Abstract
Extracellular matrix (ECM) plays a dynamic role during tissue development and re-growth. Body part regeneration efficiency relies also on effective ECM remodelling and deposition. Among invertebrates, echinoderms are well known for their striking regenerative abilities since they can rapidly regenerate functioning complex structures. To gather insights on the involvement of ECM during arm regeneration, the brittle star Amphiura filiformis was chosen as experimental model. Eight ECM genes were identified and cloned, and their spatio-temporal and quantitative expression patterns were analysed by means of whole mount in situ hybridisation and quantitative PCR on early and advanced regenerative stages. Our results show that almost none of the selected ECM genes are expressed at early stages of regeneration, suggesting a delay in their activation that may be responsible for the high regeneration efficiency of these animals, as described for other echinoderms and in contrast to most vertebrates. Moreover, at advanced stages, these genes are spatially and temporally differentially expressed, suggesting that the molecular regulation of ECM deposition/remodelling varies throughout the regenerative process. Phylogenetic analyses of the identified collagen-like genes reveal complex evolutionary dynamics with many rounds of duplications and losses and pinpointed their homologues in selected vertebrates. The study of other ECM genes will allow a better understanding of ECM contribution to brittle star arm regeneration.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Regeneration —the replacement of lost body parts— is widespread in the animal kingdom occurring in both invertebrates (Bosch 2007; Saló et al. 2009) and vertebrates (Brockes and Kumar 2002; Gemberling et al. 2013). However, the efficiency and extent of this ability vary between species (Tsonis 2000; Brockes and Kumar 2008; Bely and Nyberg 2009). Wound healing, tissue remodelling, cell apoptosis and proliferation, differentiation and dedifferentiation, and morphogenesis are just a few of the many different events that need to be finely regulated during the regenerative process (King and Newmark 2012).
Extracellular matrix (ECM) plays a pivotal and dynamic role at different levels of animal body part re-growth, providing structural and mechanical stability during the formation of new tissues and organs and supporting cell migration, adhesion and proliferation (Bonnans et al. 2014; Swinehart and Badylak 2016; Keane et al. 2018). The interactions between ECM and cells control tissue development and structural integrity and guarantee maintenance and self-renewal (Gelse et al. 2003). As evidence for this, in tissue engineering and regenerative medicine ECM scaffolds are commonly used to support the correct cell proliferation and differentiation of regenerating tissues (Yi et al. 2017; Sheehy et al. 2018).
ECM comprises several types of macromolecules which form, among the others, “filling tissues” and basal laminae. These molecules include, for example, fibrillar and non-fibrillar collagens, fibronectins, elastins, tenascins, integrins, fibrillins, glycosaminoglycans, proteoglycans and laminins, which all play different structural and functional roles (Alberts et al. 2002; Hynes 2009).
During regeneration, the remodelling of pre-existing and newly formed ECM, which involves secreted enzymes, such as metalloproteases (MMPs) and tissue inhibitors of the MMPs (TIMPs), is a fundamental event (Lu et al. 2011; Bonnans et al. 2014; Arpino et al. 2015). Among the most extensively remodelled ECM molecules are laminins, one of the main components of basal laminae; these molecules are known to be involved in the regenerative process of different tissues (Govindan and Iovine 2015; Rousselle et al. 2018).
In vertebrates, ECM is deposited during the early phase of tissue repair usually prior to cell proliferation (Diegelmann and Evans 2004; Godwin et al. 2014). Importantly, over-deposition of ECM in the connective tissue is known to impair or greatly reduce proper tissue regeneration (Bock and Mrowietz 2002; Rahban and Garner 2003; Diegelmann and Evans 2004; Ben Amar and Bianca 2016).
Among invertebrates, echinoderms are non-chordate deuterostomes that possess striking regenerative abilities, being able to regenerate nearly all tissue types and large body parts, such as the nervous system, appendages and more (Candia Carnevali 2006). Brittle stars, starfish and crinoids have been used to investigate mainly whole body part (i.e. arm) regenerative processes (Ben Khadra et al. 2015a, b, 2018; Czarkwiani et al. 2016) and to identify cellular and molecular mechanisms underpinning their extensive regenerative ability which contrasts with the limited ability of almost all vertebrates (Nye et al. 2003).
In echinoderm regeneration after traumatic amputation, the ECM of both stump and newly formed tissues is actively involved (Quiñones et al. 2002; Cabrera-Serrano and García-Arrarás 2004; Miao et al. 2017; Ferrario et al. 2018). In contrast to mammals, delay in collagen deposition has been observed during the sea cucumber repair phase (Quiñones et al. 2002). Furthermore, neither scar formation nor fibrosis have been detected during wound healing stages in any echinoderm so far investigated (Quiñones et al. 2002; Ferrario et al. 2018). Therefore, it has been suggested that the delay in ECM deposition and absence of scar tissue may partly explain the regenerative efficiency of echinoderms (Quiñones et al. 2002; Ferrario et al. 2018) as also described for the few regeneration-competent vertebrates (Erickson and Echeverri 2018). Recent morphological and ultrastructural studies comparing arm regeneration in a starfish, Echinaster sepositus, and in a brittle star, Amphiura filiformis, have confirmed the absence of collagen over-deposition in the repair phase (Ferrario et al. 2018). Furthermore, in A. filiformis, the initial collagen deposition after arm injury occurs at the end of the repair phase, 72 hours post amputation (hpa) (Ferrario et al. 2018), which coincides with the beginning of cell proliferation detected by EdU staining (Czarkwiani et al. 2016).
A. filiformis has been used to investigate mainly complex body part regeneration, due to its ability to re-grow a functional arm in a short time. Little is known about the contribution of ECM to regeneration at the molecular level in this species. A collagen gene (Afi-αcoll) has been identified as a marker of skeletal differentiation (Czarkwiani et al. 2013). Another study of the wound healing stage (1–3 days post-amputation (dpa)) using transcriptomic and proteomic approaches has shown that a collagen IV-like (the main component of the vertebrate basal lamina) and the glycoprotein fibronectin are down-regulated (Purushothaman et al. 2015) in this early phase, whereas collagen transcripts are present at high level during early stages of regeneration (7 dpa; Burns et al. 2011). These findings suggest an overall modulation of ECM genes during regeneration that needs to be further investigated in detail.
To this end, in this study, we identify eight ECM genes from an A. filiformis reference transcriptome (Dylus et al. 2018) and we analyse their spatial-temporal and quantitative expression during arm regeneration through whole mount in situ hybridisation (WMISH) and quantitative PCR (Q-PCR). Our results show an overall delay of ECM gene activation and a differential spatial gene expression pattern at advanced regenerative stages.
Materials and methods
No specific permits were required since the brittle star Amphiura filiformis is not an endangered or protected species.
Animal collection and maintenance
Adult specimens of A. filiformis were collected at the Sven Lovén Centre for Marine Sciences in Kristineberg (Sweden) and transported to London where they were kept at 14°C in tanks of filtered artificial sea water (ASW) with 34‰ salinity (Instant Ocean®). Animals were left to acclimatise around 1 week before regeneration tests were conducted (see below). Specimens were fed twice a week with Microvore Microdiet (Brightwell Aquatics) and ASW parameters were constantly checked and adjusted when necessary.
Arm regeneration tests
Specimens were anaesthetised in 3.5% MgCl2(6H2O) solution (pH 8.3) in a 1:1 mix of filtered ASW and milliQ water. Two arms per animal were amputated at 1 cm from the disc by cutting with a sharp scalpel between two subsequent segments under a stereomicroscope. Animals were then left to regenerate until they reached the desired stages (Dupont and Thorndyke 2006; Czarkwiani et al. 2016) when they were collected together with two or three segments of the stump for further analyses (see below).
Identification of A. filiformis extracellular matrix genes
Collagen-like and other ECM genes of interest were selected in the following ways:
- gene name/word search in sea urchin Strongylocentrotus purpuratus and starfish Patiria miniata genomes using EchinoBase (www.echinobase.org; Cameron et al. 2009; Kudtarkar and Cameron 2017);
- recent publications on other echinoderms (Ortiz-Pineda et al. 2009; Sun et al. 2011; Czarkwiani et al. 2013; Mashanov et al. 2014; Purushothaman et al. 2015).
Selected sequences from other echinoderms were used to search in the A. filiformis reference transcriptome (Dylus et al. 2018) using BLAST-X. The best-BLAST-hit of A. filiformis was then used in a BLAST-X search on EchinoBase and on the non-redundant (NR) National Center for Biotechnology Information (NCBI) database (http://blast.ncbi.nlm.nih.gov/) to confirm their identity as ECM genes.
Phylogenetic analyses of collagen genes
Homologous relationships for five collagen-like genes (called Afi-col-L A, Afi-col-L B, Afi-col-L C, Afi-col-L D and Afi-αcoll, see below) were identified using reciprocal BLAST of A. filiformis against S. purpuratus (Dylus et al. 2018). In-depth gene families were obtained by querying the five collagen candidates in hierarchical orthologous groups (HOGs) of an orthologous matrix algorithm (OMA) run consisting of 33 species (17 echinoderms) (Altenhoff et al. 2018). For each group, a tree was built using a MAFFT alignment (v7.305b) and FastTree (2.1.9). Homology was defined by proximity on the tree to closest vertebrate species.
Cloning and probe synthesis
To isolate fragments containing the desired genes, total A. filiformis RNA was extracted from different embryonic and/or adult arm regenerating stages and first-strand cDNA was synthesised as described in Czarkwiani and co-workers (Czarkwiani et al. 2013). This was used to amplify specific fragments by PCR using specific primers (Table S1) designed with the PRIMER3 Software version 4.0.0 (http://primer3.ut.ee/). The following parameters from default setting were implemented: max Poly-X = 3 and max 3’ Stability = 8. Purified PCR products for each gene of interest were subsequently ligated in pGEM®-T Easy Vector System I (Promega) and then transformed in Subcloning Efficiency Invitrogen DH5α Competent Cells (Life Technologies). Colonies containing the correct recombinant plasmids were selected by PCR and confirmed by sequencing (Source BioScience). Table S2 summarises clone information.
Digoxigenin (DIG) labelled RNA antisense probes were transcribed in vitro from each clone using Sp6/T7 Transcription Kit (Roche) and DIG-labelling mix (Roche) according to manufacturer’s instructions.
Whole mount in situ hybridisation (WMISH)
Samples of A. filiformis at various regenerative stages were fixed in 4% paraformaldehyde (PFA) in 1× phosphate buffer saline (PBS with 0.1% Tween-20; PBT) overnight at 4°C and stored in 100% methanol at −20°C until use.
WMISH was performed with the antisense probes newly synthesised along with positive control (Afi-c-lectin; see Czarkwiani et al. 2016 and Supplementary Materials) as described by Ferrario and co-workers (Ferrario et al. 2018). For each regenerative stage, at least three different individuals in independent biological replicas were used to test each RNA antisense probe with the following parameters: hybridisation time of 5–7 days, temperature of 50–55°C and probe concentration of 0.02–0.04 ng/μl. WMISH parameters are detailed in figure captions. After WMISH, samples were stored in 50% glycerol at 4°C and observed under a Zeiss AxioImager M1 microscope equipped with a Zeiss AxioCam HRc camera.
Post in situ sectioning
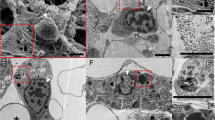
After whole mount imaging, hybridised samples were treated as described by Ferrario and co-workers (Ferrario et al. 2018) to gain a better resolution of the tissue-specific expression. Briefly, samples were decalcified for 1–2 days at 4°C in 0.5 M EDTA in 1× PBS (pH 8) or in 1:1 solution (v/v) of 2% L-ascorbic acid and 0.3 M NaCl in distilled water, then embedded in paraffin wax following standard procedures and sectioned using a rotary microtome (Leitz 1512). Xylene was used to remove paraffin from thick sections (10 μm), which were then mounted with Eukitt® and observed under a Jenaval light microscope provided with a DeltaPix Invenio 3S 3M Pixel CMOS camera and a DeltaPix ViewerLE Software.
Histology
Non-regenerating and regenerating samples were fixed, paraffin embedded, sectioned and stained with Milligan’s trichrome technique (Milligan 1946) as described in Czarkwiani and co-workers (Czarkwiani et al. 2016). Slides were mounted and observed as previously mentioned.
Results
Identification of collagen and other ECM genes in A. filiformis
To obtain a better understanding of collagen gene expression in brittle star arm regeneration, we collected more than 30 collagen-like genes from the echinoderm literature and from EchinoBase (see “Materials and methods”) and cross-referenced them with an A. filiformis transcriptome (Dylus et al. 2018). In this way, we identified a total of 9 sequences. In parallel, to be as comprehensive as possible, we searched the translated proteome with HMM (hidden Markov model) profile from Pfam (http://pfam.xfam.org/family/PF01391). This search identified 135 different transcripts of which 112 included either an orthologue or homologue to a total of 38 sea urchin genes, indicating the complexity of the collagen gene complement encoded in the A. filiformis genome and potentially involved in regeneration.
In this study, we focused on one previously identified collagen gene (Afi-αcoll; Czarkwiani et al. 2013) and on 4 as yet uncharacterised genes (here referred to as collagen-like domain containing genes: Afi-col-L A, Afi-col-L B, Afi-col-L C, Afi-col-L D), which were successfully cloned from the identified 9 selected genes and further analysed (see below). To characterise this subset of genes in terms of architecture and homology to known collagen genes, we used a bioinformatics approach (Table 1). We employed InterProScan (Jones et al. 2014) and NCBI Conserved Domain Architecture Retrieval Tool (cDART; Geer et al. 2002) to identify conserved domain architecture. We also performed a hierarchical orthologous group (HOG) analysis to better understand the homology relationships of these four genes and the Afi-αcoll previously described (Czarkwiani et al. 2013). Orthology matrix algorithm (OMA; Altenhoff et al. 2018) was run for 33 species including at least two species from each echinoderm class.
It is important to underline that Afi-col-L B, Afi-col-L C, Afi-col-L D and Afi-αcoll were classified as fibril-forming collagens (Fig. S1) and that all sequences but Afi-col-L C contained a collagen triple helix region containing multiple repeats of collagen-like domain (coll; Fig. S1). Other information about these genes is presented in the Supplementary Materials.
The overall results of the phylogenetic analyses shown in Fig. S2 highlight the complex clade-specific gene duplication and loss occurring in the evolution of collagen genes, which make it difficult to unequivocally identify collagen genes orthologous to those of the established mammalian classes.
In addition to collagen, other molecules are important for ECM formation and remodelling during regeneration, such as laminins, tissue inhibitors of metalloproteases (TIMPs) and others. Therefore, we searched for other ECM genes using a similar strategy as for collagen-like genes (see above). We collected more than 50 ECM genes from the echinoderm literature and from EchinoBase (see “Materials and methods”) and cross-referenced them with an A. filiformis transcriptome (Dylus et al. 2018). We identified a total of 26 sequences and we focused on the 3 as yet uncharacterised genes that have been successfully cloned and further analysed (2 laminin subunits and 1 TIMP encoding genes). The best-BLAST-hits in EchinoBase (SPU Best BLAST) and in NCBI (NCBI Best BLAST) are listed in Table 1.
Taken together, this initial analysis is in agreement with what described on the complexity of ECM genes in other echinoderms.
ECM gene expression during A. filiformis arm regeneration
To determine if the identified ECM genes are expressed in the process of arm regeneration, we studied their spatial and quantitative expression at different regenerative stages and in the stump tissues using WMISH and Q-PCR. Presence/absence of expression in the stump tissues was evaluated since the stump differentiated tissues are comparable to the normal tissues and they can therefore provide a good indication of the expression of the selected genes in non-regenerating conditions. Furthermore, we performed post in situ sectioning to gain a better resolution of the tissues expressing the selected genes.
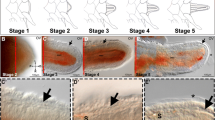
We initially performed a basic histological analysis to characterise the main tissues and structures of the brittle star arm that will regenerate post injury. Figure 1 shows the experimental model (Fig. 1a–d), anatomical diagrams (Fig. 1e–g) and thick paraffin sections of non-regenerating arms of A. filiformis (Fig. 1h–p). As in all ophiuroids, the arm is subdivided into repetitive segments (Hyman 1955; Czarkwiani et al. 2013, 2016). Skeletal elements (i.e. vertebrae, arm plates and spines) and muscle bundles are sequentially repeated in each segment, whereas three main structures run longitudinally along the whole arm, namely the aboral coelomic cavity (ACC), the radial water canal (RWC), with its associated podia and accessory vesicles, and the orally located radial nerve cord (RNC) with its adjacent sinuses. The distal arm-tip has a terminal cap (or ossicle) and terminal podium (Fig. 1d, e).
Amphiura filiformis anatomy. a–d Stereomicroscopy; e–g diagrams (see colour legend embedded in the figure for the labelling of the different tissues; the epidermis is shown in black); h–p thick paraffin sections (Milligan’s trichrome staining) of non-regenerating arms. Connective tissue (collagen) is stained green/blue, cells are stained pink/violet. a Aboral view of an adult regenerating specimen of the brittle star Amphiura filiformis (arrows = regenerating arms). b Aboral view of the proximal region of a non-regenerating arm. c Oral view of the proximal region of a non-regenerating arm. d Aboral view of the distal tip of a non-regenerating arm (arrow = terminal podium). e–g Diagrams of a non-regenerating arm of A. filiformis. e Sagittal (longitudinal) section diagram (arrow = terminal podium). f Diagrammatic cross section at the level of the proximal region of the arm. g Diagrammatic cross section at the level of the distal tip in the growth zone underneath the terminal ossicle. h Sagittal section showing the gross anatomy of an arm with its main tissues and structures. i Sagittal section of the oral region of an arm showing the epidermis (arrow). j Frontal section of a spine in which the central nerve is visible. k Cross section of a vertebra. l Frontal section of a lateral arm plate (arrow). m Sagittal section of the aboral coelomic cavity (arrow). n Sagittal section of the radial nerve cord (arrow) with its adjacent sinuses. o Sagittal section of the radial water canal (arrow) with an accessory vesicle. p Frontal section of the podium. Abbreviations: acc-aboral coelomic cavity; ap-aboral arm plate; av-aboral view; ct-connective tissue; cs-cross section; d-distal region; dcs-distal cross section; fs-frontal section; m-muscle; ov-oral view; op-oral arm plate; p-proximal region; p-podium; pcs-proximal cross section; rnc-radial nerve cord; rwc-radial water canal; ss-sagittal section; v-vertebra. Scale bars: h 100 μm; i–p 50 μm
All these structures are reformed during different regenerative stages (Dupont and Thorndyke 2006; Biressi et al. 2010; Czarkwiani et al. 2016). Specifically, after a wound healing phase (stage 1), around 5 days post-amputation (dpa), the early regenerative phase begins (stage 2), characterised by the appearance of the regenerative bud with the first outgrowths of the three main longitudinal structures (Fig. S3a, c). At stage 3/4 (around 8 dpa), complex structures regenerate and first signs of new segments at the proximal region of the new arm-tip are visible (Fig. S3b, d). Stage > 50% DI (after 2–3 weeks post-amputation (wpa)) is an advanced regenerative phase with new segments regenerated following a proximal-distal gradient as explained by the distalisation-intercalary regeneration model (Czarkwiani et al. 2016).
Expression of ECM genes during early arm regeneration (stage 2)
At stage 2, when the regenerative bud starts to appear and cells are already actively proliferating (Czarkwiani et al. 2016), only Afi-col-L B is expressed and localised in the ACC epithelium (Fig. 2a, f, k), whereas none of the other collagen-like genes show positive staining (Afi-col-L A (Fig. 3a, e, j); Afi-col-L C (Fig. 4a, f, k); Afi-col-L D (Fig. 5a, f, j); Afi-αcoll (Fig. 6a, f, j)).
Afi-col-L B expression pattern at different regenerative stages. 1st row: WMISH; 2nd row: post in situ sectioning; 3rd row: diagrams. Stage 2 (a, f, k). Afi-col-L B is expressed in the regenerative bud in the aboral coelomic cavity epithelium (arrowhead). Stage 3/4 (b, g, h, l). Afi-col-L B is expressed in the aboral connective tissue of the regenerate (arrowhead). Stage > 50% DI (c, d, e, i, j, m, n). Afi-col-L B is expressed in the proximal region in the aboral coelomic cavity epithelium, the rim layer of the aboral intervertebral muscles, the vertebrae, the lateral arm plates and the podia wall (arrowheads), whereas in the distal region, it is detectable only in the aboral coelomic cavity epithelium (arrowhead). Violet = presence of signal. Red dotted lines = amputation plane. Abbreviations: av-aboral view; cs-cross section; lv-lateral view; ov-oral view; ss-sagittal section. Scale bars: a, b, e, f, g 50 μm; c 100 μm; i, j 25 μm. WMISH parameters for all samples: 5 days of hybridisation and 0.02 ng/μl probe concentration
Afi-col-L A expression pattern at different regenerative stages. 1st row: WMISH; 2nd row: post in situ sectioning; 3rd row: diagrams. Stage 2 (a, e, j). Afi-col-L A is not expressed in the regenerative bud. Stage 3/4 (b, f, g, k). Afi-col-L A is expressed in the epidermis (arrowhead). Stage > 50% DI (c, d, h, i, l). Afi-col-L A is expressed in the aboral coelomic cavity epithelium (arrowheads) along the whole regenerating arm. Violet = presence of signal. Red dotted lines = amputation plane. Abbreviations: av-aboral view; cs-cross section; lv-lateral view; ss-sagittal section. Scale bars: a, b, d, e, f, h 50 μm; c 100 μm; i 25 μm. WMISH parameters for all samples: 5 days of hybridisation and 0.02 ng/μl probe concentration
Afi-col-L C expression pattern at different regenerative stages. 1st row: WMISH; 2nd row: post in situ sectioning; 3rd row: diagrams. Stage 2 (a, f, k). Afi-col-L C is not expressed in the regenerative bud. Stage 3/4 (b, g, h, l). Afi-col-L C is expressed in the regenerate in the aboral connective tissue (arrowhead). Stage > 50% DI (c, d, e, i, j, m, n). Afi-col-L C is expressed in the proximal region in the lateral arm plates, at the base of the spines, in the aboral coelomic cavity epithelium and in the rim layer of the aboral intervertebral muscles, whereas in the distal region, it is expressed in the developing lateral arm plates but not in the distal tip. Violet = presence of signal. Red dotted lines = amputation plane. Abbreviations: av-aboral view; cs-cross section; ov-oral view; ss-sagittal section. Scale bars: a, b, e, f, g, j 50 μm; h, i 25 μm; c 100 μm. WMISH parameters for all samples: 5 days of hybridisation and 0.02 ng/μl probe concentration
Afi-col-L D expression pattern at different regenerative stages. 1st row: WMISH; 2nd row: post in situ sectioning; 3rd row: diagrams. Stage 2 (a, f, j). Afi-col-L D is not expressed in the regenerative bud (2 individuals). Stage 3/4 (b, g, k). Afi-col-L D is not expressed in the regenerate (4 individuals). Stage > 50% DI (c, d, e, h, i, l, m). Afi-col-L D shows expression in the proximal region of the late regenerate in the aboral coelomic cavity epithelium and in the lateral arm plates, whereas in the distal tip, the signal is detectable in the aboral connective tissue and in the tip of the terminal podium (4 individuals). Violet = presence of signal. Red dotted lines = amputation plane. Abbreviations: av-aboral view; cs-cross section; lv-lateral view; ss-sagittal section. Scale bars: a, b, d, f, g, h, i 50 μm; c 200 μm. WMISH parameters for samples of stages 2 and 3/4: 5 days of hybridisation and 0.02 ng/μl probe concentration. WMISH parameters for samples of stage > 50% DI: 7 days of hybridisation and 0.04 ng/μl probe concentration
Afi-αcoll expression pattern at different regenerative stages. 1st row: WMISH; 2nd row: post in situ sectioning; 3rd row: diagrams. Stage 2 (a, f, j). Afi-αcoll is not expressed in the regenerative bud. Stage 3/4 (b, c, g, k). Afi-αcoll is expressed in the aboral connective tissue of the regenerate. Stage > 50% DI (d, e, h, i, l, m). Afi-αcoll is detectable in the proximal region in the lateral arm plates, the spines, the oral arm plate and the aboral coelomic cavity epithelium (arrowhead), whereas in the distal tip, it is visible in the aboral coelomic cavity epithelium (arrowheads). Violet = presence of signal. Red dotted lines = amputation plane. Abbreviations: av-aboral view; cs-cross section; lv-lateral view; ss-sagittal section. Scale bars: a, b, e, f, g, h, i 50 μm; d 100 μm. WMISH parameters for samples of stage 3/4: 5 days of hybridisation and 0.02 ng/μl probe concentration. WMISH parameters for samples of stages 2 and > 50% DI: 7 days of hybridisation and 0.04 ng/μl probe concentration
Afi-Lamα-L is localised in the epidermis (Fig. 7a, g, s). On the contrary, Afi-Lamβ-L is not detectable in neither regenerative nor stump tissues (Fig. S4). Afi-TIMP3 also does not show expression (Fig. 7k, o, s). Overall, WMISH data are in agreement with Q-PCR results (see Supplementary Materials).
Afi-Lamα-L and Afi-TIMP3 expression pattern at different regenerative stages. a–j Afi-Lamα-L; k–r Afi-TIMP3; s–u diagrams representing expression patterns of both genes where violet represents the Afi-Lamα-L signal and yellow represents the Afi-TIMP3 signal. 1st and 3rd rows: WMISH; 2nd and 4th rows: post in situ sectioning; 5th row: diagrams. Afi-Lamα-L: stage 2 (a, g, s). Afi-Lamα-L is expressed in the epidermis (arrowheads) of the regenerative bud. Stage 3/4 (b, h, t). Afi-Lamα-L is expressed in the epidermis (arrowheads) of the regenerate. Stage > 50% DI (c, d, e, f, i, j, u). Afi-Lamα-L is expressed in the proximal region in the radial water canal epithelium (arrowhead), the aboral coelomic cavity epithelium and at the tip of the spines (arrow), whereas it is expressed in the epidermis in the distal region of the late regenerate. Afi-TIMP3: Stage 2 (k, o, s). Afi-TIMP3 is not expressed in the regenerative bud. Stage 3/4 (l, p, t). Afi-TIMP3 is not expressed in the regenerate. Stage > 50% DI (m, n, q, r, u). Afi-TIMP3 is detectable in the aboral coelomic cavity epithelium in the proximal region of the late regenerate, whereas no expression is visible in the distal tip. Note that the blueish staining visible in the late regenerate tip (q, r) cannot be considered a specific signal. Violet (and yellow in u) = presence of signal. Red dotted lines = amputation plane. Abbreviations: av-aboral view; cs-cross section; lv-lateral view; ov-oral view; ss-sagittal section. Scale bars: a, b, f, h, i, j, k, o, p 50 μm; c, g, l, m, n, q 100 μm; r 25 μm. WMISH parameters for all samples of stages 2, 3/4 and Afi-Lamα-L > 50% DI: 5 days of hybridisation and 0.02 ng/μl probe concentration. WMISH parameters for Afi-TIMP3 samples of stage > 50% DI: 7 days of hybridisation and 0.04 ng/μl probe concentration
Expression of ECM genes during advanced arm regeneration (stages 3/4 and > 50% DI)
At stage 3/4, when the advanced regenerative phase begins, almost all the five collagen-like genes are expressed with the exception of Afi-col-L D (Fig. 5b, g, k). In particular, Afi-col-L A is localised in the epidermis of the regenerate as visible from both whole mount samples (Fig. 3b) and post in situ sections (Fig. 3f, g, k). Afi-col-L B (Fig. 2b, g, h, l), Afi-col-L C (Fig. 4b, g, h, l) and Afi-αcoll (Fig. 6b, c, g, k) are present in the aboral dermal layer of the regenerate, with the first one more localised in the aboral dermal layer of the regenerate tip.
Afi-Lamα-L is expressed in the epidermis (Fig. 7b, h, t), whereas Afi-Lamβ-L (Fig. S4b, f, j) and Afi-TIMP3 (Fig. 7l, p, t) are not detectable in neither regenerating nor stump tissues.
In the late regenerates (> 50% DI), when all structures are well differentiated, the identified collagen-like genes show different expression patterns along the proximal-distal axis. Afi-col-L A is confined to the ACC epithelium along the whole regenerate (Fig. 3c, d, h, i, l). Afi-col-L B is detectable at the proximal end (Fig. 2c, d, i, m) in the inner lining of the podia, in the coelomic lining of the aboral intervertebral muscles, in the lateral arm plates, in the vertebrae and in the ACC epithelium, whereas in the distal tip (Fig. 2e, j, n), it is visible only in the ACC epithelium. The latter and the RWC epithelium of the non-regenerating stump tissues also display a signal (Fig. 2f; Fig. S5a–d). Afi-col-L C shows a strong expression at the proximal end (Fig. 4c, d, i, m) in the lateral arm plates, the base of the spines, the ACC epithelium and the coelomic lining of the aboral intervertebral muscles, whereas it is confined to the lateral arm plates only in the distal end of the late stage regenerating arm (Fig. 4e, j) and no expression is detectable in the distal-most tip immediately proximal to the terminal ossicle (Fig. 4e, n). In the non-regenerating stump tissues (Fig. S5e–g), a signal is detectable in the lateral arm plates and the RWC epithelium. Afi-col-L D shows a clear expression localised in the proximal region (Fig. 5c, h, l) of the late regenerate in the ACC epithelium and the lateral arm plates, whereas in the distal tip (Fig. 5d, e, i, m), the signal is detectable in the aboral dermal layer and in the tip of the terminal podium. No staining is visible in the non-regenerating stump tissues (Fig. S6e, f). Afi-αcoll expression pattern at stage > 50% DI was described by Czarkwiani and co-workers (Czarkwiani et al. 2013) and it was used as positive control in our experiments along with the Afi-c-lectin gene (Czarkwiani et al. 2016; Fig. S7). Our results confirm a signal for Afi-αcoll in the lateral arm plates and in the spines in the proximal region of the regenerate but reveal an expression also in the oral arm plates and in the ACC epithelium (Fig. 6d, h, l) and in the latter also in the distal tip of the regenerate (Fig. 6e, i, m). A similar expression pattern in the lateral and oral arm plates and in the spines is also visible in the stump tissues (Fig. S6a–d). Post in situ sectioning shows that a signal is present also in the stump water vascular system and in the ACC epithelium (Fig. S6a–d).
Afi-Lamα-L is expressed in the RWC epithelium, in the spine tips of the proximal segments (Fig. 7c, d, e, i, u), and in the distal part, it is visible in the epidermis but absent in the terminal podium (Fig. 7f, j, u). In the stump tissues, expression is visible in the epidermis and in the RWC epithelium as well as in the ectoneural component of the RNC (Fig. S8). On the contrary, Afi-Lamβ-L is not detectable in neither regenerative nor stump tissues (Fig. S4). Of course, we cannot exclude the presence of technical problems in the detection of the expression of Afi-Lamβ-L during A. filiformis regeneration; however, this datum is supported by the Q-PCR analysis that shows Afi-Lamβ-L expression levels 100–200 times less than the internal standard (Log2 from − 6.3 to − 8.5; Fig. S9) and up to 1000 times less than Afi-αcoll (Log2 from − 8.5 to − 10.5; Fig. S9). Therefore, it is likely that this ECM gene is either not expressed or expressed at non-detectable levels by chromogenic WMISH. Afi-TIMP3 displays a faint signal in the ACC epithelium from both whole mount and post in situ sectioning in the proximal region of the late regenerate (Fig. 7m, q, u), whereas no signal is present in the distal tip (Fig. 7n, r, u). Overall, the two positive controls Afi-αcoll and Afi-c-lectin run in each experiment show consistent and reproducible staining pattern and the Q-PCR results are largely consistent with the WMISH (see Supplementary Materials and Fig. S9). This supports the expressions of the newly studied ECM genes.
Discussion
ECM macromolecules, especially collagens, are structural supporting molecules important for remodelling, deposition and differentiation of pre-existing and developing structures (i.e. skeletal elements and muscles) (Okazaki and Inoué 1976; Blankenship and Benson 1984). A better understanding of ECM gene activation and expression will help us to shed light on the possible relationships between echinoderm ECM remodelling/deposition pattern and their regenerative abilities in comparison with the limited equivalents displayed by most vertebrates (Erickson and Echeverri 2018). For this purpose, in this work, we identified eight ECM genes and analysed their spatial-temporal and quantitative expression patterns during brittle star arm regeneration.
Firstly, we focused on collagen-like genes. We performed a bioinformatics and phylogenetic analysis to better characterise them in relation to collagens of other echinoderms and metazoans. From the domain search analysis, only Afi-col-L A cannot be considered unambiguously a collagen-like gene due to the presence of a C1q domain (Fig. S1). Indeed, this is typical of proteins of the complement system (Kishore and Reid 2000), which are mainly involved in immune functions. Therefore, we should consider the possibility that this gene has immunological rather than structural functions (which are typical of collagens). Further phylogenetic and functional studies should address this issue.
Overall, the phylogenetic analyses revealed the complexity in the evolution of collagen genes (Fig. S2). All genes identified in the OMA run are part of very large HOGs (> 500 genes). Considering that only 33 species were used, this indicates a high incidence of duplication and loss. Moreover, echinoderm collagens grouped mostly together indicating the separate evolutionary paths for echinoderms and chordates. This, together with the high incidence of duplication and loss, makes direct evaluation of orthology very difficult. However, the overall lower fibrillar collagen type diversity described so far in echinoderms in comparison with vertebrates (Whittaker et al. 2006) needs to be further investigated to strengthen the phylogenetic results and implement data already present in the literature (Exposito et al. 2010). From our conserved domain analysis, only Afi-col-L C can be clearly assigned to the A clade of fibrillar collagens identified in vertebrates due to the presence of the VW domain (Exposito et al. 2010).
Considering their expression patterns during regeneration, the activation of almost all the identified collagen-like genes from stage 3/4 is in agreement with Burns and co-workers (Burns et al. 2011). Their activation starts after cell proliferation begins (Czarkwiani et al. 2016), suggesting that collagen deposition follows and supports cell proliferation and migration, facilitating regeneration efficiency (Quiñones et al. 2002; Cabrera-Serrano and García-Arrarás 2004). Overall, data on regeneration-competent invertebrate and vertebrate models (González-Rosa et al. 2011; Satoh et al. 2012; Yun 2014) suggest that a delay in collagen gene activation and protein production/deposition helps to avoid scar/fibrotic tissue formation and promote subsequent regeneration.
It is noteworthy that different tissues are involved in collagen deposition: the epidermis, the coelomic lining and the dermal tissue (including that of the skeletal elements). Epithelia of all animals usually produce collagen (i.e. type IV) for their basal laminae, and skeletal elements of echinoderms include a conspicuous collagenous component within the lacunae formed by the calcitic trabeculae (as in the typical dermal skeleton; Hyman 1955; Byrne 1994). Czarkwiani and co-workers (Czarkwiani et al. 2013) have described Afi-αcoll expression in some skeletal elements, thus highlighting that some cells filling or associated with the trabecular stereom produce collagen during regeneration. The expression in the aboral dermal layer, particularly at stage 3/4, suggests that the selected fibril-forming collagen genes are expressed at the beginning of the advanced regenerative phase where spicule formation and biomineralisation occur and collagen fibrils are needed to create a well-organised scaffold for skeletal re-growth (Czarkwiani et al. 2016).
A significant feature is that all selected genes are expressed in the coelomic lining, even if at different regenerative stages and in different tissues of coelomic origin (i.e. ACC epithelium or inner lining of the podia). This may indicate that the coelomic lining of both well-differentiated (i.e. proximal end of the late regenerate) and undifferentiated (i.e. re-growing area of the distal tip of the late regenerate) tissues is actively involved in collagen production. It is known that the coelomic epithelium is one of the most likely sources of coelomocytes during regeneration (Hernroth et al. 2010), and spherule cells (one of the coelomocyte sub-population; Smith 1981; Karp and Coffaro 1982) produce collagenous materials to help wound closure and the following regenerative process (Chia and Xing 1996). Cell tracking experiments will help to confirm this hypothesis.
The absence of expression of some collagen-like genes in the stump is in agreement with Q-PCR results (Fig. S9) and can suggest either that they are not activated when the tissues are already well differentiated, as in the stump, or that they are expressed at such a low level as to be undetectable by chromogenic in situ techniques. Also, a probe penetration issue cannot be excluded, although not for all tissues.
Overall, our molecular results are in agreement with microscopic analyses recently performed on collagen deposition during regeneration in this same species (Fig. S8; Ferrario et al. 2018) as well as immunolabelling of the sea cucumber gut (Quiñones et al. 2002) and RNC (Mashanov et al. 2014) regeneration.
Together with collagen-like genes, we focused on other ECM genes. Afi-Lamα-L and Afi-TIMP3 show a delay in their activation.
In particular, laminin, one of the main components of basal laminae, is involved in ECM repair and re-growth after injury (Tassava et al. 1996; Govindan and Iovine 2015; Iorio et al. 2015). Since laminin normally forms a heterotrimer composed of one α, one β and one γ chain, the absence of β subunit gene expression at any regenerative stage was surprising. The A. filiformis gene analysed in this study is similar to subunit β-2, but in the S. purpuratus genome, two different β subunits (1-like and 2-like) are described. Hence, it may be possible that the identified gene in A. filiformis is encoding for the other subunit or it is not expressed in the regenerative stages here investigated. On the contrary, the expression pattern of the α subunit gene better fitted our expectations. Indeed, laminin is usually synthesised by the epithelial cells and a signal in the epidermis at all regenerative stage is detected. However, its presence in only stages 2 and 3/4 and only in the distal tip at advanced stages may suggest that this gene is not or little expressed in the epidermis when the arm is already well differentiated. Its expression in the RWC epithelium only in late stages of regeneration and in the stump suggests that this tissue is also involved in basal lamina production and differentiation. This is consistent with ultrastructural analysis showing that the RWC basal lamina is the most developed among epithelia of different structures (personal observation). The signal in the stump RNC and in the tips of the new spines in the late regenerate is understandable when it is remembered that laminin is known to be involved also in nervous system development, remodelling, regeneration and cell migration (Liesi et al. 1984; Barros et al. 2011), and that the spines of brittle stars possess sensory functions (Delroisse et al. 2014). Considering other echinoderms, laminin α subunits have been shown to be up-regulated during Holothuria glaberrima gut (Ortiz-Pineda et al. 2009) and Apostichopus japonicus gut and body wall regeneration (Sun et al. 2011), suggesting its involvement during this process, as described for A. filiformis.
Among ECM molecules, tissue inhibitors of metalloproteinases (TIMPs) are particularly important for tissue remodelling and regeneration (Stevenson et al. 2006; Mashanov et al. 2014). The absence of expression of Afi-TIMP3 during the first ~ 8 days after injury suggests its delayed activation. Therefore, high levels of ECM remodelling may take place via metalloproteinase (MMP) activity. Depending on the species, three to six TIMPs are present in ophiuroids (Clouse et al. 2015), but until now, no molecular data have been available on their expression patterns during arm regeneration. From our protein domain analysis, Afi-TIMP3 contains an NTR-like domain (also called netrin module), typical of this protein class. So far, TIMP expression during regeneration has been evaluated only in the sea cucumber H. glaberrima, where TIMPs are up-regulated in the radial organ complex (i.e. RNC, RWC epithelium and longitudinal muscle bands) throughout the whole regenerative process (Mashanov et al. 2014). Afi-TIMP3 is homologous to the TIMP3 described as being up-regulated at days 2 and 20 in the sea cucumber. Therefore, our gene expression diverges partially from that of the radial organ complex. This could be explained by the great difference in tissue complexity between the two experimental models. Hence, a similar expression to that detected in sea cucumber could be shown by a different A. filiformis TIMP that was not investigated here.
Conclusion
ECM gene expression patterns during arm regeneration in the brittle star Amphiura filiformis are summarised in Fig. 8. Overall, our results, supported by quantitative analyses, reveal that almost all the identified ECM genes show a delayed activation since they are not detectable in the early stage of regeneration (stage 2). It is noteworthy that, once activated, they display differential spatial expression patterns, suggesting that diverse tissues are involved in ECM remodelling and deposition during the whole regenerative process and that they are all expressed in at least one regenerative stage in the ACC epithelium. Moreover, the fact that the majority of the ECM genes is activated after cell proliferation has occurred (Czarkwiani et al. 2016) may be connected to the high efficiency of regeneration, a process typical of many other echinoderms (Quiñones et al. 2002; Cabrera-Serrano and García-Arrarás 2004; Ferrario et al. 2018) and that seems to have been lost by several vertebrates, including mammals (Mescher and Neff 2005; Zhao et al. 2016).
Summary of the expression patterns of the eight identified genes during the whole regenerative process in the regenerates (the expression pattern detected in the stump tissues is not considered). For colour legend of the diagrams, see the caption of Fig. 1. Red dotted lines = amputation plane. White square = absence of expression. Abbreviations: ACC-aboral coelomic cavity; CT-connective tissue; E-epidermis; RNC-radial nerve cord; SE-skeletal elements; WVS-water vascular system
The selection of more genes (both other collagens and ECM molecules, such as MMPs, fibronectins, integrins, etc.) will help to detail the overall involvement of all ECM components during A. filiformis arm regeneration and to confirm the hypothesis that delayed ECM gene activation and consequently ECM deposition are strictly connected to the remarkable regenerative abilities of echinoderms.
References
Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P (2002) Molecular biology of the cell, Ath edn. Garland Science, New York, pp 1065–1125
Altenhoff AM, Glover NM, Train C-M, Kaleb K, Vesztrocy AW, Dylus D, de Farias TM, Zile K, Stevenson C, Long J, Redestig H, Gonnet GH, Dessimoz C (2018) The OMA orthology database in 2018: retrieving evolutionary relationships among all domains of life through richer web and programmatic interfaces. Nucleic Acids Res 46:D477–D485
Arpino V, Brock M, Gill SE (2015) The role of TIMPs in regulation of extracellular matrix proteolysis. Matrix Biol 44-46:247–254
Barros CS, Franco SJ, Müller U (2011) Extracellular matrix: functions in the nervous system. Cold Spring Harb Perspect Biol 3:a005108
Bely AE, Nyberg KG (2009) Evolution of animal regeneration: re-emergence of a field. Trends Ecol Evol 25:161–170
Ben Amar M, Bianca C (2016) Towards a unified approach in the modeling of fibrosis: a review with research perspectives. Phys Life Rev 17:61–85
Ben Khadra Y, Ferrario C, Di Benedetto C, Said K, Bonasoro F, Candia Carnevali MD, Sugni M (2015a) Wound repair during arm regeneration in the red starfish Echinaster sepositus. Wound Repair Regen 23:611–622
Ben Khadra Y, Ferrario C, Di Benedetto C, Said K, Bonasoro F, Candia Carnevali MD, Sugni M (2015b) Re-growth, morphogenesis, and differentiation during starfish arm regeneration. Wound Repair Regen 23:623–634
Ben Khadra Y, Sugni M, Ferrario C, Bonasoro F, Oliveri P, Martinez P, Candia Carnevali MD (2018) Regeneration in stellate echinoderms: Crinoidea, Asteroidea and Ophiuroidea. In: Kloc M, Kubiak JZ (eds) Marine organisms as model systems in biology and medicine ©Springer International Publishing AG, part of Springer Nature 2018, vol Chapter 14. https://doi.org/10.1007/978-3-319-92486-1_14
Biressi ACM, Zou T, Dupont S, Di Benedetto C, Bonasoro F, Thorndyke MC, Candia Carnevali MD (2010) Wound healing and arm regeneration in Ophioderma longicaudum and Amphiura filiformis (Ophiuroidea, Echinodermata): comparative morphogenesis and histogenesis. Zoomorphology 129:1–19
Blankenship J, Benson S (1984) Collagen metabolism and spicule formation in sea urchin micromere. Exp Cell Res 152:98–104
Bock O, Mrowietz U (2002) Keloids. A fibroproliferative disorder of unknown etiology. Hautarzt 53:515
Bonnans C, Chou J, Werb Z (2014) Remodelling the extracellular matrix in development and disease. Nat Rev Mol Cell Biol 15(12):786–801
Bosch TCG (2007) Why polyps regenerate and we don't: towards a cellular and molecular framework for Hydra regeneration. Dev Biol 303:421–433
Brockes JP, Kumar A (2002) Plasticity and reprogramming of differentiated cells in amphibian regeneration. Mol Cell Biol 3:566–574
Brockes JP, Kumar A (2008) Comparative aspects of animal regeneration. Ann Rev Cell Develop Biol 24:525–549
Burns G, Ortega-Martinez O, Thorndyke MC, Peck LS, Dupont S, Clark MS (2011) Dynamic gene expression profiles during arm regeneration in the brittle star Amphiura filiformis. J Exp Mar Biol Ecol 407:315–322
Byrne M (1994) Ophiuroidea. In: Microscopic anatomy of invertebrates, vol 14. Echinodermata, pp 247–343
Cabrera-Serrano A, García-Arrarás JE (2004) RGD-containing peptides inhibit intestinal regeneration in the sea cucumber Holothuria glaberrima. Dev Dyn 231:171–178
Cameron RA, Samanta M, Yuan A, He D, Davidson E (2009) SpBase: the sea urchin database and web site. Nucleic Acids Res D:750–754
Candia Carnevali MD (2006) Regeneration in echinoderms: repair, regrowth and cloning. Invert Surv J:364–376
Chia F-S, Xing J (1996) Echinoderm coelomocytes. Zool Stud 35:231–254
Clouse RM, Linchangco GV Jr, Kerr AM, Reid RW, Janies DA (2015) Phylotranscriptomic analysis uncovers a wealth of tissue inhibitor of metalloproteinases variants in echinoderms. R Soc Open Sci 2:150377
Czarkwiani A, Dylus DV, Oliveri P (2013) Expression of skeletogenic genes during arm regeneration in the brittle star Amphiura filiformis. Gene Expr Patterns 13:464–472
Czarkwiani A, Ferrario C, Dylus DV, Sugni M, Oliveri P (2016) Skeletal regeneration in the brittle star Amphiura filiformis. Front Zool 13:18
Delroisse J, Ullrich-Lüter E, Ortega-Martinez O, Dupont S, Arnone MI, Mallefet J, Flammang P (2014) High opsin diversity in a non-visual infaunal brittle star. BMC Genomics 15:1035
Diegelmann RF, Evans MC (2004) Wound healing: an overview of acute, fibrotic and delayed healing. Front Biosci 9:283–289
Dupont S, Thorndyke MC (2006) Growth or differentiation? Adaptive regeneration in the brittle star Amphiura filiformis. J Exp Biol 209:3873–3881
Dylus DV, Blowes LM, Czarkwiani A, Elphick MR, Oliveri P (2018) Developmental transcriptome of the brittle star Amphiura filiformis reveals gene regulatory network rewiring in echinoderm larval skeleton evolution. Genome Biology 19:26
Erickson JR, Echeverri K (2018) Learning from regeneration research organisms: the circuitous road to scar free wound healing. Dev Biol 433(2):144–154
Exposito J, Valcourt U, Cluzel C, Lethias C (2010) The fibrillar collagen family. IJMS 11(2):407–426
Ferrario C, Ben Khadra Y, Czarkwiani A, Zakrzewski A, Martinez P, Colombo G, Bonasoro F, Candia Carnevali MD, Oliveri P, Sugni M (2018) Fundamental aspects of arm repair phase in two echinoderm models. Dev Biol Spec Issue Regen 433(2):297–309
Geer LY, Domrachev M, Lipman DJ, Bryant SH (2002) CDART: protein homology by domain architecture. Genome Res 12(10):1619–1623
Gelse K, Pöschl E, Aigner T (2003) Collagens-structure, function, and biosynthesis. Adv Drug Deliv Rev 55:1531–1546
Gemberling M, Bailey TJ, Hyde DR, Poss KD (2013) The zebrafish as a model for complex tissue regeneration. Trends Genet 29:611-620
Godwin J, Kuraitis D, Rosenthal N (2014) Extracellular matrix considerations for scar-free repair and regeneration: insights from regenerative diversity among vertebrates. Int J Biochem Cell Biol 56:47–55
González-Rosa JM, Martín V, Peralta M, Torres M, Mercader N (2011) Extensive scar formation and regression during heart regeneration after cryoinjury in zebrafish. Development 138:1663–1674
Govindan J, Iovine MK (2015) Dynamic remodeling of the extra cellular matrix during zebrafish fin regeneration. Gene Expr Patterns 19(1–2):21–29
Hernroth B, Farahani F, Brunborg G, Dupont S, Gejmek A, Sköld HN (2010) Possibility of mixed progenitor in sea star arm regeneration. J Exp Zool B Mol Dev Evol 314B:1–12
Hyman LH (1955) The invertebrates. Vol. 4. Echinodermata. Mc Graw Hill Book Company Inc. New York, Toronto, London 34-131, 245–412
Hynes RO (2009) The extracellular matrix: not just pretty fibrils. Science 326:1216–1219
Iorio V, Troughton LD, Hamill KJ (2015) Laminins: roles and utility in wound repair. Adv Wound Care (New Rochelle) 4(4):250–263
Jones P, Binns D, Chang H-Y, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, Quinn AF, Sangrador-Vegas A, Scheremetjew M, Yong S-Y, Lopez R, Hunter S (2014) InterProScan 5: genome-scale protein function classification. Bioinformatics. DOI:https://doi.org/10.1093/bioinformatics/btu031
Karp RD, Coffaro KA (1982) Cellular defense systems of the Echinodermata. Phylogeny and Ontogeny 257–282
Keane TJ, Horejs C-M, Stevens MM (2018) Scarring vs. functional healing: matrix-based strategies to regulate tissue repair. Adv Drug Deliv Rev 129:407–419
King RS, Newmark PA (2012) The cell biology of regeneration JCB: review. J Cell Biol 196:553–562
Kishore U, Reid KB (2000) C1q: structure, function, and receptors. Immunopharmacology 49(1–2):159–170
Kudtarkar P, Cameron RA (2017) Echinobase: an expanding resource for echinoderm genomic information. Database 2017 DOI: https://doi.org/10.1093/database/bax074
Liesi P, Kaakkola S, Dahl D, Vaheri A (1984) Laminin is induced in astrocytes of adult brain by injury. EMBO J 3:683–686
Lu P, Takai K, Weaver VM, Werb Z (2011) Extracellular matrix degradation and remodeling in development and disease. Cold Spring Harb Perspect Biol 3(12):a005058
Mashanov VS, Zueva OR, García-Arrarás JE (2014) Transcriptomic changes during regeneration of the central nervous system in an echinoderm. BMC Genomics 15:357
Mescher AL, Neff AW (2005) Regenerative capacity and the developing immune system. Adv Biochem Eng Biotechnol 93:39–66
Miao T, Zixuan W, Sun L, Li X, Xing L, Bai Y, Wang F, Yang H (2017) Extracellular matrix remodeling and matrix metalloproteinases (ajMMP-2 like and ajMMP-16 like) characterization during intestine regeneration of sea cucumber Apostichopus japonicus. Comp Biochem Physiol B: Biochem Mol Biol 212:12–23
Milligan M (1946) Trichrome stain for formalin-fixed tissue. Am J Clin Pathol 10:184
Nye HLD, Cameron JA, Chernoff EAG, Stocum DL (2003) Regeneration of the Urodele limb: a review. Dev Dyn 226:280–294
Okazaki K, Inoué S (1976) Crystal property of the larval sea urchin spicule. Develop Growth Differ 18:413–434
Ortiz-Pineda PA, Ramírez-Gómez F, Pérez-Ortiz J, González-Díaz S, Santiago-De Jesús F, Hernández-Pasos J, Del Valle-Avila C, Rojas-Cartagena C, Suárez-Castillo EC, Tossas K, Méndez-Merced AT, Roig-López JL, Ortiz-Zuazaga H, García-Arrarás JE (2009) Gene expression profiling of intestinal regeneration in the sea cucumber. BMC Genomics 10:262. DOI:https://doi.org/10.1186/1471-2164-10-262
Purushothaman S, Saxena S, Meghah V, Brahmendra Swamy CV, Ortega-Martinez O, Dupont S, Idris M (2015) Transcriptomic and proteomic analyses of Amphiura filiformis arm tissue-undergoing regeneration. J Proteome 112:113–124
Quiñones JL, Rosa R, Ruiz DL, García-Arrarás JE (2002) Extracellular matrix remodeling and metalloproteinase involvement during intestine regeneration in the sea cucumber Holothuria glaberrima. Dev Biol 250:181–197
Rahban SR, Garner WL (2003) Fibroproliferative scars. Clin Plast Surg 30:77
Rousselle P, Montmasson M, Garnier C (2018) Extracellular matrix contribution to skin wound re-epithelialization. Matrix Biol 75-76:12-26
Saló E, Abril JF, Adell T, Cebrià F, Eckelt K, Fernàndez-Taboada E, Handberg-Thorager M, Iglesias M, Molina MD, Rodrìguea-Esteban G (2009) Planarian regeneration: achievements and future directions after 20 years of research. Int J Dev Biol 53:1317–1327. https://doi.org/10.1387/ijdb.072414es
Satoh A, Hirata A, Makanae A (2012) Collagen reconstitution is inversely correlated with induction of limb regeneration in Ambystoma mexicanum. Zool Sci 29:191–197
Sheehy EJ, Cunniffe GM, O’Brien FJ (2018) Collagen-based biomaterials for tissue regeneration and repair. Peptides and proteins as biomaterials for tissue regeneration and repair 127-150 DOI:https://doi.org/10.1016/B978-0-08-100803-4.00005-X
Smith VJ (1981) The echinoderms. In: Ratcliffe NA, Rowley AF (eds) Invertebrate blood cells. Academic Press, London, pp 513–562
Stevenson TJ, Vinarsky V, Atkinson DL, Keating MT, Odelberg SJ (2006) Tissue inhibitor of metalloproteinase 1 regulates matrix metalloproteinase activity during newt limb regeneration. Dev Dyn 235(3):606–616
Sun L, Chen M, Yang H, Wang T, Liu B, Shu C, Gardiner DM (2011) Large scale gene expression profiling during intestine and body wall regeneration in the sea cucumber Apostichopus japonicus. Comp Biochem Physiol D. DOI:https://doi.org/10.1016/j.cbd.2011.03.002
Swinehart IT, Badylak SF (2016) Extracellular matrix bioscaffolds in tissue remodeling and morphogenesis. Dev Dyn 245:351–360
Tassava RA, Nace JD, Wei Y (1996) Extracellular matrix protein turnover during salamander limb regeneration. Wound Repair Regen 4(1):75–81
Tsonis PA (2000) Regeneration in vertebrates. Dev Biol 221:273–284
Whittaker CA, Bergeron KF, Whittle J, Brandhorst BP, Burke RD, Hynes RO (2006) The echinoderm adhesome. Dev Biol 300:252–266
Yi S, Ding F, Gong L, Gu X (2017) Extracellular matrix scaffolds for tissue engineering and regenerative medicine. Curr Stem Cell Res Ther 12(3):233–246
Yun S (2014) The role of extracellular matrix in planarian regeneration. PhD thesis (University of Hong Kong)
Zhao A, Qin H, Fu X (2016) What determines the regenerative capacity in animals? BioScience 66(9):735–746
Acknowledgements
The authors are grateful to the Sven Lovén Centre for Marine Sciences in Kristineberg (Sweden) for help during the collection of experimental animals. We thank Wendy Hart (University College London) and Fraser Simpson (University College London) for help with gene cloning. We are grateful to Iain C. Wilkie for his suggestions for improving the manuscript. This work was partly funded by the KVA fund SL2015-0048 from the Royal Swedish Academy of Science. CF and LP were funded by an Erasmus Placement fellowship. AC was funded by a Wellcome Trust PhD studentship (099745/Z/12/Z). MS was funded by a Young Researcher Grant of the University of Milan.
Funding
This work was partly funded by the KVA fund SL2015-0048 from the Royal Swedish Academy of Science. CF and LP were funded by an Erasmus Placement fellowship. AC was funded by a Wellcome Trust PhD studentship (099745/Z/12/Z). MS was funded by a Young Researcher Grant of the University of Milan.
Author information
Authors and Affiliations
Contributions
CF, MS and PO conceived the study and wrote the manuscript. CF and AC carried out the molecular experiments and the histological sections. AC provided the experimental animal images. LP helped with in situ hybridisation experiments and performed Q-PCR with PO. DVD performed phylogenetic analysis. CF, AC, DVD, MS, PO and MDCC analysed the data. All authors contributed to and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All applicable international, national and/or institutional guidelines for the care and use of animals were followed.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 14.9 mb)
Rights and permissions
About this article
Cite this article
Ferrario, C., Czarkwiani, A., Dylus, D.V. et al. Extracellular matrix gene expression during arm regeneration in Amphiura filiformis. Cell Tissue Res 381, 411–426 (2020). https://doi.org/10.1007/s00441-020-03201-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00441-020-03201-0