Abstract
Nitrate and phenol often co-occur in wastewater because of the complex industrial and agricultural processes, while the impacts of phenol on autotrophic denitrification remain unclear. Here, a sulfur and hydrogen-oxidizing autotrophic denitrification reactor was established, and the effects of different concentrations of phenol on the nitrate removal performance, kinetics, microbial communities, and functional genes were investigated. Increasing concentrations of phenol significantly decreased the denitrification efficiency in the reactor. The kinetic data indicate the limitation of nitrate diffusion may be one of reasons. Increasing phenol concentrations declined the activities of nitrate and nitrite reductases and induced the production of reactive oxygen species (ROS) and the release of lactate dehydrogenase (LDH), suggesting potential toxicity to the denitrifying consortium. Denitrifying gene nirK was most sensitive to phenol stresses in the reactor. In addition, Thauera was the predominant genus in system with and without phenol, Bacillus was enriched under high phenol concentrations.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
With the rapid development of agricultural and industrial production, large quantities of chemicals have been released to the environment due to inappropriate waste management [36]. Among these chemicals, nitrate and phenol have been reported to cause environmental damage and public health problems [7, 9, 14]. Nitrate and phenol often coexist in wastewater discharged from pharmaceutical production, textile and petroleum refinery factory [17, 19]. 10 mg/L of nitrate (USEPA, 2010) and 2 mg/L of phenol are permitted in water considering human health [8].
Nitrate removal through microbial denitrification is widely used in wastewater treatment plant, and this process has been broadly studied [22, 38]. Autotrophic denitrification relies on inorganic sources of carbon and electron donors (e.g., hydrogen, ferrous iron, sulfur, bivalent manganese, etc.) for denitrifying bacteria to reduce nitrate to nitrogen gas [27, 31] while heterotrophic denitrification uses organic carbon and electron donors. Compared with heterotrophic denitrification, autotrophic denitrification has the advantage of low sludge production and avoiding secondary pollution caused by residual organics.
Presence of phenol causes complex effects on heterotrophic denitrification. At low concentrations, presence of phenol can promote the metabolism of heterotrophic denitrification process [20], whereas at high concentrations, it inhibits the denitrification process [15]. Toxicity of phenol to denitrifying consortium may be partially responsible [6]. However, the underlying inhibitory mechanisms have not been fully resolved. Further, the impacts of phenol stresses on the performance of autotrophic denitrification system and the community structure of the microbial consortium inside the reactors were rarely studied.
Accordingly, to improve understanding of how phenol stresses impact the nitrate removal efficiency and the microbial community structure in the reactor, we conducted a 6 months study with a custom-built autotrophic denitrification reactor and stepwisely increasing the concentrations of phenol in the system. The finding of this study not only facilitate the understanding of the effect of phenol on the autotrophic nitrate treatment process when phenol and nitrate coexist in the wastewater, but also provide possible strategy for removing nitrate and phenol simultaneously.
Materials and methods
Experimental setup
Custom-built autotrophic denitrification reactors were developed to study the effects of phenol on autotrophic denitrification process. The denitrification system was a continuous up-flow reactor, using a combination of a sulfur autotrophic unit and a bioelectrochemical hydrogen autotrophic unit to remove nitrate from the feeding influent (Fig. 1). The denitrification reactor was cylindrical, with a diameter of 16 cm and height of 15 cm (3 L), containing sulfur granules (5–8 mm) to a depth of 7.0 cm. A carbon fiber felt cathode (50.5 cm × 7.0 cm × 3.0 cm) and a carbon rod anode were also deployed in the upper part of the reactor and were connected to a DC power supply (0–30 V, 0–5 A). The reactor was inoculated with microorganisms from a similar autotrophic reactor used in our previous study [3]. Synthetic influent solution (sparged with Ar for 30 min to deoxygenate), composed of 30 mg/L of NO3−–N, 60 mg/L of HCO3−, 10 mg/L of MgCl2, 0.50 mg/L of ZnCl2, 2.00 mg/L of CoCl2, 1.00 mg/L of MnSO4, 0.30 mg/L of NiCl2, 0.30 mg/L of CuCl2, 0.20 mg/L of FeSO4, 0.50 mg/L of CaCl2, and 0.30 mg/L of Na2MoO4, was deoxygenated and continuously fed into the system throughout the experiment.
Experimental conditions
Once initiated, the denitrification system was stabilized under the following condition for a period of at least 60 days until 95% removal of nitrate in the effluent solution was achieved: hydrodynamic retention time (HRT) of 20 h, pH of 7.0, NO3−–N concentration of 30 mg/L, and a direct current of 50 mA. Following the stabilization stage, the operation conditions were adjusted, with a HRT of 16 h (selected by reviewing and comparing the HRT from previous related studies (in the range of 7–24 h) [5, 11, 12, 40] and our preliminary results), an applied current of 100 mA, a NO3−–N concentration of 100 mg/L, HCO3− concentration of 500 mg/L, while other parameters remained the same. Phenol was also added into the influent solution in a stepwise fashion—0 mg/L (Phase I), 20 mg/L (Phase II), 50 mg/L (Phase III), and 100 mg/L of (Phase IV)—and each phase was maintained for a period of 20 days. To further elucidate the fate of phenol in the reactor, inorganic carbon source HCO3− was removed from the influent while other conditions remained unchanged from Phase IV, and experiments were running for an additional 20 days. All experiments were run in triplicates and one-way analysis of variance (ANOVA) was applied to assess the statistical difference of the results at a significance level of p = 0.05.
Sampling and analysis
Sulfate concentrations and pH in the effluent, and concentrations of NO3−–N, NO2−–N, and phenol in the feeding influent and effluent were measured daily. 10 mL aqueous sample was collected and filtered by 0.45 µm membrane before measurement. At the end of each phase, biofilm samples on the surface of cathodes and sulfur granules were also obtained and mixed for pyrosequencing analysis (referred to M1, M2, M3, and M4 respectively). The concentrations of NO3−–N, phenol, NO2−–N, and SO42− were measured colorimetrically following the standard methods [1]. The pH of the effluent was determined using a pH meter (PHS-3C, Kexiao Instrument, China).
Nitrate removal and phenol removal kinetics
Previous studies demonstrated the Zero-order and Half-order kinetic models could effectively interpret the nitrate removal data in continuous up-flow biofilm reactors [13, 29]. As such, the nitrate and phenol removal data from Phase I to IV were fitted by these two models in this study.
Zero-order model: this kinetic model assumes no limitations for substrates and no mass transfer limitation on the biofilm in the reactor.
Half-order model: this kinetics model assumes the substrates concentrations are too high to penetrate the whole biofilm in the reactor (diffusion limitation of substrates).
Where, T is the time and T = 0 represents the commencement of each phase, C is the concentrations of nitrate or phenol in the effluent at time T, C0 is the initial nitrate/phenol concentrations, and K0v and K(1/2)v are the zero-order reaction and half-order reaction rate constants.
Pyrosequencing of biofilm samples
Community structure of biofilm mixture from cathodes and sulfur granules were measured at the end of each phase via high-throughput sequencing of 16S rRNA fragments. 1 mL of Genomic DNA in biofilm samples was extracted using Power Soil DNA isolation kit (MOBIO, USA). The quality and concentrations of DNA were examined using Qubit 2.0 DNA detection kit. The V3–V4 region of the bacterial 16S rRNA fragments was amplified using primers 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′). The PCR protocol was described previously [4]. The high-throughput sequencing were carried out on the Illumina Miseq platform by a commercial service (Shanghai Majorbio Bio-Pharm Technology Co., Ltd, China). The raw reads have been deposited into the NCBI sequencing reads archive (accession number: PRJNA476022).
Key enzyme activities and functional gene abundances of the biofilm samples
Lactate dehydrogenase (LDH) and reactive oxygen species (ROS) as indicators for cell toxicity were measured by Nanjing Jiancheng Bioengineering Institute and Rochen Pharma Co., Ltd, China, respectively. To quantifying potential toxicity of phenol for denitrification, activities of nitrate reductase (NR) and nitrite reductase (NIR), and the absolute abundances of denitrifying functional genes—nitrate reductase gene (narG and napA) and nitrite reductase gene (nirS and nirK) were determined by use of the method described in [4]. The experiments were performed in triplicates and the results were expressed as mean ± standard deviation.
Results and discussion
Reactor performance of nitrate removal under different phenol concentrations
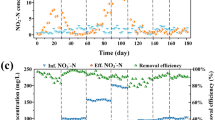
Increasing phenol concentrations in the system inhibited the denitrification and lowered the nitrate removal efficiency (Fig. 2a). Over the period of Phase I (without phenol), NO3−–N concentrations in the effluent were 3.53 ± 1.99 mg/L and this rendered nitrate removal efficiency of 99.64 ± 3.06% [corresponding rates for denitrification (DR): 146.46 ± 4.28 mg/(L·d)], indicating complete removal of nitrate from the feeding solution when phenol was absent. The DR was comparable with that of similar studies that using sulfur-based autotrophic reactor for denitrification, 150.72 mg/(L·d) [25] or 200 mg/(L·d) [37]. When phenol was present (Phase II, 20 mg/L of phenol in the feeding solution), NO3−–N concentrations in the effluent were slightly elevated (13.49 ± 6.80 mg/L), leading to nitrate removal efficiency of 95.51 ± 0.77% (DR: 139.57 ± 4.35 mg/(L·d)). Further increases in the phenol concentrations to 50 and 100 mg/L over the period of Phase III and IV increased both NO3−–N and NO2−–N concentrations in the effluent and decreased nitrate removal efficiency to 88.14 ± 1.58% (DR:127.39 ± 3.70 mg/(L·d)) and 73.80 ± 1.25% (DR:107.38 ± 3.50 mg/(L·d)), respectively. The strong negative correlation between nitrate removal efficiency (or DR) in the system and phenol concentrations in the feeding solution suggests presence of phenol inhibited the denitrification process in the system.
Concurrent removal of phenol was also observed in the denitrification system (Fig. 2b). Phenol concentrations were substantially lower in the effluent solution relative to the influent solution. The degradation rates of phenol increased from 21.73 ± 9.95 to 36.86 ± 8.44 and to 45.01 ± 11.34 mg/(L·d) with increasing phenol concentrations in the influent solution over the periods of Phase II, III, and IV, respectively.
Removal of HCO3− from the system significantly decreased the denitrification efficiency, but did not affect the removal of phenol (Fig. 2, Fig S1). In system without inorganic carbon source, phenol may also function as electron donor and provide carbon to the microbial denitrification (Supporting information). This hypothesis is further supported by decreasing sulfate concentrations in the effluent when HCO3− was removed from the system (Fig. S1c, Fig. 2c), which suggest that heterotrophic denitrification processes compete with sulfur-based autotrophic denitrification in the system.
Nitrate and phenol removal kinetics
Table 1 presents the Zero-order and Half-order kinetics results of nitrate and phenol removal from Phase I to IV. The nitrate removal rate constant K(1/2)v decreased from 7.62 (Phase I) to 3.89 (Phase IV) mg1/2/L1/2·h, suggesting phenol addition inhibited the nitrate removal. The calculated correlation coefficients of nitrate removal rates were better fitted by Half-order than Zero-order kinetics model, implying the decreased autotrophic denitrification rates may be restricted by the substrates diffusion in biofilm, probably caused by increased mass transfer resistance under stepwise increased phenol stresses [29].
Biofilm community succession in the denitrification reactor
Generally, a total of 1,629,333 high-quality sequences corresponding to 1230 OTUs were obtained (Table 2). All the coverage values of the samples were above 99%, indicating that the obtained sequences covered all the species in the reactor. The rarefaction curves (Fig. S3) of the biofilm samples suggested the sequencing samples were representative in the reactor. As indicated by the Venn diagram (Fig. S4), the biofilm samples M1, M2, M3 and M4 shared 174 OTUs, which accounted for 40.56% of the total OTUs obtained from all biofilm samples, suggesting that the microbial community composition shifts under different phenol stresses.
The OTU numbers of M2, M3 and M4 were substantially higher than that of M1, suggesting that the presence of phenol in the system enhanced the richness of microbial community. However, the OTUs number in Phase IV (100 mg/L of phenol) was slightly lower relative to that in Phase II and III (20 and 50 mg/L of phenol). The diversity index also showed consistent trends—the Shannon index increased with increasing phenol concentrations from Phase I to III, but declined at much higher phenol concentration (Phase IV).
The microbial communities in the denitrification system were analyzed at the phyla, class and genus levels. The top five most-abundant microbial phyla and the top 12 most-abundant bacterial classes under increasing phenol stresses are shown in Fig. 3. The predominant phyla of the biofilm communities were Proteobacteria, followed by Firmicutes, Bacteroidetes and Tenericutes, which comprised autotrophic and heterotrophic denitrifying bacteria [28, 33, 34]. Specifically, Proteobacteria and Firmicutes accounted for 69.58% and 13.73% in Phase I (without phenol), 80.86% and 7.68% in Phase II (20 mg/L of phenol), 56.89% and 33.06% in Phase III (50 mg/L of phenol), and 68.24% and 27.33% for Phase IV (100 mg/L of phenol). These results suggested that relative moderate phenol concentration (20 mg/L) enriched Proteobacteria, while Firmicutes was enriched at higher phenol concentrations (50–100 mg/L). Since Proteobacteria and Firmicutes were reported as denitrification and phenol degradation-related bacteria [21, 41], these two phyla identified in the current system may be responsible for denitrification and degradation of phenol in the reactor.
At class level, Betaproteobacteria, Clostridia, Epsilonproteobacteria, Alphaproteobacteria, Gammaproteobacteria, Bacilli, Deltaproteobacteria and Flavobacteriia were the most-abundant bacteria classes (Fig. 3b). Betaproteobacteria was the most predominant class in the system throughout the experiment, and did not show significant variations with increasing phenol concentrations. However, the relative abundance of the classes Bacilli, Clostridia, Alphaproteobacteria and Deltaproteobacteria increased with increasing phenol concentrations, suggesting that these classes were tolerant to phenol and may also be the potential phenol degradation contributors in the reactor [28, 33]. In contrast, the relative abundances of Epsilonproteobacteria and Gammaproteobacteria decreased with increasing phenol concentrations, suggesting that these two classes were vulnerable to the phenol stresses and may be out selected in the system.
The microbial community structure at the genus level under different phenol stresses is shown via a heatmap (Fig. 4) The dendrograms indicated that the community structure of biofilm not exposed to phenol (M1) was distinctly different from that of the biofilm exposed to phenol (M2–M4), while biofilm exposed to high phenol concentrations (M3 and M4) had similar community structure. Thauera was the most predominant genus, which was reported to be autotrophic/mixotrophic denitrification bacteria genus [24, 39]. When phenol concentrations increased stepwisely from 0 to 20, 50 and 100 mg/L, the relative abundance of Thauera decreased from 32.12 to 11.13%, and then increased to 18.43% and 29.36%, suggesting that the denitrification reactor may have potential adaption to phenol stresses. In addition, Thiobacillus, Azoarcus, Pseudomonas and Sulfurimonas were also the predominant genera in the denitrification reactor. These genera comprised mixotrophic denitrifying bacteria or phenol degradation bacteria [39, 41]. Enrichment of the genera Pseudomonas and Sulfurimonas, which were reported as autotrophic/heterotrophic denitrifying genera, were favored under lower phenol concentrations [28, 39]. However, Bacillus was enriched with increasing phenol concentrations, which might be responsible for the phenol degradation [41].
Enzyme activities, ROS production and LDH release under phenol stresses
NR and NIR are important enzymes involved in the denitrification process, which are related to the reduction of nitrate and nitrite in the system respectively. Overall, the specific activities of NR and NIR enzymes declined with increasing phenol concentrations (R2 = 0.99, p = 0.0008; R2 = 0.95, p = 0.016) (Fig. 5a), suggesting that presence of phenol in the system inhibited denitrification processes by decreasing the activities of NR and NIR enzymes. The activities of NIR in M3 (Phase III, 50 mg/L of phenol) and M4 (Phase IV, 100 mg/L of phenol) were significantly lower relative to the NIR activity in M1 (Phase I, 0 mg/L of phenol). In contrast, the activity of NR was significantly lower in M4 relative to that in M1. These may suggest that the reduction of nitrite was more sensitive to the presence of phenol in the system than the reduction of nitrate. This was consistent with the observation that NO2−–N was detected in the effluent solution at Phase III and IV, which indicated incomplete removal of nitrite in the system at higher phenol concentrations (Fig. 2a).
ROS production and LDH release are indicators frequently associated with biotoxicity and the integrity of cytomembrane, respectively [23, 30, 32]. Compared to M1 in Phase I without phenol, the ROS production in M2, M3, and M4 increased by 1.67 ± 0.68, 3.04 ± 0.32 and 5.02 ± 0.49 times (p < 0.05), indicating increasing phenol concentrations in the system facilitated the production of ROS in the microorganisms. The LDH release also showed similar trends - the LDH release increased by 19.05 ± 2.48%, 36.21 ± 4.96% and 75.34 ± 3.47% in M2, M3, and M4 relative to the LDH release in M1 (Fig. 5b). This indicated that increasing phenol concentrations facilitated the release of LDH and caused destruction in the integrity of the cytomembrane.
Denitrifying functional gene abundances under phenol stresses
The quantitative real time PCR revealed the variations in absolute abundances of four important denitrifying genes—nitrate reductase gene (narG and napA) [2, 26] and nitrite reductase gene (nirS and nirK) [18, 35] under different phenol stresses in the reactor (Fig. 6). The abundance of narG decreased from 5.37 ± 0.31 × 104 to 2.02 ± 0.23 × 104 copies/ng DNA, while the abundance of napA did not decline. The results differed with Miao et al. [16], in which the abundance of napA significantly decreased with increasing Cd(II) stress, probably because the napA-gene containing bacteria could withstand phenol stress to some extent (present study). Additionally, the abundances of nirS and nirK declined from 5.62 ± 0.18 × 106 and 7.82 ± 0.30 × 105 to 9.02 ± 0.61 × 105 and 2.52 ± 0.25 × 105 copies/ng DNA, respectively, with increasing phenol concentrations. The decreased copy number of nitrite reductase gene would probably hinder the nitrite reduction process [10], as Fig. 2a shown, the nitrite started accumulating from the phase III. Furthermore, the strongest negative correlation between phenol concentration and the abundance of nirK indicated nirK was most sensitive to phenol stress.
Conclusions
The present study investigated the effects of different phenol concentrations on the performance of denitrification and degradation of phenol in an autotrophic denitrification system. The microbial community structure at phyla, class and genus levels under different phenol stresses were also determined. Increasing phenol concentrations decreased the efficiency of denitrification. Increasing in the ROS production and the LDH release with increasing phenol concentrations suggested that increasing phenol concentrations in the system exerted potential toxicity to the microorganism. Shifting from predominantly autotrophic denitrification to mixotrophic denitrification compromised the removal efficiency of nitrate with the capability of phenol degradation. Microbial community analyses results supported this observation by showing enrichment of phenol degradation bacteria in the system with increasing phenol stresses.
References
APHA (2005) Standard methods for the examination of water and wastewater, 21st edn. American Public Health Association, Washington DC
Bru D, Sarr A, Philippot L (2007) Relative abundances of proteobacterial membrane-bound and periplasmic nitrate reductases in selected environments. Appl Environ Microb 73:5971–5974
Chen D, Wang H, Yang K, Ma F (2018) Performance and microbial communities in a combined bioelectrochemical and sulfur autotrophic denitrification system at low temperature. Chemosphere 193:337–342
Chen D, Yang K, Wei L, Wang H (2016) Microbial community and metabolism activity in a bioelectrochemical denitrification system under long-term presence of p-nitrophenol. Bioresour Technol 218:189–195
Demirel S, Uyanık 0, Yurtsever A, Çelikten H, Uçar D (2014) Simultaneous bromate and nitrate reduction in water using sulfur-utilizing autotrophic and mixotrophic denitrification processes in a fixed bed column reactor. Clean Soil Air Water 42:1185–1189
Eiroa M, Vilar A, Amor L, Kennes C, Veiga MC (2005) Biodegradation and effect of formaldehyde and phenol on the denitrification process. Water Res 39:449–455
El-Naas MH, Al-Zuhair S, Alhaija MA (2010) Removal of phenol from petroleum refinery wastewater through adsorption on date-pit activated carbon. Chem Eng J 162:997–1005
EPA (2014) Draft update of human health ambient water quality criteria: Phenol 108-95-2, 2014, Office of Water & Office of Science and Technology, United States Environmental Protection Agency, EPA 820-D-14-060
García-Martínez Y, Chirinos J, Bengoa C, Stüber F, Font J, Fortuny A, Fabregat A (2017) Nitrate removal in an innovative up-flow stirred packed-bed bioreactor. Chem Eng Process Process Intensif 121:57–64
Guo G, Deng H, Qiao M, Yao H, Zhu Y (2013) Effect of long-term wastewater irrigation on potential denitrification and denitrifying communities in soils at the watershed scale. Environ Sci Technol 47:3105–3113
Hwang JH, Cicek N, Oleszkiewicz JA (2009) Inorganic precipitation during autotrophic denitrification under various operating conditions. Environ Technol 30:1475–1485
Kim I, Oh S, Bum M, Lee J, Lee S (2002) Monitoring the denitrification of wastewater containing high concentrations of nitrate with methanol in a sulfur-packed reactor. Appl Microbiol Biot 59:91–96
Koenig A, Liu LH (2001) Kinetic model of autotrophic denitrification in sulphur packed-bed reactors. Water Res 35:1969–1978
Li M, Li R, Feng C, Xi B, Chen N, Jiang Y, Zhao Y, Zhao B, Dang Q (2017) Nitrate removal efficiency of a mixotrophic denitrification wall for nitrate-polluted groundwater in situ remediation. Ecol Eng 106:523–531
Miao Y, Wang Z, Liao R, Shi P, Li A (2017) Assessment of phenol effect on microbial community structure and function in an anaerobic denitrifying process treating high concentration nitrate wastewater. Chem Eng J 330:757–763
Miao Y, Zhang X, Jia S, Liao R, Li A (2018) Comprehensive analyses of functional bacteria and genes in a denitrifying EGSB reactor under Cd(II) stress. Appl Microbiol Biot 102:8551–8560
Nowee SM, Taherian M, Salimi M, Mousavi SM (2017) Modeling and simulation of phenol removal from wastewater using a membrane contactor as a bioreactor. Appl Math Model 42:300–314
Oakley BB, Francis CA, Roberts KJ, Fuchsman CA, Srinivasan S, Staley JT (2007) Analysis of nitrite reductase (nirK and nirS) genes and cultivation reveal depauperate community of denitrifying bacteria in the Black Sea suboxic zone. Environ Microbiol 9:118–130
Pan GT, Pan KL, Chen DL, Chong S, Chang MB (2017) Removal of phenol from gas streams via combined plasma catalysis. J Ind Eng Chem 52:108–120
Pereira AD, Leal CD, Dias MF, Etchebehere C, Chernicharo CAL, de Araújo JC (2014) Effect of phenol on the nitrogen removal performance and microbial community structure and composition of an anammox reactor. Bioresour Technol 166:103–111
Ren L, Chen R, Zhang X, Shao J, He Y (2017) Phenol biodegradation and microbial community dynamics in extractive membrane bioreactor (EMBR) for phenol-laden saline wastewater. Bioresour Technol 244:1121–1128
Roques C, Aquilina L, Boisson A, Vergnaud-Ayraud V, Labasque T, Longuevergne L, Laurencelle M, Dufresne A, de Dreuzy J, Pauwels H, Bour O (2018) Autotrophic denitrification supported by biotite dissolution in crystalline aquifers: (2) transient mixing and denitrification dynamic during long-term pumping. Sci Total Environ 619–620:491–503
Rossi SP, Windschüttl S, Matzkin ME, Rey-Ares V, Terradas C, Ponzio R, Puigdomenech E, Levalle O, Calandra RS, Mayerhofer A, Frungieri MB (2016) Reactive oxygen species (ROS) production triggered by prostaglandin D2 (PGD2) regulates lactate dehydrogenase (LDH) expression/activity in TM4 Sertoli cells. Mol Cell Endocrinol 434:154–165
Rungkitwatananukul P, Nomai S, Hirakata Y, Pungrasmi W, Puprasert C, Hatamoto M, Yamaguchi T (2016) Microbial community analysis using MiSeq sequencing in a novel configuration fluidized bed reactor for effective denitrification. Bioresour Technol 221:677–681
Sahinkaya E, Dursun N, Kilic A, Demirel S, Uyanik S, Cinar O (2011) Simultaneous heterotrophic and sulfur-oxidizing autotrophic denitrification process for drinking water treatment: control of sulfate production. Water Res 45:6661–6667
Smith CJ, Nedwell DB, Dong LF, Osborn AM (2007) Diversity and abundance of nitrate reductase genes (narG and napA), nitrite reductase genes (nirS and nrfA), and their transcripts in estuarine sediments. Appl Environ Microb 73:3612–3622
Su J, Liang D, Lian T (2018) Comparison of denitrification performance by bacterium Achromobacter sp. A14 under different electron donor conditions. Chem Eng J 333:320–326
Su JF, Cheng C, Huang T, Wei L (2017) Performance of the dominant bacterial species and microbial community in autotrophic denitrification coupled with iron cycle in immobilized systems. Mar Pollut Bull 117:88–97
Wan D, Liu Y, Wang H, Wang Y, Xiao S (2017) Simultaneous bio-autotrophic reduction of perchlorate and nitrate in a sulfur packed bed reactor: Kinetics and bacterial community structure. Water Res 108:280–292
Wang S, Wang X, Li Z, Gao F, Gao M, She Z, Guo L, Zheng D, Zhao Y, Ma B (2017) Long-term effects of nickel oxide nanoparticles on performance, microbial enzymatic activity, and microbial community of a sequencing batch reactor. Chemosphere 169:387–395
Wei Y, Dai J, Mackey HR, Chen G (2017) The feasibility study of autotrophic denitrification with iron sludge produced for sulfide control. Water Res 122:226–233
Wu Q, Huang K, Sun H, Ren H, Zhang X, Ye L (2018) Comparison of the impacts of zinc ions and zinc nanoparticles on nitrifying microbial community. J Hazard Mater 343:166–175
Xu D, Xu D, Xu P, Xiao E, Lin L, Zhou Q, Wu Z (2017) Bacterial community and nitrate removal by simultaneous heterotrophic and autotrophic denitrification in a bioelectrochemically-assisted constructed wetland. Bioresour Technol 245:993–999
Xu D, Xu D, Xu P, Xiao E, Zhou Y, Zhou Q, He F, Wu Z (2017) Performance and microbial communities of completely autotrophic denitrification in a bioelectrochemically-assisted constructed wetland system for nitrate removal. Bioresour Technol 228:39–46
Yan T, Fields MW, Wu L, Zu Y, Tiedje JM, Zhou J (2003) Molecular diversity and characterization of nitrite reductase gene fragments (nirK and nirS) from nitrate- and uranium-contaminated groundwater. Environ Microbiol 5:13–24
Yavuz Y, Ögütveren ÜB (2018) Treatment of industrial estate wastewater by the application of electrocoagulation process using iron electrodes. J Environ Manag 207:151–158
Zeng H, Zhang TC (2006) Development of a response surface for prediction of nitrate removal in sulfur–limestone autotrophic denitrification fixed-bed reactors. J Environ Eng 132:1068–1072
Zhai S, Ji M, Zhao Y, Zhao Q, Pavlostathis SG (2018) Effects of salinity and COD/N on denitrification and bacterial community in dicyclic-type electrode based biofilm reactor. Chemosphere 192:328–336
Zhang C, Zhang L, Hu C, Liu H, Bai Y, Qu J (2015) Sulfur-based mixotrophic denitrification corresponding to different electron donors and microbial profiling in anoxic fluidized-bed membrane bioreactors. Water Res 85:422–431
Zhao Y, Feng C, Wang Q, Yang Y, Zhang Z, Sugiura N (2011) Nitrate removal from groundwater by cooperating heterotrophic with autotrophic denitrification in a biofilm–electrode reactor. J Hazard Mater 192:1033–1039
Zhou H, Wang G, Wu M, Xu W, Zhang X, Liu L (2018) Phenol removal performance and microbial community shift during pH shock in a moving bed biofilm reactor (MBBR). J Hazard Mater 351:71–79
Acknowledgements
The present study was financially supported by the Major Project of the Ministry of Science and Technology of China (the 13th 5-year Plan) (2018ZX07208-004), and Natural Science Foundation of Jiangsu Province, China (BK20180716 and BK20180719).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xia, T., Xie, M., Chen, D. et al. Impact of phenol on the performance, kinetics, microbial communities and functional genes of an autotrophic denitrification system. Bioprocess Biosyst Eng 42, 1105–1114 (2019). https://doi.org/10.1007/s00449-019-02108-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-019-02108-7