPS (2013) Molecular marker-based characterization and genetic diversity of pomegranate genotypes. Indian J Hortic 70:469–474
Ravishankar KV, Anand L, Dinesh MR (2000) Assessment of genetic relatedness among mango cultivars of India using RAPD markers. J Hort Sci Biotechnol 75:198–201. https://doi.org/10.1080/14620316.2000.11511223
Ravishankar KV, Chaturvedi K, Puttaraju N, Gupta S, Pamu S (2015) Mining and characterization of SSRs from pomegranate (Punica granatum L.) by pyrosequencing. Plant Breed 134:247–254. https://doi.org/10.1111/pbr.12238
Sarkhosh A, Zamani Z, Fatahi R, Wiedow C, Chagne D, Gardiner SE (2012) A pomegranate (Punica granatum L.) linkage map based on AFLP markers. J Hortic Sci Biotechnol 87:1–6. https://doi.org/10.1080/14620316.2012.11512821
Sarkhosh A, Zamani Z, Fatahi R, Hassani M, Wiedow C, Buck E, Gardiner S (2011) Genetic diversity of Iranian soft-seed pomegranate genotypes as revealed by fluorescent-AFLP markers. Physiol Mol Biol Plants 17:305–311. https://doi.org/10.1007/s12298-011-0070-x
Shishodia S, Adams L, Bhatt ID, Aggarwal BB (2006) Anticancer potential of pomegranate. In: Seeram NP, Schulman RN, Heber D (eds) Pomegranates: ancient roots to modern medicine. CRC Press Taylor and Francis Group, Boca Raton, pp 107–116
Simsek O, Donmez D, Imrak B, Ozguven AI, Kaçar YA (2018) Development of SSR markers by using Next Generation Sequencing technology in pomegranate (Punica granatum L.). Int J Agric Wildl Sci 4:161–167. https://doi.org/10.24180/ijaws.469748
Singh NK, Gupta DK, Jayaswal PK, Mahato AK, Dutta S, Singh S, Bhutani S, Dogra V, Singh BP, Kumawat G, Pal JK, Pandit A, Singh A, Rawal H, Kumar A, Prashat GR, Khare A, Yadav R, Raje RS, Singh MN, Datta S, Fakrudin B, Wanjari KB, Kansal R, Dash PK, Jain PK, Bhattacharya R, Gaikwad K, Mohapatra T, Srinivasan R, Sharma TR (2012) The first draft of the pigeonpea genome sequence. J Plant Biochem Biotechnol 21:98–112. https://doi.org/10.1007/s13562-011-0088-8
Singh H, Deshmukh RK, Singh A, Singh AK, Gaikwad K, Sharma TR, Mohapatra T, Singh NK (2010) Highly variable SSR markers suitable for rice genotyping using agarose gels. Mol Breed 25:359–364. https://doi.org/10.1007/s11032-009-9328-1
Singh NV, Abburi V, Ramajayam D, Kumar R, Chandra R, Sharma K, Sharma J, Babu KD, Pal R, Mundewadikar D, Saminathan T, Cantrell R, Nimmakayala P, Reddy U (2015) Genetic diversity and association mapping of bacterial blight and other horticulturally important traits with microsatellite markers in pomegranate from India. Mol Genet Genomics 290:1393–1402. https://doi.org/10.1007/s00438-015-1003-0
Singh SK, Meghwal PR, Pathak R, Gautam R, Kumar S (2013) Genetic diversity in Punica granatum revealed by nuclear rRNA, internal transcribed spacer and RAPD polymorphism. Natl Acad Sci Lett 36:115–124. https://doi.org/10.1007/s40009-013-0120-8
Soleimani MH, Talebi M, Sayed-Tabatabaei BE (2012) Use of SRAP markers to assess genetic diversity and population structure of wild, cultivated, and ornamental pomegranates (Punica granatum L.) in different regions of Iran. Plant Syst Evol 298:1141–1149. https://doi.org/10.1007/s00606-012-0626-4
Soriano JM, Zuriaga E, Rubio P, Llacer G, Infante R, Badenes ML (2011) Development and characterization of microsatellite markers in pomegranate (Punica granatum L.). Mol Breed 27:119–128. https://doi.org/10.1007/s11032-010-9511-4
Temnykh S, DeClerck G, Lukashova A, Lipovich L, Cartinhour S, McCouch S (2001) Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genet Res 11:1441–1452. https://doi.org/10.1101/gr.184001
Varshney RK, Graner A, Sorrells ME (2005) Genic microsatellite markers in plants: features and applications. Trends Biotechnol 23:48–55. https://doi.org/10.1016/j.tibtech.2004.11.005
Wang Q, Fang L, Chen J, Hu Y, Si Z, Wang S, Chang L, Guo W, Zhang T (2015) Genome-wide mining, characterization, and development of microsatellite markers in Gossypium Species. Sci Rep 5:10638. https://doi.org/10.1038/srep10638
Xue H, Zhang P, Shi T, Yang J, Wang L, Wang S, Su Y, Zhang H, Qiao Y, Li X (2018) Genome-wide characterization of simple sequence repeats in Pyrus bretschneideri and their application in an analysis of genetic diversity in pear. BMC Genomics 19:473. https://doi.org/10.1186/s12864-018-4822-7
Zarei A, Sahraroo A (2018) Molecular characterization of pomegranate (Punica granatum L.) accessions from Fars Province of Iran using microsatellite markers. Hort Env Biotech 59:239–249. https://doi.org/10.1007/s13580-018--001
Acknowledgements
The authors are highly grateful to the Indian Council of Agricultural Research (ICAR), New Delhi for extended financial support through ICAR-Notational Research Centre on Pomegranate, Solapur. Authors are also thankful to the Ex-Director of ICAR-NRCP, Solapur Dr. R.K Pal for their encouragement and support to carry out this work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they do not have any conflict of interest.
Data archiving statement
This research contains no data that requires submission to a public database. The details of SSRs markers designed and used for validation in this study were listed in supplementary files 1 & 2. All the relevant information on Class I SSR markers designed here will be freely available to any scientist wishing to use them for their research purposes.
Additional information
Communicated by Carlson.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Patil, P.G., Singh, N.V., Parashuram, S. et al. Genome-wide characterization and development of simple sequence repeat markers for genetic studies in pomegranate (Punica granatum L.). Trees 34, 987–998 (2020). https://doi.org/10.1007/s00468-020-01975-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00468-020-01975-y