Abstract
Glioblastoma multiforme (GBM) is the most common and aggressive malignant primary brain tumor in humans, whose invasiveness and proliferation are associated with poor prognosis. Matrix metalloproteinases (MMPs) and the related family of “a disintegrin and metalloproteinase” (ADAM) both contribute to increase cell invasion, and its substrate N-cadherin is involved in proliferation and metastatic capacities of tumor cells. However, these molecular determinants of aggressiveness have not been adequately characterized in GBM. In an attempt to better define these pathogenetic signatures, in the present study we evaluated the comparative expression of two main MMPs (MMP-2 and -9), as well as of ADAM-10 and N-cadherin in surgical samples from patients diagnosed with WHO grade IV GBM (n = 25) and in cortical tissue specimens obtained from untreatable epileptic patients (controls, n = 8) through a series of histopathological, immunohistochemical and biochemical tests. Our studies revealed that both MMP-2 and -9 immunoreactivities (IRs) were upregulated in 13 of 25 (52 %) and 19 of 25 (76 %) GBMs, respectively, and the extent of the increase was highly significant with respect to controls (p < 0.001). ADAM-10 IR was also found to be increased (p < 0.001) in 16 of 25 GBM specimens (64 %). Conversely, N-cadherin IR was remarkably decreased (p < 0.001) in almost the totality of tumor samples (22 of 25, 88 %). A similar trend was also obtained at the mRNA and protein level by qPCR and western blot analyses, respectively. Collectively, the current study provides a comprehensive molecular portrayal of some of the major pathological hallmarks of GBM aggressiveness, which could be exploitable as potential targets for a new therapeutic approach.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glioblastoma multiforme (GBM) is the most common and aggressive malignant primary brain tumor in humans, classified as grade IV in the World Health Organization (WHO) classification of tumors of the central nervous system (CNS) (Louis et al. 2007). The annual incidence is about 5 per 100,000 population or 17,000 new diagnoses per year (Omuro and DeAngelis 2013). These malignant tumors are highly invasive, and easily infiltrate into the surrounding brain parenchyma by taking advantage of the facilitating microenvironment found in the close proximity of the tumor periphery (Payne and Huang 2013). Because of the high invasiveness of these types of tumor, the current therapeutic approach for GBM patients is very aggressive and consists of a combination of surgery, radiotherapy and chemotherapy. Despite these efforts, however, to date less than 5 % of diagnosed patients survive more than 5 years after the initial diagnosis. One of the reasons that contribute to such a high mortality rate of GBM patients is that complete surgical resection of these infiltrative tumors is virtually impossible, due to the delicate location of the tumor and also because neoplastic cells can easily escape surgical removal and are relatively resistant to the standard care regimen with radiation and chemotherapy (Drappatz et al. 2009; Sturm et al. 2014). It is thus important to find alternative strategies of intervention that can effectively arrest/prevent tumor growth and increase patient life-span. Interestingly, a large number of studies have identified that multiple mediators of tumor invasion are deregulated in GBM (Rich et al. 2005), suggesting that molecules involved in cell migration may be attractive targets for treatment. Considering that part of the process that regulates glioma cells’ infiltrative capacities in the surrounding brain tissue is dictated by proteolytic enzymes, the timely and accurate characterization of these key molecular determinants may provide the tools necessary to develop more efficacious and less invasive approaches for treatment.
Members of the matrix metalloproteinase (MMP) family are known to be involved in the breakdown of extracellular matrix (ECM) in normal physiological processes, such as embryonic development, reproduction, and tissue remodeling, as well as in several diseases (Nagase et al. 2006; Surgucheva et al. 2010). In particular, MMP-2 and -9 act as important oncogenes that improve invasiveness of cancer cells (Giannelli et al. 1997; Egeblad and Werb 2002), thereby conferring poor prognosis to tumors in which high expression levels and activities are detected (Folgueras et al. 2004; Liu et al. 2010b). In GBM, high levels of the active forms of both MMP-2 and -9 have been associated with primary GBM subtype, but their relationship with prognosis remains to be partly clarified, as are the molecular mechanisms that drive MMP transcriptional regulation. In addition to MMPs, another related family of proteinases known as “a disintegrin and metalloproteinases” (ADAMs) is involved in ECM degradation, and in particular ADAM-10 is known to also regulate other aspects of tumorigenesis, including cell proliferation, adhesion, migration and metastasis, acting by initiating the cleavage of cell surface proteins such as N-cadherin (Huovila et al. 2005; Kohutek et al. 2009).
N-cadherin, a type I transmembrane glycoprotein that forms adherent junctions between cells by interacting with other cadherin molecules via its extracellular domain, is a direct substrate of ADAM-10 (Kohutek et al. 2009). Upon cleavage by ADAM-10, N-cadherin promotes cell migration and metastasis (Cavallaro 2004; Qi et al. 2005). Interestingly, in some studies, N-cadherin mRNA and protein levels have been directly correlated with glial tumor grade (Shinoura et al. 1995; Utsuki et al. 2002), findings corroborated by microarray data (Asano et al. 2004). In other studies, however, investigators were not able to identify any differences in N-cadherin protein levels between low- and high-grade gliomas or, conversely, their studies revealed an inverse relationship with glioma grade (Asano et al. 2000). While this contrasting evidence suggests the involvement of the glycoprotein in the pathogenesis of GBM, they also imply that further studies to better elucidate how N-cadherin expression is regulated are still needed.
In the present study, we hypothesized that the expression levels of two major MMPs, namely MMP-2 and -9, as well as of ADAM-10 and of its substrate N-cadherin, could be differentially expressed in GBMs in comparison to normal non-tumoral brain tissue, and therefore we performed comparative analyses on both tissues in an effort to provide a comprehensive panel of biomarkers associated with aggressiveness that are specifically altered in WHO grade IV GBMs. In addition, we also assessed whether or not the mRNA expression profiles of other “indirect players” associated with poor prognosis, including the cytokine interleukin-6 (IL-6) and the pro-angiogenic factor vascular endothelial growth factor (VEGF), were deregulated in GBM samples.
Materials and methods
Patients and tissue samples
Twenty-five samples of glioblastoma multiforme (GBM), diagnosed by magnetic resonance imaging (MRI), were collected from patients who had undergone surgery at two clinical centers (Neurosurgery Unit, Fondazione GB Morgagni, Catania, Italy, and Neurosurgery Unit, Cannizzaro Hospital, Catania, Italy). All the patients underwent surgical resection between June 2012 and May 2014. The median age of these patients was 68 years (range 51–76). They had undergone macroscopic total or near-total resection of tumors, as confirmed by postoperative MRI, and had a postoperative Karnofsky Performance Scale (KPS) score ≥70. The patients received conventional therapy consisting of maximal surgical resection, followed by radiotherapy and/or chemotherapy. The diagnosis of primary GBM was established on a strict morphological criteria (grade IV), according to the classification of 2007 from the World Health Organization (WHO) (Louis et al. 2007) with the relative code of the International Classification of Diseases for Oncology (ICD-O 94440/3). The glial nature of the tumors was also confirmed by immunohistochemical analyses, showing a diffuse and strong cytoplasmic staining for GFAP (glial fibrillary acidic protein) in all cases (data not shown). Non-tumor brain tissue samples (controls, n = 8) were represented by small portions of the anterior temporal cortex resected from another group of age-matched patients who had undergone surgery for intractable epilepsy. The reason why we chose brain tissue samples from epileptic patients as “normal” controls rather than the peritumoral tissues was based on preliminary evidence obtained in our laboratories showing variability in the levels of MMP-9 and stemness cell markers in tissues surrounding glioblastomas (data not shown). Furthermore, different evidence suggested the presence of tumor cells infiltrating the peritumoral tissue or changes in the gene expression profile of cells surrounding the tumor (Stevenson et al. 2008; Nowacki and Kojder 2001; Lama et al. 2007; Sica et al. 2011) that are compatible with a transition state defined as “precancerous state” or “quiescent cancer cells”, which showed apparently normal morphology (Mangiola et al. 2013). The investigation was performed following the approval of the institutional review board. Informed consent was obtained from each patient, and the research was approved by the Local Medical Ethical Committee and conformed to the ethical guidelines of the Declaration of Helsinki.
Histology
Samples were rinsed in phosphate-buffered saline (PBS) fixed in 10 % buffered formalin as previously described (Martinez et al. 2007; Castorina et al. 2013). After an overnight wash, specimens were dehydrated in graded ethanol, cleared in xylene, and paraffin-embedded, preserving their anatomical orientation. Sections (4–5 μm thick) were cut from paraffin blocks using a microtome, mounted on sialinate-coated slides, and stored at room temperature. The sections were then stained with hematoxylin and eosin (H&E) and examined using a Zeiss Axioplan light microscope (Carl Zeiss, Oberkochen, Germany) for general morphological characterization and to highlight the presence or absence of structural alterations. Finally, representative photomicrographs were captured using a digital camera (AxioCam MRc5; Carl Zeiss).
Immunohistochemistry (IHC)
For immunohistochemical analyses, brain tissues were processed as previously described (Puzzo et al. 2014). Briefly, slides were dewaxed in xylene, hydrated using graded ethanols, and incubated for 30 min in 0.3 % H2O2/methanol to quench endogenous peroxidase activity before being rinsed for 20 min with PBS (Bio-Optica, Milan, Italy). The sections were heated (5 min × 3) in capped polypropylene slide-holders in citrate buffer (10 mM citric acid, 0.05 % Tween 20, pH 6.0; Bio-Optica), using a microwave oven (750 W) to unmask antigenic sites as previously described (Loreto et al. 2013). The blocking step was performed before application of the primary antibody with 5 % bovine serum albumin (BSA; Sigma, Milan, Italy) in PBS for 1 h in a moist chamber. BSA was used as a blocking agent to prevent non-specific binding of the antibody to the tissue sections. Following blocking, the sections were incubated overnight at 4 °C with a mouse monoclonal anti-MMP-2 antibody (ab86607; Abcam, Cambridge, UK) diluted 1:200 in PBS (Sigma), a mouse polyclonal anti-MMP-9 antibody (ab38898; Abcam) diluted 1:100 in PBS, a mouse monoclonal anti-N-cadherin antibody clone 13A9 (05-915; EMD Millipore, Billerica, MA, USA) diluted 1:400 in PBS, and a rabbit polyclonal anti-ADAM10 antibody (ab1997; Abcam) diluted 1:200 in PBS. Immune complexes were then treated with a biotinylated link antibody (HRP-conjugated anti-mouse and anti-rabbit were used as secondary antibody) and then detected with peroxidase-labelled streptavidin, both incubated for 10 min at room temperature (LSAB+ System-HRP, K0690; Dako, Glostrup, Denmark). Immunoreactivity was visualized by incubating the sections for 2 min in a 0.1 % 3,3′-diaminobenzidine and 0.02 % hydrogen peroxide solution (DAB substrate Chromogen System; Dako). The sections were faintly counterstained with Mayer’s hematoxylin (Histolab Products, Göteborg, Sweden) mounted in GVA (Zymed Laboratories, San Francisco, CA, USA) and observed under an Axioplan Zeiss light microscope (Carl Zeiss) and then photographed with a digital camera (AxioCam MRc5; Carl Zeiss).
Evaluation of immunohistochemistry
MMP-2, MMP-9, N-cadherin and ADAM10 staining was defined as either negative or positive. Immunohistochemical positive staining was determined by the presence of a brown chromogen on the edge of the H&E-stained cell nucleus, distributed along the cytoplasm or in the cell membrane via evaluation by light microscopy as previously described (Musumeci et al. 2013a). Intensity of staining (IS) and the proportion of immunopositive cells (defined here as extent score, ES) was also assessed by light microscopy. IS was ranked on a scale ranging from 0 to 4, according to the following criteria: no detectable staining (0); weak staining (1); moderate staining (2); strong staining (3); and very strong staining (4). The percentage of MMP-2, MMP-9, N-cadherin and ADAM-10 immunopositive cells (ES) was independently evaluated by 3 investigators (2 anatomists/morphologists and one surgical pathologist) and was scored empirically as the mean percentage of stained cells over 100 cells, according to the following classification: no staining (0); 1–5 % (+); 5–50 % (++); 51–75 % (+++); and >75 % (++++). Counting was performed at ×200 magnification. Sections treated with PBS without the primary antibodies served as negative controls. Throughout our analyses, no IRs were observed in any of the negative controls (ES = 0; IS = 0) that were treated with PBS in place of the primary antibody (data not shown).
Computerized morphometric measurements and image analysis
Fifteen fields, randomly selected from each section, were analyzed and the percentage of area stained with MMP-2, MMP-9, N-cadherin and ADAM-10 antibodies was calculated using an image analysis software (AxioVision Release 4.8.2, SP2 Software; Carl Zeiss Microscopy, Jena, Germany), which allows the creation of macros to reliably quantify the levels of staining intensity of immunopositive areas in a fixed field, as described previously (Musumeci et al. 2013a). Digital photomicrographs were taken using the Zeiss Axioplan light microscope (Carl Zeiss), using objective lens of magnification ×20 (i.e. final magnification ×400) equipped with a digital camera (AxioCam MRc5; Carl Zeiss). Parameters for acquisition, including the threshold levels to detect stained areas, were kept constant throughout the experiments to reduce technical biases. The formula employed to calculate the percentage of stained area was the following: percentage of area with positive staining = (mean of above-threshold stained area per field / total area per field) × 100.
Quantitative real time polymerase chain reaction
Total RNA extracts obtained from non-tumor brain samples (brain tissue specimens from patients with untreatable epilepsy; n = 4) and resected GBM tissue samples (n = 8) were isolated using 1 ml TRIzol reagent (Invitrogen) and 0.2 ml chloroform and precipitated with 0.5 ml isopropanol. Pellets were then washed with 75 % ethanol and air-dried. cDNAs were synthesized by incubating total RNA (5 μg) with SuperScript III RNase H-reverse transcriptase (200 U/μl) (Invitrogen, Carlsbad, CA, USA); Oligo-(dT)20 primer (100 nM) (Invitrogen); 1 mM dNTP mix (Invitrogen), dithiothreitol (DTT, 0.1 M), recombinant RNase-inhibitor (40 U/μl) at 42 °C for 1 h in a final volume of 20 μl. The reaction was terminated by incubation of samples at 70 °C for 10 min. Aliquots of cDNA (100 ng) from non-tumor and GBM tumor samples and external standards at known amounts (purified PCR products, ranging from 102 to 108 copies) were amplified in parallel reactions, using primer pairs indicated in Table 1. mRNA levels of the reference gene, the S18 ribosomal protein subunit, were measured in each amplification. Each PCR reaction contained 0.5 μM primers, 1.6 mM MgCl2+, 1× Light Cycler-FastStart DNA Master SYBR Green I (Roche Diagnostic). Amplifications were performed using the Light Cycler 1.5 instrument (Roche Diagnostic) with the following program setting: (1) cDNA denaturation (1 cycle: 95 °C for 10 min); (2) quantification (45 cycles: 95 °C for 10 s, 60 °C for 30 s, 72 °C for 7 s); (3) melting curve analysis (1 cycle: 95 °C for 0 s, 65 °C for 15 s, 95 °C for 0 s); and (4) cooling (1 cycle: 40 °C for 30 s). Quantification was obtained by comparing the fluorescence emitted by PCR products at unknown concentration with the fluorescence emitted by external standards at known concentration. For this analysis, fluorescence values, measured in the log-linear phase of amplification, were estimated with the second derivative maximum method using Light Cycler Data Analysis software. PCR product specificity was evaluated by melting curve analysis.
To assess the different expression levels, we analyzed the mean fold change values of each sample, calculated using the ΔΔCt method as previously described (Castorina et al. 2011). The Ct represents the number of cycles needed to detect a fluorescence above a specific threshold level and is inversely correlated to the amount of nucleic acids in the reaction. The ΔCt was calculated by normalizing the mean Ct of each sample to the mean Ct of the reference gene measured in the same experimental condition. For the quantification of each gene, we considered the cDNA from non-tumor samples as the positive sample (calibrator sample). The ΔΔCt of each sample was then calculated by subtracting calibrator ΔCt to target sample (GBM) ΔCt. The formula 2–ΔΔCt was used to calculate fold changes. Baseline measurements for each calibrator sample were set to 1.
Protein extraction procedure
Protein lysates from non-tumoral (control) and GBM tissue samples were obtained as described previously (Castorina et al. 2014; Giunta et al. 2014). Briefly, tissues were homogenized in ice-cold RIPA buffer containing 20 mM Tris (pH 7.4), 2 mM EDTA, 0.5 mM EGTA; 50 mM mercaptoethanol, 0.32 mM sucrose, a protease inhibitor cocktail (Roche Diagnostics) and a phosphatase inhibitor (PhosSTOP; Roche Diagnostic) using a Teflon-glass homogenizer and then sonicated twice for 20 s using an ultrasonic probe, followed by centrifugation at 10,000g for 10 min at 4 °C. Protein concentrations were determined using the Quant-iT Protein Assay Kit (Invitrogen) and then stored at −80 °C until further analyses.
Western blot analysis
Western blot analysis was performed according to a well-established procedure in our laboratories (Castorina et al. 2012). Briefly, solubilized proteins (40 μg) were diluted in 2× Laemmli buffer (Invitrogen), heated at 70 °C for 10 min and then separated on a Biorad Criterion XT 4–15 % Bis-tris gel (Invitrogen) by gel electrophoresis. Thereafter, proteins were transferred to a nitrocellulose membrane (Invitrogen). Blots were then blocked using the Odyssey Blocking Buffer (Li-Cor Biosciences). Effective transfer was monitored using a prestained protein molecular weight marker (BioRad Laboratories). Immunoblot analyses for MMP-2, MMP-9, N-cadherin and ADAM-10 were performed employing the same antibodies used for immunohistochemical studies, but diluted as follows: anti-MMP-2 (1:1000), anti-MMP-9 (1:500), N-cadherin (1:2000) and ADAM-10 (1:1000). A rabbit anti-β-tubulin antibody (H-235, cat. no. sc-9104, 1:500; Santa Cruz Biotechnologies) was used as loading control. To detect phosphorylated forms of JAK2 and STAT3, we used a rabbit anti-phospho JAK2 antibody (Tyr1007/1008 residue) (cat. no. #3771, 1:1000; Cell Signaling) and a rabbit anti-phospho STAT3 antibody (Tyr705 residue) (D3A7 XP®, cat no. 4093, 1:2000; Cell Signaling), respectively. Related total unphosphorylated proteins were detected using: a rabbit anti-total JAK2 (ab108596, 1:1000; Abcam) and anti-total STAT3 antibody (ab119352, 1:3000; Abcam), respectively.
Secondary antibodies were used at a dilution of 1:20,000 and 1:30,000: the goat anti-rabbit IRDye 800CW, (cat. no. 926-32211; Li-Cor Biosciences) and the goat anti-mouse IRDye 680CW (cat. no. 926-68020D; Li-Cor Biosciences), respectively. Blots were scanned using an Li-Cor Odyssey Infrared Imaging System (Li-Cor Biosciences). Densitometric analyses of bands were performed at non-saturating exposures using ImageJ software (NIH, Bethesda, MD, USA; available at http://rsb.info.nih.gov/ij/index.html). Values of non-phosphorylated proteins were normalized to β-tubulin, which served as loading control, whereas values from phosphorylated proteins were normalized to their respective total protein levels.
Zymography of matrix metalloproteinase-9 (MMP-9) activity
Control and GBM ECM/tissue from healthy and diseased patients (n = 4 × group) were collected with 1 % Triton X-100 in PBS buffer. The activity of matrix metalloproteinase-9 (MMP-9) was then determined by zymographic analysis using a procedure recently established in our laboratories (Castorina et al. 2015). Equal amounts of protein were loaded onto 12 % polyacrylamide gel containing 1 mg/ml gelatin. After electrophoresis, enzyme reaction was initiated by incubating the gel in 0.1 M glycine-NaOH (pH 8.3) at 37 °C for 18 h, and lytic areas were identified after staining of the gel with a solution containing 30 % methanol, 10 % glacial acetic acid, and 0.5 % Coomassie blue G250. Gels were then destained in the same solution in the absence of the dye and scanned with a Li-Cor Odyssey Imaging system. Images were assessed semiquantitatively using ImageJ software. Gels that did not contain gelatin were also run and failed to produce lytic areas corresponding to pro- and active MMP-9 (data not shown). Bands were identified based on their relative molecular weight. In parallel experiments, equal amounts of the same proteins (40 μg) were separated under denaturing conditions and blotted against a β-tubulin antibody, which was used to determine even sample loading.
Statistical analysis
Statistical analyses were performed using SPSS software (SPSS® release 16.0; Chicago, IL, USA). The Pearson correlation matrix and linear regression analyses were computed using GraphPad Prism v.5.03 for Windows; GraphPad Software, San Diego, CA, USA, www.graphpad.com. Datasets were tested for normal distribution with the Kolmogorov–Smirnov test. All variables were normally distributed. Comparisons between Control and GBM group means were performed using the upaired two-tailed Student’s t test. P values of less than 0.05 were considered statistically significant. All data are presented as the mean ± SEM. Cohen’s kappa was applied to measure the agreement between the three observers and averaged to evaluate overall agreement as previously described (Musumeci et al. 2013b).
Results
Histological findings
Histological examination (H&E staining) revealed no pathological changes in brain tissue samples from intractable epileptic patients (here used as controls) (Fig. 1a).
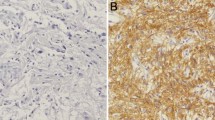
Histological findings. a Representative hematoxylin & eosin (H&E)-stained tissue section showing normal morphology in brain tissue biopsies obtained from intractable epileptic patients (Control). Magnification ×40; scale bar 50 μm. b Representative H&E-stained tissue section (viewed at lower magnification) showing the characteristic histological features of glioblastoma multiforme (GBM): malignant cells with high nuclear grade pleomorphism are seen around necrotic foci (palisading necrosis). Magnification ×10; scale bar 200 μm
Conversely, GBMs were composed of highly atypical glial cells, variable in size and shape (from polygonal to spindle-shaped cells), set in a fine fibrillary background (Fig. 1b). Notably numerous mitoses, including atypical mitoses, were observed. Characteristically, in some tumor areas, neoplastic cells tended to aggregate around necrotic foci, with configuration of the so-called “palisading necrosis” (Fig. 1b). In addition, a diffuse neoangiogenesis, mainly in the form of glomeruloid microvascular hyperplasia, was identified. Based on these histopathological features, diagnosis of GBM, World Health Organization (WHO) grade IV, was rendered. As mentioned in “Materials and Methods”, the glial nature of each tumor was immunohistochemically confirmed by immunostaining of neoplastic cells with GFAP antibodies.
Immunohistochemical analyses
To assess whether a differential expression of MMP-2, MMP-9, N-cadherin and ADAM-10 immunoreactivities (IR) does exist between non-tumoral brain tissues (controls) and GBM tumors, immunohistochemical analyses, using specific antibodies followed by computer-aided image quantifications, were carried out. In this regard, we were able to detect significant differences in terms of staining extension and intensity between healthy (control) and GBM tissues. Specifically, no MMP-2 staining (IS = 0; ES = 0) was found in control specimens (Fig. 2a, a’), whereas a strong and diffuse (>75 % of neoplastic cells) MMP-2 IR (IS = 4; ES = ++++) was observed in GBM specimens (Fig. 2b, b’). Image quantification confirmed such significant increase in the percentage of area with MMP-2 IR in GBM (77.42 ± 7.50 %) with respect to controls (0.82 ± 0.41 %) (t 8 = 10.19; ***p < 0.001 vs. control; unpaired Student’s t test).
Immunohistochemical analyses showing the differential expression of MMP-2 and MMP-9 in Control vs. GBM tissue samples. a Representative photomicrograph showing the absence of MMP-2 staining (IS = 0; ES = 0) in control specimens, where no cells appeared to be immunostained. b Representative photomicrograph showing very strong MMP-2 staining (IS = 4; ES = ++++) in GBM tissue. a’, b’ Insets of (a, b) showing stained control and GBM tissue at higher magnification. c Representative photomicrograph showing focal and weak MMP-9 staining (IS = 0; ES = 0) in control specimens. d Representative photomicrograph showing very strong MMP-9 staining (IS = 4; ES = ++++) in GBM tissue. c’, d’ Insets of (c, d) showing stained control and GBM tissue at higher magnification. Magnification ×20; scale bar 100 μm. Magnification in insets ×100; scale bar 30 μm. b”, d” Representative bar graphs showing the mean (%) of MMP-2 (b”) and MMP-9 (d”) stained areas over the mean total field area ± SEM in control and GBM tissue samples, measured using an image analysis software (AxioVision Release 4.8.2, SP2 Software; Carl Zeiss Microscopy, Jena, Germany) according to the procedures detailed in “Materials and Methods”. Each data point was calculated by analyzing the % of stained area in three randomly selected fields from the same tissue section. Afterwards, the same procedure was repeated in four other sections taken from distinct biological samples in controls and GBM (n = 5 per group) to produce the reported mean values. ***p < 0.001 vs. Control, as determined using the unpaired two-tailed Student’s t test
MMP-9 IR was focally and weakly detected in both isolated neurons and glial cells (IS = 1; ES = +) in control specimens (Fig. 2c, c’), while it was strongly and diffusely obtained in GBM specimens (IS = 4; ES = ++++) (Fig. 2d, d’). Inter-observer agreement, measured as Kappa coefficient for both MMP-2 and -9, was 0.96 (almost perfect). The percentage of area with MMP-9 IR was 4.39 ± 1.47 % in controls versus 80.21 ± 7.01 % in GBM samples (t 8 = 10.58; ***p < 0.001 vs. control), consistent with the observational appraisals.
Regarding the expression of N-cadherin, weak to moderate staining was observed in the control specimens analyzed, where glial positive cells appeared homogenously distributed (IS = 2; ES = ++) (Fig. 3a, a’). This finding, in line with results obtained by image quantification, showed that the percentage of area N-cadherin+ was about 48.45 ± 3.84 %, indicative of constitutive expression. By contrast, focal weak or absent N-cadherin staining (IS = 1; ES = +) was seen in GBM specimens, where apparently none or only a few cells seemed immunopositive (Fig. 3b, b’). Consistently, quantification demonstrated that only 4.65 ± 1.91 % of the area was positive for N-cadherin in GBM, being significantly reduced in comparison to controls (t 8 = 10.21; ***p < 0.001 vs. control). The difference between ADAM-10 staining scores in controls (IS = 1; ES = +) and GBM specimens (IS = 4; ES = ++++) was found to be the most remarkable (Fig. 3c, c’, d, d’). Indeed, unpaired two-tailed Student’s t test revealed a highly significant difference when comparing the two tissue types (control vs. tumoral; t 8 = 14.68; ***p < 0.001), with a % of ADAM-10+ area of 3.70 ± 0.87 % in control tissues and 85.38 ± 5.39 % in GBM samples, suggesting that ADAM-10 expression is dramatically induced in GBM.
Immunohistochemical analyses showing the differential expression of N-cadherin and ADAM-10 in Control vs. GBM tissue samples. a Representative photomicrograph showing moderate N-cadherin staining (IS = 2; ES = ++) in control specimens, mostly in glial cells. b Representative photomicrograph showing a notable reduction of N-cadherin staining (IS = 1; ES = +) in GBM tissue samples. a’, b’ Insets of (a, b) showing stained control and GBM tissue at higher magnification. c Representative photomicrograph showing focal and very weak ADAM-10 staining (IS = 1; ES = +) in control specimens with respect to the very strong MMP-9 staining (IS = 4; ES = ++++) observed in GBM tissue samples (d). c’, d’ Insets of (c, d) showing stained control and GBM tissue at higher magnification. Magnification ×20; scale bar 100 μm. Magnification in insets ×100; scale bar 30 μm. b”, d” Representative bar graphs showing the mean (%) of N-cadherin (b”) and ADAM-10 (d”) stained areas over the mean total field area ± SEM in control and GBM tissue samples, measured using image analysis software (AxioVision Release 4.8.2) according to the procedures detailed in “Materials and Methods”. Each data point and mean values were obtained as described in caption to Fig. 2. ***p < 0.001 vs. control, as determined using the unpaired two-tailed Student’s t test
Tissues incubated with PBS in place of the primary antibody were used as negative controls (data not shown) and thoroughly failed to produce immunoreactivity signals (ES = 0; IS = 0). Inter-observer agreement for N-cadherin and ADAM-10 scorings, measured as Kappa coefficient, was 0.92 (almost perfect).
Correlation among MMP-2, MMP-9, N-cadherin and ADAM-10 immunoreactivities in GBM tissues
Having established that the immunoreactivities of these potential markers of aggressiveness were significantly altered in GBM versus control tissues, we opted to further investigate whether and the extent at which each of these molecules correlated with each other. To this aim, the percentage of staining for MMP-2, MMP-9, N-cadherin and ADAM-10 from each GBM sample (n = 25) was analyzed using GraphPad Prism software, which was used to compute a Pearson correlation matrix. Through these analyses, we identified three positive and three negative correlations, although none of the latter reached statistical significance (please refer to the supplementary Table S1). To highlight results from each direct comparison, pairwise IHC values are depicted in scatter plots showing the correlation line (Fig. 4a–f). We found that MMP-2 and MMP-9 were highly correlated with each other in GBM samples (Fig. 4a; r = 0.843, p < 0.0001). Unexpectedly, we also detected a good correlation level between both MMP-2 and ADAM-10 and MMP-9 with ADAM-10 (Fig. 4b, c, r = 0.633 and 0.587, p < 0.0007 and p < 0.002, respectively), suggesting that, despite these proteolytic enzymes involved in ECM degradation belonging to different families, a positive association among these molecules could also relate to the degree of invasiveness in GBMs. An inverse correlation was found between MMP-9 and N-cadherin, but the association was weak and not statistically relevant (Fig. 4d, r = −0.368, p = 0.07). Finally, two further weak associations were identified: ADAM-10 and N-cadherin (Fig. 4e, r = −0.334, p = 0.1022) and MMP2 and N-cadherin (Fig. 4f, r = −0.233, p = 0.2618), none of which were statistically significant.
Correlation among MMP-2, MMP-9, N-cadherin and ADAM-10 immunoreactivities in GBM tissues. Scatter plots and correlation lines of IHC results were computed to show the correlation level among the indicated biomarkers of GBM aggressiveness. The percentage of staining for MMP-2, MMP-9, N-cadherin and ADAM-10 from each GBM sample (n = 25) was analyzed using GraphPad Prism software, which was used to compute a Pearson correlation matrix (shown in Table S1), followed by pairwise correlation comparisons (a–f)
Interleukin-6 (IL-6), vascular endothelial growth factor (VEGF), MMP-2, MMP-9, N-cadherin and ADAM-10 transcript levels in GBM tissues
In order to investigate whether or not transcript levels of the previously detected pathological markers were also differentially regulated in GBM compared to healthy tissues, we performed a series of real-time quantitative PCR analyses using cDNA templates obtained from controls (n = 4) and GBMs (n = 8). Additionally, we tested whether the constitutive mRNA expression of IL-6, a cytokine involved in glioblastoma tumor growth and survival (Liu et al. 2010a), and of VEGF, a growth factor known to take part in tumor angiogenesis whose expression has been correlated with MMP levels (Munaut et al. 2003; Edwards et al. 2008), were also affected. Our data showed that IL-6 mRNA levels were about 2.80-fold that of controls in GBM tissues (t 4 = 3.233; *p < 0.05 vs. control; Unpaired two-tailed Student’s t test) (Fig. 5a). The extent of the increase was even higher when comparing the relative mRNA expression of VEGF between controls and GBM → ↑3.43-fold (t 4 = 6.228; **p < 0.01 vs. control) (Fig. 5b). In line with IHC data, MMP-2 transcript levels were also upregulated in GBM (↑2.18-fold; t 4 = 4.838; **p < 0.01 vs. control), and so was MMP-9, although to a greater extent (↑4.64-fold;t 4 = 4.839; **p < 0.01 vs. control) (Fig. 5c, d). Surprisingly, N-cadherin mRNA expression did not show significant changes between control and GBM tissues (t 4 = 0.434; p > 0.05, not significant) (Fig. 5e), signifying that altered protein expression may be the result of post-transcriptional processing rather than the consequence of a deregulated transcription. As expected, ADAM-10 mRNA levels were strikingly increased in GBM samples, reaching levels about 5.31-fold higher than in healthy brain tissues (t 4 = 11.64; ***p < 0.001 vs. control) (Fig. 5f).
Interleukin-6 (IL-6), vascular endothelial growth factor (VEGF), MMP-2, MMP-9, N-cadherin and ADAM-10 mRNA expression in Control vs. GBM tissue samples. Differential expression of transcripts encoding for the indicated genes in non-tumor brain tissue biopsies (Control, n = 4) and in GBM tissue samples (n = 8) was determined by quantitative real time PCR analyses (a–f). Amplifications were performed using selected primers optimized for qPCR analyses (<150 bp length) and recognizing fragments within the coding sequence of the gene of interest (for details, refer to Table 1). Results are presented as mean fold changes with respect to Control ± SEM. Fold changes of each gene were obtained after normalization to the endogenous ribosomal protein S18 (reference gene) and then calculated using the comparative ΔΔCt method. Baseline expression levels of the control groups (Control) were set to 1. Bar graphs show the mean results from at least three independent determinations. n.s. = not significant. *p < 0.05, **p < 0.01, ***p < 0.001 vs. Control, as determined using the unpaired two-tailed Student’s t test
MMP-2, MMP-9, N-cadherin and ADAM-10 protein expression in GBM tissues
To further complement results obtained by IHC and mRNA studies, we performed additional western blot experiments using specific primary antibodies on three distinct biological samples per group from either controls or GBM (n = 3). Representative blots are shown in Fig. 6a. As depicted in the different bar graphs (Fig. 6b–e), densitometric analyses of bands showed high correspondence with results obtained through mRNA and IHC studies. In more detail, the overall MMP-2 protein expression was significantly increased in GBM tissues as compared to controls (t 4 = 5.091, **p < 0.01 vs. Control, unpaired Student’s t test) (Fig. 6b). Similarly, MMP-9 expression was also significantly increased in tumor specimens (t 4 = 4.853, **p < 0.01 vs. Control) (Fig. 6c). N-cadherin levels were found to be substantially downregulated in GBM samples (t 4 = 2.864, *p < 0.05 vs. Control) (Fig. 6d), although not to the same extent as obtained by IHC studies (***p < 0.001; Fig. 3a, b). Immunoblots using an ADAM-10 antibody identified two distinct bands, one with a molecular weight corresponding to the precursor of ADAM-10 (namely proADAM-10, 80–85 kDa) and the other corresponding to the mature form (namely mADAM-10, 65–70 kDa). Since both the pro- and mADAM-10 forms seemed to be regulated in a similar manner between control and GBM tissues, densitometry was calculated considering both bands as a whole. The differences observed in ADAM-10 expression between non-neoplastic and GBM samples clearly showed a significant increase in tumor tissue samples (t 4 = 4.752, **p < 0.01 vs. Control) (Fig. 6e).
MMP-2, MMP-9, N-cadherin and ADAM-10 protein expression in Control vs. GBM tissue samples. Representative immunoblots (a) and densitometric analyses (b–e) showing the differential protein expression of MMP-2, MMP-9, N-cadherin and ADAM-10 between non-tumoral and GBM samples. Protein extracts (40 μg) obtained from three separate controls and three GBM samples (n = 3 per group) were separated by SDS-PAGE and transferred to nitrocellulose membranes. Thereafter, membranes were incubated using appropriate antibodies (listed in “Materials and Methods”) and blots were scanned with Odyssey Infrared Imaging System. Densitometric analyses were then performed using the ImageJ software and values obtained were normalized to β-tubulin, which was used as loading control. Results are expressed as the average ratios ± S.E.M. from at least three independent determinations. *p < 0.05, **p < 0.01 vs. Control, as assessed using the unpaired two-tailed Student’s ttest
Identification of a persistently activated JAK2/STAT3 signaling pathway in GBM tissue samples
Signal transducer and activator of transcription 3 (STAT3) represents a point of convergence for multiple oncogenic signaling cascades, since it acts as a potent regulator of gliomagenesis, able to trigger angiogenesis, host immunosuppression, and tumor invasion (recently reviewed by Kim et al. 2014). A major upstream activator of STAT3 is known to be the Janus kinase 2 (JAK2), which phosphorylates specific residues on STAT3 to trigger its transcriptional activity (Iwamaru et al. 2007). Therefore, in the light of current advances showing that STAT3 directly up-regulates the expression of several matrix metalloproteinases, including MMP-2 and MMP-9 (Huang 2007), we thought it was of interest to also investigate the constitutive activity/phosphorylation state of the JAK2/STAT3 axis. Activation state was measured as the amount of phosphorylated substrate over its total non-phosphorylated protein levels. Western blot analyses (Fig. 7a, b) followed by densitometric measurements (Fig. 7a’ b’) revealed a persistently heightened expression of both phospho-JAK2Tyr1007 (t 4 = 5.945, **p < 0.01 vs. Control, unpaired Student’s t test) and phospho-STAT3Tyr705 (t 4 = 3.87, *p < 0.05 vs. Control) in GBM samples, identifying a potential mechanism responsible for the transcriptional regulation of metalloproteinase expression.
Persistent activation/phosphorylation of JAK2Tyr1007 and STAT3Tyr705 in GBM tissue samples. Representative immunoblots (a, b) and densitometric analyses (a’, b’) showing the phosphorylation/activation state of the upstream kinase JAK2 and of its main target, the transcription factor STAT3, in Control (n = 3) and GBM samples (n = 3), using protein lysates (40 μg) obtained as described in “Materials and Methods”. Persistent activation of the JAK2/STAT3 axis was established by measuring the relative band intensities of phosphorylated proteins at the indicated phospho-sites, which were then normalized to the corresponding densitometric values from unphosphorylated/total protein, which also served as loading controls. Each plotted result represents the mean ± SEM from three separate experiments. *p < 0.05, **p < 0.01 vs. Control, as assessed using the unpaired two-tailed Student t-test
MMP-9 activity is constitutively heightened in tissue samples from primary GBM
Primary and secondary GBMs have distinct clinical and molecular features. Indeed, primary GBMs are de novo high grade tumors that exhibit aberrant activation of growth factor signalling (Smith et al. 2001), whereas secondary GBMs are usually defined so when they arise as the result of malignant progression of lower grade gliomas (Watanabe et al. 1996). In agreement with previous studies, which have identified MMP-9 activation and not the merely increased expression as a signature to distinguish these two types of GBM (Choe et al. 2002), we strived to provide a direct molecular evidence of the primary nature of the tested tumors beside the histological appraisal reported above. Using gelatin zymography as a means to assess MMP-9 proteolytic activity, we found two distinct lytic bands of different molecular weight in both control and GBM tissues (Fig. 8a), which were subsequently identified as the proteolytically inactive (pro-MMP-9, higher MW, about 97 kDa) and active forms (lower MW, 88–90 kDa). Densitometric analyses of the zymograms showed a remarkable increase in the intensity of both the pro-MMP-9 lytic bands (t 4 = 6.673, **p < 0.01 vs. Control, unpaired Student’s t test; Fig. 8b) and the active MMP-9 bands in GBM tissues when compared to controls (t 4 = 6.002, **p < 0.01 vs. Control; Fig. 8c), in line with Choe and co-workers studies and our histopathological determinations. In these experiments, even protein loading in zymograms was established by blotting equal amounts of the corresponding denatured samples with β-tubulin by western blot (Fig. 8a, lower blot) and then by normalizing lytic band intensities over β-tubulin values in the corresponding tissue types.
Zymographic assessment of MMP-9 activity in Control vs. GBM tissue samples. Representative zymogram (a) and related densitometric analyses showing the relative levels of the proteolytically inactive (b) and active form of MMP-9 (c) in non-tumor (Control, n = 2) and glioblastoma multiforme (GBM, n = 2) samples. MMP-9 activity was determined by zymography as detailed in “Materials and Methods”. The representative picture shows two distinct lytic areas (white bands) on a Coomassie-stained gel (blue background) that correspond to the predicted molecular weight for the inactive precursor of MMP-9 (pro-MMP-9, ~97 kDa) and the active enzyme (active-MMP-9, 88–90 kDa). To ensure even protein loading in zymograms, equal amounts of the corresponding denatured protein samples were incubated with β-tubulin and blotted by western blot (a, lower blot). The resulting band densities were measured and used to normalize lytic band intensities in zymograms. The whisker plots indicate the relative amount of pro-MMP9 in each tissue (b) as well as the activity state of MMP-9 (c), indicated as the normalized mean values ± SEM obtained from at least three separate determinations, measured using the ImageJ software. **p < 0.01 vs. Control; unpaired two-tailed Student’s t test
Discussion
One of the main reasons why, especially in the last decade, continuous efforts have been made to identify new therapeutic strategies to defeat GBM relies on the fact that these types of tumors are resistant to currently available treatment options and their growth causes devastating effects to affected patients. It should be considered that, despite the efforts, median life expectancy has been estimated to be around 14–15 months after the initial diagnosis of GBM (Stupp et al. 2006; Signore et al. 2014). Therefore, there is a real need to find a valid cure. However, a step ahead in this process is the timely and accurate characterization of potential molecular targets that are aberrantly regulated in these glial-derived cancers as well as the ability to establish whether the tumor is of primary origin or whether it arises from a previously unidentified lower grade glioma (secondary GBM), especially important for the choice of the appropriate therapeutical approach.
The present study was devoted to offer a contribution in both directions by providing a portrayal of several distinct molecular targets that have been previously associated with aggressiveness, invasiveness, tumor growth and vascularization as well as metastatic properties of GBM. For the purpose, we carried out comparative histological and biochemical analyses on 25 GBM tissue samples and in 8 brain biopsies from patients with untreatable epilepsy, which were used as non-tumoral controls. After establishing the histological diagnosis of GBM, we performed a battery of biochemical evaluations to establish whether the distribution pattern, mRNA and protein expression of MMP-2, MMP-9, ADAM-10 and N-cadherin, but also of IL-6 and VEGF (at the transcript level) were effectively altered in GBM tissues in comparison to non-tumor controls. Lastly, we strived to identify the predominant cell types that characterize these malignant tumors. Immunohistochemical (IHC) analyses, in strict convergence with mRNA and protein expression experiments, reported that both MMP-2 and MMP-9 transcripts and proteins were strikingly augmented in GBM versus controls (Figs. 2, 4, 5). Such evidence led us to further investigate MMP-9 proteolytic activity, since it has been correlated with primary GBM subtype derivation (Choe et al. 2002). Indeed, in that study, Choe and coworkers defined a neat difference between primary and secondary GBM, in that the former is a de novo neoplastic formation whereas the latter arises from the malignant progression of a lower grade glioma. Further, the authors indicate a high occurrence of epidermal growth factor (EGF) receptor amplification, which may lead to increased MMP-9 activation. Interestingly, similar data have been reported previously by Watanabe et al. (1996), where the researchers also identified p53 mutations as a potential signature of secondary GBM origin, hence supporting the idea that overexpression of the EGF receptor and mutations of the p53 tumor suppressor gene are mutually exclusive events defining two different genetic pathways in the evolution of glioblastoma as the common phenotypic endpoint. Our zymographic analyses confirmed that metalloproteinase activity, in agreement with expression levels, was also significantly higher in GBM in comparison to control biopsies (Fig. 8). However, it should be mentioned, as elegantly reviewed by Urbańska et al. (2014), that the mere genetic typing of tumors may not be sufficient to establish GBM tumor origin. In fact, more subtypes of glioblastomas may exist with intermediate clinical and genetic profiles, a factor that has been exemplified by the giant-cell glioblastoma, that clinically and genetically occupies a hybrid position between primary (de novo) and secondary glioblastomas. In previous reports, inhibition of the JAK2/STAT3 signaling pathway has been reported to drive reduced invasive potential of glioblastoma cells (Senft et al. 2011), proposing it as a key transduction mechanism to control cancer cell motility in GBM. In the light of these data, we tried to perform more in depth investigations by also assessing whether or not the heightened MMP-2 and -9 levels were, at least in part, due to a deregulated upstream JAK2/STAT3 signaling. We found that GBM tissue displayed persistent activation of this signaling cascade, as indicated by the heightened phosphorylation state of both the kinase JAK2 and the transcription factor STAT3 (Fig. 7), which might be responsible for the increased proteolytic activity of the tumor. However, considering the existence of other possible factors that could contribute to the activation of STAT3, we also analyzed the mRNA levels of another known upstream activator of STAT3, namely the cytokine interleukin-6 (IL-6) (Goswami et al. 1998; Liu et al. 2010a), and of a substrate directly regulated by STAT3 transcriptional activity, VEGF, a pro-angiogenic factor also known to directly correlate with MMP activities in human glioblastomas (Munaut et al. 2003). Interestingly, we found that transcript levels for both molecules were substantially increased in GBM specimens versus controls (Fig. 5), implying that multiple factors besides JAK2 might contribute to determine persistent STAT3 activation and, in turn, cause abnormal downstream signaling events to drive, among others, tumor angiogenesis through the increased VEGF expression.
Although MMPs are notoriously known for their potential to increase tumor invasiveness, other emerging targets endowed with similar properties in GBM are still arising. Among these, members of the “a disintegrin and metalloproteinase” have been demonstrated to be critical in increasing GBM infiltrative capacities, especially ADAM-10, that acts as a cleaving enzyme on the cell-adhesion molecule N-cadherin to favour tumor cells mobility (Kohutek et al. 2009). With this in mind, we sought to examine ADAM-10 and N-cadherin expression in our GBM biopsies samples and compared the expression to that of controls. Results showed that ADAM-10 protein expression was up-regulated whereas N-cadherin expression was down-regulated in tumor samples (Fig. 3, 6). However, at the mRNA level, N-cadherin transcripts were only marginally decreased, and never reached statistical significance (Fig. 5e). Such a discrepancy between transcript and protein levels could not be explained in terms of transcriptional activity, but suggested that post-transcriptional or most likely post-translational modifications might occur during GBM tumorigenesis. This idea was further corroborated by the study conducted by Kohutek et al. (2009), showing that, upon cleavage by ADAM-10, a sizeable pool of N-cadherin forms protein products that may or may not be recognized by commonly available antibodies, while the intact uncleaved portion is easily detectable and quantifiable. Therefore, in our opinion, the reason why in our study we obtained decreased protein but not mRNA levels suggests that the antibody employed recognizes the intact protein and not the cleaved fragment/s, implicitly suggesting that transcript measurements are the most reliable assessment of N-cadherin in this study. Therefore, the current findings, in agreement with Koutek’s evidence, identify a limit in the immunoblot procedure as a mean to efficaciously assess the expression of this adhesion protein, hence suggesting the use of a panel of selected antibodies recognizing several different epitopes to properly assess how the protein is functionally regulated in GBM and/or in other tumor types.
Although the sample population used in this study was limited because of the difficulties associated with the retrieval of biopsy samples (two distinct centers were involved), we still succeeded in providing a comprehensive histological and molecular portrayal of important markers of glioblastoma aggressiveness through comparative assessments between human GBM versus control brain tissue samples. Our findings appear to be consistent with literature data, although the involvement of N-cadherin in GBM tumor invasion has still not reached unanimous consensus (Table 2). In addition, unifying studies addressing how MMP-2, MMP-9, N-cadherin and ADAM-10 are regulated in such a high-grade malignancy are currently missing. In light of the continuous scientific commitment to further the current knowledge of the pathogenetic mechanisms lying behind this devastating disease, the present study offers a further support by indicating potential targets that could be used to aid in the development of new therapies against GBM.
References
Asano K, Kubo O, Tajika Y, Takakura K, Suzuki S (2000) Expression of cadherin and CSF dissemination in malignant astrocytic tumors. Neurosurg Rev 23:39–44
Asano K, Duntsch CD, Zhou Q, Weimar JD, Bordelon D, Robertson JH, Pourmotabbed T (2004) Correlation of N-cadherin expression in high grade gliomas with tissue invasion. J Neurooncol 70:3–15
Castorina A, Leggio GM, Giunta S, Magro G, Scapagnini G, Drago F, D’Agata V (2011) Neurofibromin and amyloid precursor protein expression in dopamine D3 receptor knock-out mice brains. Neurochem Res 36:426–434
Castorina A, Giunta S, Scuderi S, D’Agata V (2012) Involvement of PACAP/ADNP signaling in the resistance to cell death in malignant peripheral nerve sheath tumor (MPNST) cells. J Mol Neurosci 48:674–683
Castorina A, D’Amico AG, Scuderi S, Leggio GM, Drago F, D'Agata V (2013) Dopamine D3 receptor deletion increases tissue plasminogen activator (tPA) activity in prefrontal cortex and hippocampus. Neuroscience 250:546–556
Castorina A, Scuderi S, D’Amico AG, Drago F, D’Agata V (2014) PACAP and VIP increase the expression of myelin-related proteins in rat schwannoma cells: involvement of PAC1/VPAC2 receptor-mediated activation of PI3K/Akt signaling pathways. Exp Cell Res 322:108–121
Castorina A, Waschek JA, Marzagalli R, Cardile V, Drago F (2015) PACAP interacts with PAC1 receptors to induce tissue plasminogen activator (tPA) expression and activity in Schwann cell-like cultures. PLoS ONE 10:e0117799
Cavallaro U (2004) N-cadherin as an invasion promoter: a novel target for antitumor therapy? Curr Opin Investig Drugs 5:1274–1278
Choe G, Park JK, Jouben-Steele L, Kremen TJ, Liau LM, Vinters HV, Cloughesy TF, Mischel PS (2002) Active matrix metalloproteinase 9 expression is associated with primary glioblastoma subtype. Clin Cancer Res 8:2894–2901
Drappatz J, Norden AD, Wen PY (2009) Therapeutic strategies for inhibiting invasion in glioblastoma. Expert Rev Neurother 9:519–534
Edwards LA, Woo J, Huxham LA, Verreault M, Dragowska WH, Chiu G, Rajput A, Kyle AH, Kalra J, Yapp D, Yan H, Minchinton AI, Huntsman D, Daynard T, Waterhouse DN, Thiessen B, Dedhar S, Bally MB (2008) Suppression of VEGF secretion and changes in glioblastoma multiforme microenvironment by inhibition of integrin-linked kinase (ILK). Mol Cancer Ther 7:59–70
Egeblad M, Werb Z (2002) New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer 2:161–174
Folgueras AR, Pendás AM, Sánchez LM, López-Otín C (2004) Matrix metalloproteinases in cancer: from new functions to improved inhibition strategies. Int J Dev Biol 48:411–424
Giannelli G, Falk-Marzillier J, Schiraldi O, Stetler-Stevenson WG, Quaranta V (1997) Induction of cell migration by matrix metalloproteinase-2 cleavage of laminin-5. Science 277:225–228
Giunta S, Andriolo V, Castorina A (2014) Dual blockade of the A1 and A2A adenosine receptor prevents amyloid beta toxicity in neuroblastoma cells exposed to aluminum chloride. Int J Biochem Cell Biol 54:122–136
Goswami S, Gupta A, Sharma SK (1998) Interleukin-6-mediated autocrine growth promotion in human glioblastoma multiforme cell line U87MG. J Neurochem 71:1837–1845
Hazan RB, Phillips GR, Qiao RF, Norton L, Aaronson SA (2000) Exogenous expression of N-cadherin in breast cancer cells induces cell migration, invasion, and metastasis. J Cell Biol 148:779–790
Huang S (2007) Regulation of metastases by signal transducer and activator of transcription 3 signaling pathway: clinical implications. Clin Cancer Res 13:1362–1366
Huang D, Du X, Yuan R, Chen L, Liu T, Wen C, Huang M, Li M, Hao L, Shao J (2014) Rock2 promotes the invasion and metastasis of hepatocellular carcinoma by modifying MMP2 ubiquitination and degradation. Biochem Biophys Res Commun 453:49–56
Huovila AP, Turner AJ, Pelto-Huikko M, Kärkkäinen I, Ortiz RM (2005) Shedding light on ADAM metalloproteinases. Trends Biochem Sci 30:413–422
Iwamaru A, Szymanski S, Iwado E, Aoki H, Yokoyama T, Fokt I, Hess K, Conrad C, Madden T, Sawaya R, Kondo S, Priebe W, Kondo Y (2007) A novel inhibitor of the STAT3 pathway induces apoptosis in malignant glioma cells both in vitro and in vivo. Oncogene 26:2435–2444
Jiang C, Pecha J, Hoshino I, Ankrapp D, Xiao H (2007) TIP30 mutant derived from hepatocellular carcinoma specimens promotes growth of HepG2 cells through up-regulation of N-cadherin. Cancer Res 67:3574–3582
Kim JE, Patel M, Ruzevick J, Jackson CM, Lim M (2014) STAT3 activation in glioblastoma: biochemical and therapeutic implications. Cancers (Basel) 6:376–395
Kohutek ZA, diPierro CG, Redpath GT, Hussaini IM (2009) ADAM-10-mediated N-cadherin cleavage is protein kinase C-alpha dependent and promotes glioblastoma cell migration. J Neurosci 29:4605–4615
Lama G, Mangiola A, Anile C, Sabatino G, De Bonis P, Lauriola L, Giannitelli C, La Torre G, Jhanwar-Uniyal M, Sica G, Maira G (2007) Activated ERK1/2 expression in glioblastoma multiforme and in peritumor tissue. Int J Oncol 30:1333–1342
Liu L, Wu J, Ying Z, Chen B, Han A, Liang Y, Song L, Yuan J, Li J, Li M (2010a) Astrocyte elevated gene-1 upregulates matrix metalloproteinase-9 and induces human glioma invasion. Cancer Res 70:3750–3759
Liu Q, Li G, Li R, Shen J, He Q, Deng L, Zhang C, Zhang J (2010b) IL-6 promotion of glioblastoma cell invasion and angiogenesis in U251 and T98G cell lines. J Neurooncol 100:165–176
Loreto C, Leonardi R, Musumeci G, Pannone G, Castorina S (2013) An ex vivo study on immunohistochemical localization of MMP-7 and MMP-9 in temporomandibular joint discs with internal derangement. Eur J Histochem 57:e12
Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P (2007) The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol 114:97–109
Mangiola A, Saulnier N, De Bonis P, Orteschi D, Sica G, Lama G, Pettorini BL, Sabatino G, Zollino M, Lauriola L, Colabianchi A, Proietti G, Kovacs G, Maira G, Anile C (2013) Gene expression profile of glioblastoma peritumoral tissue: an ex vivo study. PLoS ONE 8:e57145
Martinez G, Musumeci G, Loreto C, Carnazza ML (2007) Immunohistochemical changes in vulnerable rat brain regions after reversible global brain ischaemia. J Mol Histol 38:295–302
Mayes DA, Hu Y, Teng Y, Siegel E, Wu X, Panda K, Tan F, Yung WK, Zhou YH (2006) PAX6 suppresses the invasiveness of glioblastoma cells and the expression of the matrix metalloproteinase-2 gene. Cancer Res 66:9809–9817
Munaut C, Noël A, Hougrand O, Foidart JM, Boniver J, Deprez M (2003) Vascular endothelial growth factor expression correlates with matrix metalloproteinases MT1-MMP, MMP-2 and MMP-9 in human glioblastomas. Int J Cancer 106:848–855
Musumeci G, Castrogiovanni P, Loreto C, Castorina S, Pichler K, Weinberg AM (2013a) Post-traumatic caspase-3 expression in the adjacent areas of growth plate injury site: a morphological study. Int J Mol Sci 14:15767–15784
Musumeci G, Trovato FM, Pichler K, Weinberg AM, Loreto C, Castrogiovanni P (2013b) Extra-virgin olive oil diet and mild physical activity prevent cartilage degeneration in an osteoarthritis model. An “in vivo” and “in vitro” study on lubricin expression. J Nutr Biochem 24:2064–2075
Nagase H, Visse R, Murphy G (2006) Structure and function of matrix metalloproteinases and TIMPs. Cardiovasc Res 69:562–573
Nowacki P, Kojder I (2001) Peritumoral angiogenesis around primary and metastatic brain neoplasms. Morphometric analysis. Folia Neuropathol 39:95–102
Omuro A, DeAngelis LM (2013) Glioblastoma and other malignant gliomas: a clinical review. JAMA 310:1842–1850
Payne LS, Huang PH (2013) The pathobiology of collagens in glioma. Mol Cancer Res 11:1129–1140
Péglion F, Etienne-Manneville S (2012) N-cadherin expression level as a critical indicator of invasion in non-epithelial tumors. Cell Adh Migr 6:327–332
Puzzo D, Loreto C, Giunta S, Musumeci G, Frasca G, Podda MV, Arancio O, Palmeri A (2014) Effect of phosphodiesterase-5 inhibition on apoptosis and beta amyloid load in aged mice. Neurobiol Aging 35:520–531
Qi J, Chen N, Wang J, Siu CH (2005) Transendothelial migration of melanoma cells involves N-cadherin-mediated adhesion and activation of the beta-catenin signaling pathway. Mol Biol Cell 16:4386–4397
Rahme GJ, Israel MA (2014) Id4 suppresses MMP2-mediated invasion of glioblastoma-derived cells by direct inactivation of Twist1 function. Oncogene 34:53–62
Rich JN, Hans C, Jones B, Iversen ES, McLendon RE, Rasheed BK, Dobra A, Dressman HK, Bigner DD, Nevins JR, West M (2005) Gene expression profiling and genetic markers in glioblastoma survival. Cancer Res 65:4051–4058
Senft C, Priester M, Polacin M, Schröder K, Seifert V, Kögel D, Weissenberger J (2011) Inhibition of the JAK-2/STAT3 signaling pathway impedes the migratory and invasive potential of human glioblastoma cells. J Neurooncol 101:393–403
Shinoura N, Paradies NE, Warnick RE, Chen H, Larson JJ, Tew JJ, Simon M, Lynch RA, Kanai Y, Hirohashi S et al (1995) Expression of N-cadherin and alpha-catenin in astrocytomas and glioblastomas. Br J Cancer 72:627–633
Sica G, Lama G, Anile C, Geloso MC, La Torre G, De Bonis P, Maira G, Lauriola L, Jhanwar-Uniyal M, Mangiola A (2011) Assessment of angiogenesis by CD105 and nestin expression in peritumor tissue of glioblastoma. Int J Oncol 38:41–49
Signore M, Pelacchi F, di Martino S, Runci D, Biffoni M, Giannetti S, Morgante L, De Majo M, Petricoin EF, Stancato L, Larocca LM, De Maria R, Pallini R, Ricci-Vitiani L (2014) Combined PDK1 and CHK1 inhibition is required to kill glioblastoma stem-like cells in vitro and in vivo. Cell Death Dis 5:e1223
Smith JS, Tachibana I, Passe SM, Huntley BK, Borell TJ, Iturria N, O’Fallon JR, Schaefer PL, Scheithauer BW, James CD, Buckner JC, Jenkins RB (2001) PTEN mutation, EGFR amplification, and outcome in patients with anaplastic astrocytoma and glioblastoma multiforme. J Natl Cancer Inst 93:1246–1256
Stevenson CB, Ehtesham M, McMillan KM, Valadez JG, Edgeworth ML, Price RR, Abel TW, Mapara KY, Thompson RC (2008) CXCR4 expression is elevated in glioblastoma multiforme and correlates with an increase in intensity and extent of peritumoral T2-weighted magnetic resonance imaging signal abnormalities. Neurosurgery 63:560–569, discussion 569–570
Stupp R, Hegi ME, van den Bent MJ, Mason WP, Weller M, Mirimanoff RO, Cairncross JG, European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups, National Cancer Institute of Canada Clinical Trials Group (2006) Changing paradigms–an update on the multidisciplinary management of malignant glioma. Oncologist 11:165–180
Sturm D, Bender S, Jones DT, Lichter P, Grill J, Becher O, Hawkins C, Majewski J, Jones C, Costello JF, Iavarone A, Aldape K, Brennan CW, Jabado N, Pfister SM (2014) Paediatric and adult glioblastoma: multiform (epi)genomic culprits emerge. Nat Rev Cancer 14:92–107
Surgucheva I, Chidambaram K, Willoughby DA, Surguchov A (2010) Matrix metalloproteinase 9 expression: new regulatory elements. J Ocul Biol Dis Inf 3:41–52
Urbańska K, Lynn RC, Stashwick C, Thakur A, Lum LG, Powell DJ Jr (2014) Targeted cancer immunotherapy via combination of designer bispecific antibody and novel gene-engineered T cells. J Transl Med 12:347
Utsuki S, Sato Y, Oka H, Tsuchiya B, Suzuki S, Fujii K (2002) Relationship between the expression of E-, N-cadherins and beta-catenin and tumor grade in astrocytomas. J Neurooncol 57:187–192
Velpula KK, Rehman AA, Chelluboina B, Dasari VR, Gondi CS, Rao JS, Veeravalli KK (2012) Glioma stem cell invasion through regulation of the interconnected ERK, integrin α6 and N-cadherin signaling pathway. Cell Signal 24:2076–2084
Watanabe K, Tachibana O, Sata K, Yonekawa Y, Kleihues P, Ohgaki H (1996) Overexpression of the EGF receptor and p53 mutations are mutually exclusive in the evolution of primary and secondary glioblastomas. Brain Pathol 6:217–223, discussion 23–24
Acknowledgments
This study was partly supported by a national grant (PON 01_00110) and by funds provided by the Faculty of Medicine and Surgery to the Department of Bio-Medical Sciences, School of Medicine, University of Catania, Catania, Italy (2009BM7LJC_005). We would like to thank Mr. Pietro Asero for his technical support.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S1
(DOCX 17 kb)
Rights and permissions
About this article
Cite this article
Musumeci, G., Magro, G., Cardile, V. et al. Characterization of matrix metalloproteinase-2 and -9, ADAM-10 and N-cadherin expression in human glioblastoma multiforme. Cell Tissue Res 362, 45–60 (2015). https://doi.org/10.1007/s00441-015-2197-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00441-015-2197-5