Abstract
The integration and structure of a transgene locus can have profound effects on the level and stability of transgene expression. We screened 28 transgenic birch (Betula platyphylla Suk.) lines transformed with an insect-resistance gene (bgt) using Agrobacterium tumefaciens. Among the transgenic plants, the copy number of transgene varied from one to four. A rearrangement or partial deletion had occurred in the process of T-DNA integration. T-DNA repeat formation, detected by reverse primer PCR, was found among randomly screened transgenic lines. Sequencing of the junctions between the T-DNA inserts revealed deletions of 19–589 bp and an additional 45 bp filler DNA sequence was inserted between the T-DNA repeats at one junction. Micro-homologous sequences (1–6 bp) were observed in the junctions between the T-DNA inserts. Using SiteFinding-PCR, a relatively high percentage of AT value was found for the flanking regions. Deletion of the right border repeat was observed in 12/18 of the T-DNA/plant junctions analyzed. The number of nucleotides deleted varied from 3 to 712. Deletions of 17–89 bp were observed in all left T-DNA/plant junctions analyzed. A vector backbone DNA sequence in the transgene loci was also detected using primer pairs outside the left and right T-DNA borders. Approximately 89.3% of the lines contained some vector backbone DNA. These observations revealed that it is important to check the specificity of the integration. A mechanism of T-DNA transport and integration is proposed for this long-lived tree species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Transformation of plants mediated by the microbe Agrobacterium tumefaciens is a widely used means of generating transgenic plants. Agrobacterium–plant cell T-DNA transport represents a unique case of transfer of genetic material between kingdoms (Stachel and Zambryski 1989). In nature, Agrobacterium T-DNA carries genes involved in the synthesis of plant growth regulators and in the production of opines that form a major carbon and nitrogen source for this microorganism. Integration and expression of the T-DNA into the host plant genome results in the formation of neoplastic growths, known as crown gall tumors, that produce and secrete opines that are then consumed by the bacterium (Hooykaas and Beijersbergen 1994; Sheng and Citovsky 1996; Zambryski 1992). The T-DNA itself is not sequence-specific; any DNA fragment placed between two 25 bp direct repeats (known as T-DNA borders) on the Ti plasmid will be transported into the plant cell and integrated into the host genome. This is followed by expression of the introduced genes in the transformed host cell (Gelvin 1998).
Several Agrobacterium chromosomal virulence (chv) genes, a series of Ti plasmid-encoded virulence (Vir) genes, and various host plant genes have been identified as participants in the different stages of the Agrobacterium–plant transfection process (Tzfira and Citovsky 2000; Gelvin 2003; Wood et al. 2001). Because Agrobacterium-mediated transformation represents a major tool for plant breeding, the molecular mechanism by which it genetically transforms the host cells has been intensively studied in recent years (Tzfira and Citovsky 2000; Zupan et al. 2000). However, in the case of long-lived tree species, although genetic modification using Agrobacterium has matured to some extent in its technical aspects, the mechanism underlying T-DNA integration has not yet been identified.
The aim of the present paper was therefore to obtain further insights into the molecular structure of the T-DNA integration sites and the mechanism of integration of transferred DNA (T-DNA) in a perennial tree system. To this end, the copy number and rearrangement of T-DNAs in transgenic birch (Betula platyphylla Suk.) trees were comprehensively studied. We also sequenced and analyzed T-DNA/plant DNA junctions and junctions between linked T-DNAs in transgenic birch lines, using modified SiteFinding-PCR and reverse primer PCR. To determine the presence of vector backbone sequences in the transgenic birch plants obtained through Agrobacterium-mediated transformation, PCR analysis was performed on DNA from these plants using specific primers for vector backbone sequence amplification. The findings have significant implications for both the basic and applied research areas that focus on long-lived tree species.
Materials and methods
Plant material, bacterial strains, and plasmids
Transgenic plants of Betula platyphylla Suk. were obtained via Agrobacterium (LBA4404 strain)-mediated transformation. The transformation vector was pCAMBIA-2301, which contained the selectable marker nptII gene and reporter gene gus, along with a fused bgt gene consisting of an insecticidal toxin gene from a spider (Atrax robustus) and the C terminal of the Cry IA(b) gene from Bacillus thuringiensis. In all, 53 independent transgenic lines were obtained, from which 28 transgenic lines were screened for this study. Transgenic plants were established by in vitro propagation and then, together with non-transformed control plants, cultivated in a greenhouse under natural daylight conditions for 5 years.
Nucleic acid isolation and southern blot analysis
Nucleic acid isolation and southern blot analysis were done as described in Zeng et al. 2009.
Reverse primer PCR for T-DNA structure
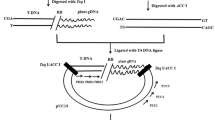
T-DNA structure for inserts with multiple copies was evaluated by Reverse primer PCR (RP-PCR) (Kumar and Fladung 2001). Six different primers (Table 1) located along the T-DNA sequence were used for PCR amplification (Fig. 1). T-DNA structure was inferred by the presence and size of amplified bands. PCR conditions were Long Taq DNA Polymerase (TaKaRa) in a volume of 20 μl containing 2 μL 10 × LA PCR Buffer II (Mg2+ plus), 0.2 mM dNTP Mixture, 0.5 μM Primer 1, 0.5 μM Primer 2, 50 ng DNA, 2 U LA Taq DNA Polymerase. The PCR cycles were: 95°C for 2 min; 30 cycles at 95°C for 20 s; 68°C 15 min; 72°C for 7 min.
PCR analysis for backbone sequence detection
PCR was used to confirm the presence of a vector backbone sequence in the transgenic birch lines. In total, 6 primer pairs were designed to check the vector backbone sequence in 28 independent lines. Three primer pairs (BSP1-3) were used to detect the 682, 708, and 920 bp fragments outside T-DNA at the left border and another three primer pairs (BSP4-6) were used to detect the sequences 870, 1,093, and 649 bp beyond the T-DNA right border. Sequences of primer pairs, annealing temperatures, and expected fragment sizes are shown in Table 2.
Flanking sequence cloning based on modified SiteFinding-PCR
The PCR mixture included 2 μl of 10 × long Taq DNA polymerase buffer, 2 μl of mixed dNTP solution (2.5 mM each of dATP, dTTP, dCTP, and dGTP), 1 U of long Taq DNA polymerase (TaKaRa), 10 pmol of SiteFinder (5′ CACGACACGCTACTCAACACACCACCTCGCACAGCGTCCTCAGCGGCCGCNNNNNNGCCT 3′) and 100 ng of template DNA. The final volume was brought to 20 μl with Milli-Q water, and then a single cycle PCR was run. Eight gene-specific primers (GSP) were designed using Oligo 6.0 software (Table 3). The distance between GSPL2 and GSPL3 was 94 bp, between GSPL3 and GSPL4 was 169 bp between GSPR2 and GSPR3 was 59 bp, and between GSPR3 and GSPR4 was 98 bp. The target DNA was amplified by nested PCR with GSPs and SiteFinder primers (SFP1, 5′ CACGACACGCTACTCAACAC 3′ and SFP2, 5′ ACTCAACACACCACCTCGCACAGC 3′). The cycling conditions, the SiteFinder, and the corresponding primers were identical to those described by Tan et al. (2005).
Sequencing and data analysis
The resulting PCR products were either sequenced directly using gene-specific primers or preferably cloned into the pGEM-T Easy vector (Promega) and sequenced using the universal primer (5′ GGAAACAGCTATGACCATG 3′) and M13 reverse primer (5′ GTAAA CGACGGCCAGT 3′).
Results
Description of transgene integration in birch
T-DNA construct used in the study was schematically depicted in Fig. 2. Southern blot analysis of the transgenic lines indicated that the bgt and gus genes had been integrated into the birch genome. Among the transgenic plants, the copy number of T-DNA varied from one to four (Zeng et al. 2009). 9.1% had two copies, 54.5% had three copies, and the remaining 9.1% contained four copies. No more than four copies of the insert were observed in any transformant. As reported in Zeng et al. (2009), the copies of bgt were inconsistent with the gus in 68.2% of the transgenic lines, indicating that rearrangement or partial deletion had occurred during the process of T-DNA integration.
Schematic presentation of T-DNA repeats and PCR primers to determine T-DNA structure. Relative positions of six PCR amplification primers (1–6) are used to detect transgene repeats and arrowheads indicate their respective 5′–3′ orientations. Larger arrows represent T-DNA and point toward the right border. LB T-DNA left border, RB T-DNA right border
Schematic representation of the T-DNA, restriction sites, and probes. PstI restriction endonuclease was used to analyze the copy number of bgt and gus. 35S-p Promoter from the cauliflower mosaic virus, 35 s-t terminator from the cauliflower mosaic virus, nos-t terminator from nopaline synthase, nptII bacterial neomycin phosphotransferase II gene, bgt the chimeric gene of a spider insecticidal peptide gene and the C peptide sequence of Bt gene, gus β-glucuronidase gene, LB T-DNA left border, RB T-DNA right border
Analysis of the genomic insertion sites
Genomic DNA fragments that flanked the T-DNA were amplified from transgenic plants by SiteFinding-PCR, with nested specific primers located near the borders and SiteFinder site. The DNA fragments were recovered from the specific band (Fig. 3), and then cloned and sequenced. We obtained 26 left border flanking sequences and 24 right border flanking sequences. However, in 10 out of 26 flanking sequences, LB connected with vector backbone sequences without any deletion. In 6 out of 24 flanking sequences, RB connected with vector backbone sequences without any deletion. The remaining flanking sequences were identified as non-vector sequences. The percentage of AT value for these sequences was between 62 and 73.5% indicating that these sequences showed a relatively high percentage of AT nucleotides. Among the 18 right T-DNA ends and 16 left T-DNA ends that were connected with birch genome sequence, the deletion of the right border repeat was observed in 12/18 of the junctions analyzed. The number of deleted nucleotides varied from 3 to 712 bp. For the left border repeat, deletions of 17–89 bp were observed in all left T-DNA/plant junctions analyzed. As shown in Fig. 4, 2 and 3 bp sequence similarity between the T-DNA left end and birch genomic DNA was found in two transgenic lines, but no micro-homologous sequence was found between the T-DNA end and birch genomic DNA in the other lines.
Products of the second round of PCR based on modified SiteFinding-PCR. Lane M, 250-bp DNA ladder (250, 500, 750, 1,000, 1,500, 2,250, 3,000, 4,500 bp); Lane C, PCR product of distilled water. Transgenic birch samples are labeled as S1–S4. Lanes II, III, and IV, the products generated by SFP2/GSPR2, SFP2/GSPR3, and SFP2/GSPR4, respectively
The vector backbone sequence is frequently co-transferred with T-DNA
To determine the presence of vector backbone sequences, DNA isolated from the control and transgenic birch plants was analyzed by PCR using specific primers for vector backbone sequence amplification (Fig. 5). Of the 28 lines analyzed, the vector backbone sequence was detected in 25 (89.3%). Among these, ten vector backbone sequences were connected with flanking sequences of LB without any deletion or filler DNA. Six vector backbone sequences were connected with flanking sequences of RB without any deletion or filler DNA.
Molecular structure of T-DNA integration sites
Southern blotting analysis revealed repeat formation for the eight events containing one to four copies of transgenes. RP-PCR was performed using eight primer pairs (3+4, 1+2, 1+5, 2+4, 2+5, 2+3, 1+6, and 1+4) in the T-DNAs (Fig. 6). Amplified bands should only result from primer pairs 3+4 in the case of complete single copy integration. Certain amplified bands will be observed when multiple copies of T-DNA are arranged as direct or inverted repeats at the same chromosomal position (see Fig. 1). The presence and size of PCR products from different primer pairs may determine the type of repeats, as well as whether the repeats are complete or incomplete (Table 4).
Molecular analysis of T-DNA repeats in transgenic line TP71. Lane M, 250-bp DNA ladder (250, 500, 750, 1,000, 1,500, 2,250, 3,000, 4,500 bp); Lanes 3+4, 1+2, 1+5, 2+4, 2+5, 2+3, 1+6, and 1+4 are PCR products amplified by each primer pair (see Table 1)
The T-DNA repeat formation in the different transgenic birch lines is summarized in Table 4. Amplified bands were obtained only with primer pair 3+4 in transgenic birch line TP46 and the copy number of bgt and gus was one. Both results indicated that T-DNA was a complete single copy integration. Result of southern blot showed that TP27 contained three copies of T-DNA. Amplified bands were obtained with primer pairs (3+4, 1+2, 1+5, and 2+4), and the sizes of the PCR products were consistent with those expected. Two T-DNAs were apparently arranged as direct repeats in one site. Another T-DNA could be integrated in other sites. The copy number of bgt in TP71 detected by southern blot was four, while the copy number of gus was three, indicating that rearrangement or partial deletion had occurred in the process of T-DNA integration. Amplified bands were obtained with primer pairs (3+4, 1+2, 1+5, 2+4, 1+6, and 1+4), and the sizes of PCR products were consistent with those expected. Three T-DNAs appeared to be linked as tandem repeats (head-to-head-to-tail).
We sequenced and analyzed junctions between linked T-DNAs. As shown in Table 4 and Fig. 7, transgenic line TP28 contained a single copy of T-DNA with 712-bp deletion at RB and 29-bp deletion at LB. Transgenic line TP73 contained an incomplete T-DNA with an 800-bp deletion of bgt. LB connected with a vector backbone sequence without any deletion. TP46 also contained a single copy of T-DNA with 21-bp deletion at RB and 7-bp deletion at LB.
The molecular structure of T-DNA integration sites was comprehensively analyzed. All left and right borders were variable at T-DNA repeat junctions. Deletion of the entire left and right border repeat, together with part of the adjacent T-DNA, was observed in transgenic lines TP23 and TP74. A 45-bp filler DNA was found at the junction region in TP74, among which 16 bp originated from coding sequence of gus, another 29 bp originated from nos poly A of nptII. 1–6 bp sequence similarity between recombining ends at the junction region between T-DNA repeats was found in transgenic lines TP23, TP27, and TP74.
Discussion
Cloning of T-DNA flanking sequences
Several PCR methods have been developed for isolating unknown DNA segments adjacent to a known DNA sequence, including inverse PCR (Keim et al. 2004), ligation-mediated PCR (Riley et al. 1990; Siebert et al. 1995; Yan et al. 2003) and randomly primed PCR (Liu et al. 1995; Antal et al. 2004). Compared to other PCR methods for isolating an unknown region adjacent to a known DNA sequence, SiteFinding-PCR does not require size optimization, biotinylation, or cDNA synthesis. This method is simple, fast, inexpensive, sensitive, and efficient, and easily produces long specific fragments.
A key point pertaining to this method is whether GCCT oligonucleotides at the 3′-end of the SiteFinder can initiate the SiteFinding reaction at a low temperature. This depends on GCCT numbers in the genome and on the choice of an appropriate annealing temperature. In fact, any 4, 5, or 6 nt oligonucleotides can be used to initialize the SiteFinding reaction at adjusted annealing temperatures (Tan et al. 2005). In order to avoid false positive results, the target DNA was amplified by nested PCR with four gene-specific primers (GSPs) and SiteFinder primers (SFPs) in our study. Using this technique, we obtained 26 left border flanking sequences and 24 right border flanking sequences.
Integration of T-DNA and vector backbone sequences in transgenic plants
Multiple copies of T-DNA can be simultaneously integrated into the host genome at single or multiple loci. Direct DNA transfer methods (e.g., electroporation or particle bombardment) often result in the integration of many copies of transgenes (Kohli et al. 1998), while Agrobacterium-mediated transformation usually results in fewer copies. Cheng et al. (1997) transformed wheat using both Agrobacterium and particle bombardment. Of 26 Agrobacterium-mediated transformants, more than one-third contained a single T-DNA insert, half contained 2–3 copies and the remainder contained 4–5 copies. In contrast, from the population of 77 bombarded transformants, only 13 (17%) contained a single copy of the transgene. In our study, southern blot analysis of the transgenic birch plants indicated that the foreign gene had been integrated into the birch genome. Among the transgenic plants, the copy number varied from one to four. We found that our Agrobacterium-mediated transformation system produced a majority of multiple copy transformants, with only 18.2% of the transformants containing a single copy. The copies of bgt were inconsistent with those of gus in 68.2% transgenic lines, indicating that rearrangement or partial deletion had occurred in the process of T-DNA integration. These results are generally consistent with previous observations of Agrobacterium-mediated transformed plants (Tang et al. 2007; Zhang et al. 2008).
Agrobacterium-mediated transformation has long been considered as a clean technology compared with other direct transformations, such as particle bombardment or electroporation, because only the sequences between the RB and LB T-DNA borders are introduced into the plant genomes. However, detailed studies of transgene integration patterns in several species have shown that vector backbone sequences outside the T-DNA are also frequently inserted (De Buck et al. 2000; Yin and Wang 2000; Olhoft et al. 2004; Lange et al. 2006). Using primer pairs outside the left and right T-DNA borders, we found that approximately 89.3% of our transgenic lines contained some vector backbone DNA. This integration frequency of vector backbone DNA was similar to that of transgenic strawberry (90%) (Abdal-Aziz et al. 2006; Lange et al. 2006), but was higher than that reported in other research (Zhai et al. 2004; Lange et al. 2006; Zhang et al. 2008).
This issue with incorporation of vector sequences during Agrobacterium-mediated transformation has been explained by two hypotheses. The first hypothesis is that the non-recognition of LB by Vir proteins would enable T-strand formation, initiated at RB, to continue at the left border, also referred to as read-through at the left border (Kuraya et al. 2004; Lange et al. 2006). In the second hypothesis, T-strand formation could start at LB and proceed towards RB, resulting in the integration of the entire vector (van der Graaff et al. 1996). In the present paper, 38.5% (10/26) T-DNA LB flanking sequences were connected with vector backbone sequences without any deletion or filler DNA and 25.0% (6/24) RB flanking sequences were connected with vector backbone sequences without any deletion or filler DNA. In contrast to a previous study, which indicated that LB read-through was quite frequent (Kim et al. 2003), our results demonstrated that vector backbone sequences could integrate not only by read-through at the left border but also at the right border. Hence, it would be useful if termination efficiency at LB and RB could be increased and its initiation efficiency decreased (Kim et al. 2003).
The transfer of vector backbone sequences to a plant genome has important consequences. First, these vector sequences may influence transgene expression (Iglesias et al. 1997). Second, regulatory authorities and consumers are demanding that commercial transgenic plants should be free of unnecessary genes, e.g., marker genes or vector backbone sequences (Zhang et al. 2008). In this study, we have shown that a high percentage of transgenic birch plants obtained through Agrobacterium-mediated transformation contain vector backbone sequences when using binary vectors. Thus, molecular analysis to detect non-T-DNA sequences should be performed routinely in transgenic birch plants, especially if these plants are going to be considered for field release and future commercialization.
Molecular mechanism of T-DNA transport and integration
T-DNA is transferred into a plant cell nucleus as a single strand molecule attached at the 5′ end to the protein VirD2 and coated with virulence protein VirE2. T-DNA integrates into the genome (Wenck et al. 1997; Ziemienowicz et al. 2000) by illegitimate recombination (Zupan et al. 2000; Brunaud et al. 2002) via a largely unknown mechanism. Various studies on T-DNA integration have indicated that there are, in principle, two different classes of integration patterns (Salomon and Puchta 1998; Kumar and Fladung 2002). In the first class, the right border (which is conserved in the recombinant) and the target locus contain no or only one base pair that shows micro homology, whereas longer homologies exist between the left border (combined with truncations) and the pre-insertion site. In the second class, sequence similarity can also be found in most cases between the right border and the pre-insertion site (Mayerhofer et al. 1991) and the right border is partly truncated. However, these integration patterns do not elucidate the origin of complex T-DNA inserts, especially multiple copies of transgenes (direct or inverted repeat) integrated at the same or tightly linked loci, which are frequently found in transgenic lines. Two studies (Chilton and Que 2003; Tzfira et al. 2003) have now suggested the intriguing possibility that T-DNA integration may be based on a double-strand break (DSB) and that the T strands are first converted to double-stranded intermediates.
In the present study, a total of 18 right T-DNA/plant junctions and 16 left T-DNA/plant junctions were obtained. Most of the T-DNA flanking genomic sequences contained a relatively high percentage of AT value (62–73.5%). T-DNA integration into AT-rich regions has also been reported in tobacco, Arabidopsis, and rice (Takano et al. 1997). AT-rich regions may act as recombination hot spots for the transgenes, possibly because of steric bending of the chromosomal DNA structure (Takano et al. 1997). The right border of the T-DNA beside the flanking sequences connected with backbone vector sequence was conserved up to the cleavage site in 6 out of 18 transgenic birch lines, whereas the left border was variable in all the lines. These results are in disagreement with some reports that have shown that the virulence protein VirD2 specifically preserves the T-DNA right border to the point of its processing (Tzfira and Citovsky 2000; Gelvin 2000).
According to our results, rearrangement or partial deletion occurred during the process of T-DNA integration. Sequencing the junctions between the T-DNA inserts revealed deletions of 19–589 bp and homologous sequences (1–6 bp) were observed in these junctions. An additional 45-bp DNA sequence, referred to as filler DNA, was inserted between the T-DNA repeats at one junction, as determined from micro-homologous sequences. These results indicate that T-DNA recombination is based on the micro-homologous sequence between T-DNA repeats. However, the micro-homologous sequence between T-DNA end and plant junction was found only in two transgenic lines. The results suggested that T-DNA strands may be randomly captured at double-strand breaks (DSBs) and integrated via a non-homologous end-joining (NHEJ) pathway.
Prior to T-DNA integration, T-DNA recombination might occur (De Buck et al. 1999), in a manner analogous to that causing the two-phase ligation for integration of exogenous DNA (Kohli et al. 1998). In the preintegration phase, ligation occurs extrachromosomally between molecules of the transforming plasmid, resulting in rearranged transgenic sequences. In the second phase, these rearranged sequences integrate at the same locus through a second round of ligations. After ligation and recombination of the T-DNA, double-stranded T-DNA could then interact with the break ends.
In summary, among the transgenic plants, the copy number of transgene varied from one to four. Only 18.2% of the transformants contained a single copy. Approximately 89.3% of the lines contained some vector backbone DNA. Thus, molecular analysis to detect the copy number of transgene and non-T-DNA sequences should be performed routinely in transgenic plants obtained through Agrobacterium-mediated transformation, especially if these transgenic plants are going to be considered for field release. We analyzed the molecular structure of the T-DNA integration sites. On the basis of the results obtained, a mechanism of T-DNA transport and integration is proposed for this long-lived tree species. This fundamental knowledge will have a profound effect on our understanding of rearrangement of T-DNAs and the presence of vector backbone sequences in plants and allow development of improved genetic engineering procedures for efficient nuclear delivery and integration of foreign genes.
References
Abdal-Aziz SA, Pliego-Alfaro F, Quesada MA, Mercado JA (2006) Evidence of frequent integration of non-T-DNA vector backbone sequences in transgenic strawberry plant. J Biosci Bioeng 6:508–510
Antal Z, Rascle C, Fevre M, Bruel C (2004) Single oligonucleotide nested PCR: a rapid method for the isolation of genes and their flanking regions from expressed sequence tags. Curr Genet 46:240–246
Brunaud V, Balzergue S, Dubreucq B, Aubourg S, Samson F, Chauvin S, Bechtold N, Cruaud C, DeRose R, Pelletier G, Lepiniec L, Caboche M, Lecharny A (2002) T-DNA integration into the Arabidopsis genome depends on sequences of pre-insertion sites. EMBO Rep 12:1152–1157
Cheng M, Fry JE, Pang S, Zhou H, Hironaka CM, Duncan DR, Conner TW, Wan Y (1997) Genetic transformation of wheat mediated by Agrobacterium tumefaciens. Plant Physiol 115:971–980
Chilton MD, Que Q (2003) Targeted integration of T-DNA into the tobacco genome at double-stranded breaks: new insights on the mechanism of T-DNA integration. Plant Physiol 133:956–965
De Buck S, Jacobs A, Van Montagu M, Depicker A (1999) The DNA sequences of T-DNA junctions suggest that complex T-DNA loci are formed by a recombination process resembling T-DNA integration. Plant J 20:295–304
De Buck S, De Wilde C, Van Montagu M, Depicker A (2000) Determination of the T-DNA transfer and the T-DNA integration frequencies upon cocultivation of Arabidopsis thaliana root explants. Mol Plant Microbe Interact 13:658–665
Gelvin SB (1998) The introduction and expression of transgenes in plants. Curr Opin Biotechnol 9:227–232
Gelvin SB (2000) Agrobacterium and plant genes involved in T-DNA transfer and integration. Annu Rev Plant Physiol Plant Mol Biol 51:223–256
Gelvin SB (2003) Agrobacterium-mediated plant transformation: the biology behind the “gene-jockeying” tool. Microbiol Mol Biol Rev 67:16–37
Hooykaas PJJ, Beijersbergen AGM (1994) The virulence system of Agrobacterium tumefaciens. Annu Rev Phytopathol 32:157–179
Iglesias VA, Moscone EA, Papp I, Neuhuber F, Michalowski S, Phelan T, Spiker S, Matzke M, Matzke AJM (1997) Molecular and cytogenetic analyses of stably and unstably expressed transgene loci in tobacco. Plant Cell 9:1251–1264
Keim M, Williams RS, Harwood AJ (2004) An inverse PCR technique to rapidly isolate the flanking DNA of dictyostelium insertion mutants. Mol Biotechnol 26:221–224
Kim SR, Lee J, Jun SH, Park S, Kang HG, Kwon S, Gynheung A (2003) Transgene structures in T-DNA-inserted rice plants. Plant Mol Biol 52:761–773
Kohli A, Leech M, Vain P, Laurie DA, Christou P (1998) Transgene organization in rice engineered through direct DNA transfer supports a two-phase integration mechanism mediated by the establishment of integration hot spots. Proc Natl Acad Sci USA 95:7203–7208
Kumar S, Fladung M (2001) Gene stability in transgenic aspen (Populus). II. Molecular characterization of variable expression of transgene in wild and hybrid aspen. Planta 213:731–740
Kumar S, Fladung M (2002) Transgene integration in aspen: structures of integration sites and mechanism of T-DNA integration. Plant J 31:543–551
Kuraya Y, Ohta S, Fukude M (2004) Suppression of transfer of non-T-DNA ‘vector backbone’ sequences by multiple left border repeats in vectors for transformation of higher plants mediated by Agrobacterium tumefaciens. Mol Breed 14:309–320
Lange M, Vincze E, Moller MG, Holm PB (2006) Molecular analysis of transgene and vector backbone integration into the barley genome following Agrobacterium-mediated transformation. Plant Cell Rep 25:815–820
Liu YG, Mitsukawa N, Oosumi T, Whittier RF (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8:457–463
Mayerhofer R, Koncz-Kalman Z, Nawrath C, Bakkeren G, Crameri A, Angelis K, Redei GP, Schell J, Hohn B, Koncz C (1991) T-DNA integration: a mode of illegitimate recombination in plants. EMBO J 10:697–704
Olhoft PM, Flagel LE, Somers DAT (2004) DNA locus structure in a large population of soybean plants transformed using the Agrobacterium-mediated cotyledonary node method. Plant Biotechnol J 2:289–300
Riley J, Butler R, Ogilvie D, Finniear R, Jenner D, Powell S, Anand R, Smith JC, Markham AF (1990) A novel, rapid method for the isolation of terminal sequences from yeast artificial chromosome (YAC) clones. Nucleic Acids Res 18:2887–2890
Salomon S, Puchta H (1998) Capture of genomic and T-DNA sequences during double-strand break repair in somatic plant cells. EMBO J 17:6086–6095
Sheng J, Citovsky V (1996) Agrobacterium-plant cell DNA transport: have virulence proteins, will travel. Plant Cell 8:1699–1710
Siebert PD, Chenchik A, Kellogg DE, Lukyanov KA, Lukyanov SA (1995) An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res 23:1087–1088
Stachel SE, Zambryski P (1989) Generic trans-kingdom sex. Nature 340:190–191
Takano M, Egawa H, Ikeda JE, Wakasa K (1997) The structures of integration sites in transgenic rice. Plant J 11:353–361
Tan G, Gao Y, Shi M, Zhang X, He S, Chen Z, An C (2005) SiteFinding-PCR: a simple and efficient PCR method for chromosome walking. Nucleic Acids Res 33:e122
Tang W, Newton RJ, Weidner DA (2007) Genetic transformation and gene silencing mediated by multiple copies of a transgene in eastern white pine. J Exp Bot 58:545–554
Tzfira T, Citovsky V (2000) From host recognition to T-DNA integration: the function of bacterial and plant genes in the Agrobacterium plant cell interaction. Mol Plant Pathol 1:201–212
Tzfira T, Frankmen L, Vaidya M, Citovsky V (2003) Site-specific integration of Agrobacterium T-DNA via double-stranded intermediates. Plant Physiol 133:1011–1023
Van Der Graaff E, Den Dulk-Ras A, Hooykaas PJJ (1996) Deviating T-DNA transfer from Agrobacterium tumefaciens to plants. Plant Mol Biol 31:677–681
Wenck A, Czako M, Kanevski I, Marton L (1997) Frequent collinear long transfer of DNA inclusive of the whole binary vector during Agrobacterium-mediated transformation. Plant Mol Biol 34:913–922
Wood DW, Setubal JC, Kaul R, Monks DE, Kitajima JP, Okura VK, Zhou Y, Chen L, Wood GE, Almeida NF et al (2001) The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294:2317–2323
Yan YX, An CC, Li L, Gui JY, Tan GH, Cen ZL (2003) T-linker-specific ligation PCR (T-linker PCR): an advanced PCR technique for chromosome walking or for isolation of tagged DNA ends. Nucleic Acids Res 31:e68
Yin Z, Wang GL (2000) Evidence of multiple complex patterns of T-DNA integration into the rice genome. Theor Appl Genet 100:461–470
Zambryski P (1992) Chronicles from the Agrobacterium-plant cell DNA transfer story. Annu Rev Plant Physiol Plant Mol Biol 43:465–490
Zeng FS, Zhan YG, Nan N, Xin Y, Qi FH, Yang CP (2009) Expression of BGT gene in Transgenic Birch (Betula platyphylla). Afr J Biotechnol 8:3392–3398
Zhai W, Chen C, Zhu X, Chen X, Zhang D, Li X, Zhu L (2004) Analysis of T-DNA Xa21 loci and bacterial blight resistance effects of the transgene Xa21 in transgenic rice. Theor Appl Genet 109:534–542
Zhang J, Cai L, Cheng J, Mao H, Fan X, Meng Z, Chan KM, Zhang H, Qi J, Ji L, Hong Y (2008) Transgene integration and organization in cotton (Gossypium hirsutum L.) genome. Transgenic Res 17:293–306
Ziemienowicz A, Tinland B, Bryant J, Gloeckler V, Hohn B (2000) Plant enzymes but not Agrobacterium VirD2 mediate T-DNA ligation in vitro. Mol Cell Biol 20:6317–6322
Zupan J, Muth TR, Draper O, Zambryski P (2000) The transfer of DNA from Agrobacterium tumefaciens into plants: a feast of fundamental insights. Plant J 23:11–28
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (NO: 30471413 and 30872045) and Specialized Research Fund for the Doctoral Program of Higher Education (NO: 200802251038) and China Postdoctoral Science Foundation (NO: 20090460071).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J. Carlson.
Rights and permissions
About this article
Cite this article
Zeng, FS., Zhan, YG., Zhao, HC. et al. Molecular characterization of T-DNA integration sites in transgenic birch. Trees 24, 753–762 (2010). https://doi.org/10.1007/s00468-010-0445-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00468-010-0445-6