Abstract
Biological systems span multiple levels of structural organisation from the macroscopic, via the microscopic, to the nanoscale. Therefore, comprehensive investigation of systems biology requires application of imaging modalities that reveal structure at multiple resolution scales. Nanomorphomics is the part of morphomics devoted to the systematic study of functional morphology at the nanoscale and an important element of its achievement is the combination of immunolabelling and transmission electron microscopy (TEM). The ultimate goal of quantitative immunocytochemistry is to estimate numbers of target molecules (usually peptides, proteins or protein complexes) in biological systems and to map their spatial distributions within them. Immunogold cytochemistry utilises target-specific affinity markers (primary antibodies) and visualisation aids (e.g., colloidal gold particles or silver-enhanced nanogold particles) to detect and localise target molecules at high resolution in intact cells and tissues. In the case of post-embedding labelling of ultrathin sections for TEM, targets are localised as a countable digital readout by using colloidal gold particles. The readout comprises a spatial distribution of gold particles across the section and within the context of biological ultrastructure. The observed distribution across structural compartments (whether volume- or surface-occupying) represents both specific and non-specific labelling; an assessment by eye alone as to whether the distribution is random or non-random is not always possible. This review presents a coherent set of quantitative methods for testing whether target molecules exhibit preferential and specific labelling of compartments and for mapping the same targets in two or more groups of cells as their TEM immunogold-labelling patterns alter after experimental manipulation. The set also includes methods for quantifying colocalisation in multiple-labelling experiments and mapping absolute numbers of colloidal gold particles across compartments at specific positions within cells having a point-like inclusion (e.g., centrosome, nucleolus) and a definable vertical axis. Although developed for quantifying colloidal gold particles, the same methods can in principle be used to quantify other electron-dense punctate nanoparticles, including quantum dots.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Biological systems span multiple levels of structural organisation from the macroscopic (organism, organ), via the microscopic (tissue, cell, organelle), to the nanoscale (organelle, membrane, molecular assemblage, molecule). Therefore, comprehensive investigation of systems biology requires the application of imaging modalities that reveal and assess structural complexes at multiple resolution scales. By light microscopy (LM), immunochemistry allows the visualisation of interesting target molecules or molecular assemblages within cells and tissues. The process involves labelling each target with the aid of a suitable marker (such as a fluorophore) and a specifically bound affinity reagent (usually a primary antibody). Ultimately, the aim of immunocytochemistry (ICC) is to localise targets more precisely in the context of fine structure and, in some instances, to quantify them. In order to achieve this, we must venture beyond the lateral resolution obtainable by conventional LM and confocal microscopy (about 250 nm). By using super-resolution microscopes, such as photoactivated localisation microscopy (PALM) and stochastic optical reconstruction microscopy (STORM), spatial resolution can be improved to 25–90 nm in order to obtain a more precise localisation of single molecules (Owen et al. 2013; Klein et al. 2014; Oddone et al. 2014; Sibarita 2014). Resolution can be improved further, to 10 nm, by means of scanning electron microscopy (SEM) and to less than 1 nm by transmission electron microscopy (TEM). This brings us into the realm of nanomorphomics, the systematic study of morphology at the nanoscale (see Lucocq et al. 2014; www.nanomorphomics.com).
For ICC at the TEM level, alternative technical procedures and visualisation markers are available (Griffiths 1993; Roth 1996; Koster and Klumperman 2003; Nickel et al. 2008; Webster et al. 2008; Killingsworth et al. 2012; Amiry-Moghaddam and Ottersen 2013; Griffiths and Lucocq 2014). Visualisation markers include colloidal gold particles and quantum dots and these can be bound to a secondary reagent (e.g., protein A, streptavidin or IgG secondary antibody). These complexes can be coupled to the primary affinity markers and attain close proximity to the target molecule. Detection and localisation depend on the target specificity of the primary antibody and the ability of TEM to resolve the various compartments and visualisation markers. The ability to identify and quantify the readout is facilitated by the availability of visualisation markers that are electron-dense and point-like and that differ in size and/or shape (see Griffiths 1993; Philimonenko et al. 2014). For instance, colloidal gold particles are available in various sizes (usually 5–20 nm). This process allows the labelling of distributions (or, in the case of multiple-labelling, codistributions) across various structural compartments and/or the labelling intensities of those compartments to be quantified. Collectively, these characteristics distinguish nanoparticle-based ICC (by using, say, colloidal gold or quantum dots) from enzyme-based techniques (such as the peroxidase–antiperoxidase method) that depend on a reaction product that is difficult to quantify and a readout that might be more diffuse.
Some ultrastructural compartments are volume-occupying (VOCs) and others surface-occupying (SOCs). Examples of VOCs include granules, mitochondria and nuclei, whereas SOCs are generally some type of membrane such as the outer mitochondrial membrane or the cisternal membrane of the rough endoplasmic reticulum. The labelling intensities of compartments on the cut surfaces of TEM sections have usually been expressed as numerical densities, e.g., as numbers of gold particles per profile area for VOCs and numbers per length of a membrane trace for SOCs (Griffiths 1993).
The observed distributions of marker particles across structural compartments (whether VOCs or SOCs) represent both specific and non-specific labelling and a qualitative assessment as to whether the distribution is random or non-random is not always possible. Specificity is an important potential source of bias in immunolabelling studies. Although it often seems to affect mitochondria and nuclei, non-specific labelling can influence any compartment and arise in various ways. It is influenced by factors such as primary antibody dilution, salt concentration, use of detergents and addition of blocking agents (Griffiths 1993).
Study designs for quantitative ICC at the TEM level should incorporate random sampling procedures and estimation tools that yield unbiased (or minimally biased) estimates of the numbers of marker particles and sizes of compartments. Although other methods of quantification are available (see Nikonenko et al. 2000; Philimonenko et al. 2000; Anderson et al. 2003; D’Amico and Skarmoutsou 2008a, b), a coherent and fairly comprehensive set of stereology-based methods has been developed for post-embedding (or on-section) labelling based on the use of colloidal gold particles (Mayhew et al. 2002, 2003, 2004; Lucocq et al. 2004; Mayhew and Desoye 2004; Mayhew and Lucocq 2008a, 2011; Lucocq and Gawden-Bone 2009, 2010; Mayhew 2011).
These stereology-based approaches permit efficient and valid analyses to be made of digital readouts from single- or multiple-labelling studies and estimates to be made of specific labelling. In the case of a single-labelling study, the distribution of marker particles across compartments can be mapped in three-dimensional (3D) space. Formulating appropriate null hypotheses permits the rigorous testing of whether labelling distributions (1) are random across compartments after single labelling, (2) shift between compartments after single-labelling and following experimental manipulation or (3) in multiple-labelling studies, are independent and not indicative of colabelling or colocalisation.
This review summarises these developments in quantitative molecular nanomorphmics and illustrates the requisite calculations by using worked examples. It updates earlier reviews on the same topic (Mayhew and Lucocq 2008b; Mayhew et al. 2009; Mayhew 2011) and provides further discussion of alternative markers including quantum dots and other nanoparticles that differ in shape from colloidal gold and extend the range of possibilities of multiple-labelling.
Preliminary issues
The aim here is not to cover the technical aspects and requirements of immunocytochemical labelling per se. These have been addressed more thoroughly elsewhere (Griffiths 1993; Webster et al. 2008; Amiry-Moghaddam and Ottersen 2013; Griffiths and Lucocq 2014). Instead, the reader can take for granted that the investigation is undertaken by paying proper regard to relevant factors including the use of effective antibodies and appropriate cell/tissue preparation procedures for TEM ultrathin sectioning. The following provides advice on the way in which to sample specimens, select structural compartments, abstract quantitative data and subject them to appropriate statistical testing.
-
[a]
Ways of sampling. Biological specimens (whether from cell replicates, inbred strains of animals or domestic/wild populations) exhibit natural differences that contribute to natural or biological variation. To account for this in study designs, more than one specimen (between 3 and 5 randomly selected specimens might be a reasonable starting point) needs to be sampled as part of the overall sampling scheme. Another important point is to remember that specimens exist in 3D space and are not, in general, homogeneous or isotropic. Rather, their external and internal morphology varies with both position and orientation in 3D space.
Physical, optical and tomographic sectioning reveal internal morphology. Two important issues arise from the process of producing ultrathin physical sections for viewing by TEM: (1) a loss of dimensional information about the specimen occurs and (2) only a miniscule fraction of each specimen can be examined. Therefore, if the final set of TEM fields of view (FOVs) is to be a fair representation of the composition of the intact specimen, it needs to be selected carefully and appropriately. However, images appearing on independent sections can be misleading in terms of the size, shape and number of the real structures that gave rise to them (Lucocq and Hacker 2013). Consequently, we need to select multiple sampling items that cover a range of positions and orientations within the specimen.
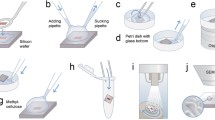
A fair and unbiased sample of the specimen can be obtained by using a multistage random sampling scheme. On such a scheme, each specimen might provide a minimum of two tissue blocks (in order to cater for between-block differences), which are sampled further by cutting ultrathin sections. Next, parts of those sections are sub-sampled to generate a minimum of two microscopical FOVs per section (in order to cater for between-FOV differences) on which quantitative assessments will be undertaken (Fig. 1). By randomly sampling at each of these stages, all positions and all orientations within the specimen can be given equal chances of being chosen. If the aim of the study involves the investigation of a set of compartments all, of which are VOCs, the randomisation of positions within the specimen is sufficient. However, when dealing with SOCs or a mixture of SOCs and VOCs, both position and orientation must be randomised. Design-based stereological sampling procedures for meeting these requirements have been described in detail elsewhere (Baddeley et al. 1986; Gundersen and Jensen 1987; Mattfeldt et al. 1990; Nyengaard and Gundersen 1992; Lucocq et al. 2004; Howard and Reed 2005; Mayhew 2008, 2011).
Fig. 1 Multistage sampling cascade for generating fields of view (FOV) for transmission electron miscroscopy (TEM). The statistical value of the final set of fields is only as good as the selection process adopted at each successive stage. Here, at the highest sampling level, a group of specimens is selected and each specimen produces a set of, say, 2–5 blocks. Each block is sampled in order to provide at least two widely separated ultrathin sections and these are sampled in turn to provide FOVs (say 20–30 per specimen). With the aid of stereological test probes and sampling tools, a coherent set of methods for quantitative immunogold cytochemistry can be applied. The test probes can be points, lines, areas (i.e., section planes) or volumes (i.e., physical disectors). Each stage of the process should involve random sampling of which an efficient variant is systematic uniform random (SUR) sampling (see also Fig. 2)
Notably, whereas all types of random sampling are unbiased, they might vary in their efficiencies. In independent or simple random sampling, the position and orientation of each item is randomised. In systematic uniform random (SUR) sampling, the position and orientation of the first item are randomised. Thereafter, a pre-determined pattern, dictated by the chosen sampling interval, decides the positions and orientations of all other items (Fig. 2). This approach produces a more even coverage of the specimen than independent random sampling and, as long as the chosen sampling interval does not correspond to some inherent periodicity in the specimen itself, this has the advantage that SUR sampling tends to provide greater precision and efficiency (Gundersen and Jensen 1987; Gundersen et al. 1999; Howard and Reed 2005; Mayhew 2008; Lucocq 2012; Lucocq and Hacker 2013). All the quantitative procedures presented in this review are based on random (preferably SUR) sampling.
Fig. 2 Illustration of SUR sampling at the stage of selecting FOVs from ultrathin sections. A ribbon of two sections lies on a copper support grid. The windows of the grid provide a convenient template for SUR sampling. Beginning at a starting point independent of the ribbon, windows are scanned in a predetermined pattern indicated by red arrows (moving to every second window in “horizontal” and “vertical” directions). The sampled windows are shown containing FOV frames (red rectangles) and, of these, only those indicated (green stars) will be recorded, because they contain areas from one of the sections. In probability terms, this SUR scheme (sampling interval 1 in 4) samples ¼ of the upper ultrathin section. SUR schemes can be applied at other stages of a multistage scheme
-
[b]
Selection of cell compartments. Although compartments can be volume-, surface- or length-occupying (e.g., organelles, membranes and filaments, respectively; see Mayhew et al. 2002), the focus here is on VOCs and SOCs. These might be heterogeneous or homogeneous. As an example of a homogeneous VOC, consider the sum total of all mitochondria within a cell (sometimes referred to as the chondriome). As an example of a heterogeneous SOC, consider the functionally and spatially distinct plasma membrane domains (apical and basolateral) of a polarised epithelial cell.
The choice of type and number of compartments depends on the aims of the individual study and prior knowledge of the biological processes being investigated. Ideally, the choice should include not only the labelled compartments of primary interest but also some classed as unlabelled or background-labelled. If necessary, these can be handled together for convenience as a natural or artificial composite compartment, e.g., different Golgi cisternae and vesicles might be described under the heading “Golgi complex” or early and late endosomes simply as “endosomes” (Mayhew et al. 2002). For statistical reasons, the number of compartments should lie somewhere between 3 and 12. Selection of too few could compromise the biological interpretation. Selection of too many will improve the precision of topological localisation but reduce estimation precision. The latter is related to the variation of gold counts within a compartment and, if estimation precision is too low, this might compromise statistical evaluations and require a rethink of the study design in order to reduce the number of compartments or to be able to count more gold particles.
-
[c]
Assignment of gold particles to compartments and their enumeration. Colloidal gold is available in various particle sizes making it attractive for multiple-labelling experiments in which two or more different target molecules are localised simultaneously (Geuze et al. 1981; Bendayan 1982; Slot and Geuze 1985). In relating gold particles to compartments, to assign a gold particle to a VOC is sensible if its centre lies on that compartment. For SOCs, a useful rule is to assign a gold particle to a membrane trace if its centre lies within a distance equivalent to twice the gold particle diameter (Mayhew and Lucocq 2008a).
Estimation of the numerical or percentage frequency distributions of gold particles offers a quick and simple way of showing where target molecules reside but these frequencies depend partly on compartment size. Larger compartments have a greater chance of being cut by section planes than smaller compartments. Even when two compartments share the same concentration of label and labelling efficiency (Lucocq 1992; Griffiths 1993), more gold particles will appear on the larger compartment. Therefore, a better approach is to estimate a labelling intensity for each compartment and this can be expressed as a labelling density (LD; Griffiths 1993) or a relative labelling index (RLI; Mayhew et al. 2002).
Another consideration is how many gold particles to count. Imagine an experimental group of cells that comprises n = 3 independent replications. A reasonable workload is to count no more than 200 gold particles per replication. To account for within-specimen variation, this total should be based on at least two ultrathin sections per replication (one section from each of two blocks) and spread across the selected microscopic FOVs and compartments from that specimen (Lucocq et al. 2004).
-
[d]
Choice of a suitable method. A coherent set of methods for quantifying gold labelling intensities and patterns in various compartments and experimental groups has been devised (Mayhew et al. 2002, 2003, 2004; Lucocq et al. 2004; Mayhew and Desoye 2004; Mayhew and Lucocq 2008a, 2011; Lucocq and Gawden-Bone 2009, 2010; Mayhew 2011). The methods are based on multistage random sampling (from specimens to FOVs), unbiased counting and/or stereological estimation and inferential statistical evaluation of a null hypothesis. Although the choice of method depends on the aim(s) of a particular study (see Fig. 3), it can be related conveniently to the following set of questions:
-
Question 1
In a single-labelling study, is the observed distribution of gold particles between compartments different from that expected by the null hypothesis of random labelling? For compartments within a given study group, this question can be answered by comparing the observed distribution of gold particles with an expected distribution calculated by randomly superimposing lattices of stereological test probes (test points for a pertinent set of VOCs or test lines for a set of SOCs). The labelling intensities are expressed as compartmental LDs or RLIs and distributions compared by chi-squared (χ 2) analysis for which, ideally, no expected value should be < 1 and no more than 20 % should be < 5 (Daly and Bourke 2000; Petrie and Sabin 2000).
-
Question 2
In a single-labelling study, what is the observed distribution of specific labelling between compartments and does it depart from the null hypothesis of random labelling? An effective way of addressing this question is to introduce a specimen-based control (Lucocq and Gawden-Bone 2010) and to alter the expression or location of the target molecule in some fashion. Examples include knockdown of protein expression (by using gene deletion, small interfering RNA or mutation), introduction of the target (by microinjection or endocytosis) and chemical modification of the target on the TEM section. The easiest outcome to interpret is if controls show little or no signal. By using separate groups of cells that show normal and reduced expression, specific labelling estimates can be abstracted from the observed distributions.
-
Question 3
In a single-labelling study, does the observed distribution of labelling between compartments change after experimental manipulation? This question can be answered by comparing directly the observed numerical frequency distributions of gold particles in two or more experimental groups by contingency table analysis (Mayhew et al. 2002, 2003; Mayhew and Desoye 2004). Ideally, no expected value should be < 5 (Petrie and Sabin 2000). The null hypothesis is that of no change in observed distributions between the groups.
-
Question 4
In a multiple-labelling study, do different sizes of gold particle colocalise in the chosen set of compartments? Evidence for colocalisation can be accepted if the distribution of labelling between compartments or the labelling of a given compartment does not change significantly for the different targets localised by using different sizes of gold particle. The question can be answered by applying the same method used to answer Question 3 (Mayhew and Lucocq 2011). Again, the null hypothesis is that of no change in observed distributions of differently sized gold particles.
-
Question 5
In a dual-labelling study, do different sizes of gold particle colabel the same set of VOC profiles? This question can be tackled by examining whether a VOC profile (e.g., a transected vesicle, granule or vacuole) identified as containing one target molecule labelled with, say, small gold particles also labels for a second target identified by large gold particles (Mayhew and Lucocq 2011). Here, individual VOC profiles must be selected by SUR sampling with the aid of a set of unbiased counting frames such as forbidden line frames (Gundersen 1977; Sterio 1984). If any expected value is < 5, a comparison of the various classes of labelled and unlabelled VOC profiles in a 2 × 2 table by Fisher’s exact probability test is preferable (Petrie and Sabin 2000; Mayhew 2011; Mayhew and Lucocq 2011). The null hypothesis is that of independence of labelling by the two sizes of gold particle marker.
-
Question 6
In a single-labelling study, can the spatial distribution of gold particles across compartments be mapped in a 3D sense? To answer this question, a stereological tool known as the vertical rotator can be applied (Vedel-Jensen and Gundersen 1993; Mironov and Mironov 1998). First, cells are sampled uniformly by using another stereological test probe, namely a volume probe known as the physical disector (Sterio 1984). Next, the distances of gold particles falling on identified compartments are measured or classified with respect to a vertical axis that passes through some identifiable cellular feature (e.g., nucleolus or centrosome/centrioles). By this approach, estimates of numbers of particles at specific locations within 3D space can be obtained (Nyengaard and Gundersen 2006; Lucocq and Gawden-Bone 2009).
Fig. 3 Workflow for quantitative molecular nanomorphomics by TEM on-section immunogold cytochemistry. Key decision steps are identified in a multistage random sampling scheme followed by stereology-based quantification. From each study group, a set of specimens is selected and taken through the appropriate preparation procedures. Sampling requirements for selecting the positions and orientations of blocks, sections and FOVs are influenced by the study aim(s) and the choice of compartments (volume-occupying [VOCs] or surface-occupying [SOCs] or a mix of both) to be investigated. Stereological sampling and estimation tools can be used to select and count items (viz. gold particles, random test points and line intersections, VOC profiles) and to classify the locations of gold-labelled molecules with respect to a fixed vertical axis of a rotator. Thereafter, outcomes are used to test an appropriate null hypothesis
-
Question 1
Worked examples of various methods
-
[a]
Tackling Question 1. The aim is to test whether the observed distribution of gold particles between compartments within a given study group is random or preferential (Mayhew et al. 2002). A potential use is in the early stages of a more detailed study in order to identify compartments that are labelled preferentially.
On the cut surface of a TEM ultrathin section, a random distribution can be simulated by applying simple geometric probes (test points and lines). Consider the example of a study in which only VOCs are of interest (i.e., compartments that are membranes or filaments are not included). Test point probes randomly positioned on randomly selected section planes will hit VOCs with probabilities determined by their fractional sectional areas and, in 3D, by their fractional volumes (Howard and Reed 2005). Hence, if a given VOC accounts for 20 % of cell volume, then, on average, 20 % of randomly applied test points should hit that VOC. By counting test points that fall on each compartment in the final set of FOVs, the resulting numerical frequency distribution can be taken to represent the expected spread of randomly positioned gold particles.
In a similar fashion, expected distributions of gold particles on a set of membranes (SOCs) can be obtained by applying random test line probes. Test lines randomly distributed and oriented on random section planes intersect various membrane domains with probabilities determined by the relative trace lengths (on the section plane) of those membranes and by their relative surface areas (in 3D).
As stated above, labelling intensities can be expressed as LDs or RLIs. LD values relate numbers of gold particles to the sizes of compartments. Although LD values can be expressed as numbers of gold particles per square micrometer (VOC profiles) or per micrometer (SOC membrane traces) on the section plane (Griffiths 1993), design-based stereology offers simpler, unbiased and efficient estimators (Mayhew 1991; Lucocq 1994; Howard and Reed 2005). These take the form of numbers of gold particles per test point (VOC profiles) or per test line intersection (SOC membrane traces) and, thereby, avoid the need to know the final FOV magnification or the lattice constants used to convert test point totals into organelle profile areas or to convert intersection counts into membrane trace lengths (Mayhew et al. 2003). Lattices of test points and lines are randomly superimposed on section planes and used to identify and count chance encounters with images of VOCs and SOCs, respectively. For a mixture of VOCs and SOCs or for SOCs alone, unbiased estimation depends crucially on randomised sampling for the position and orientation of section planes and test lattice lines (Gundersen and Jensen 1987; Gundersen et al. 1999; Mayhew 2008). For VOCs alone, only the positions of section planes and test points must be randomised.
If all compartments label randomly, they should all share the same LD value and this should correspond to that of the cell as a whole. Therefore, the ratio LDX/LDC (where X represents a compartment and C the whole cell) provides a convenient measure of the degree to which the observed labelling of a compartment deviates from the expected (random) pattern. This measure is known as the RLI (Mayhew et al. 2002) and can be calculated directly (from raw gold counts and counts of chance encounters between test probes and sectional images of compartments) or indirectly (from LD values).
The procedures about to be described were devised originally for compartments of the same type, i.e., they are either all VOCs or all SOCs (Mayhew et al. 2002). However, some target molecules might be found in other types of compartments or translocate from one to another (Mayhew and Lucocq 2008a). The following worked examples address these alternative scenarios.
-
Scenario 1
Dealing with a set of VOCs. Direct estimates of RLI can be obtained by simulating a random distribution and then by comparing it with the observed raw counts of gold particles. The random distribution of gold particles is simulated by randomly superimposing test points on randomly positioned section planes. Consider the example in Table 1 in which counts are based on a sample of labelled cells of the same type or from the same study group. For the sake of illustration, the cell is divided into a set of five VOCs.
Table 1 Testing the null hypothesis that the observed distribution of gold particles between volume-occupying compartments (VOCs) in a single group of cells is random. Relative labelling index (RLI) is estimated directly from the observed and expected numbers of gold particles before undertaking a χ 2 analysis (oN G observed number of gold particles, eN G expected number of gold particles, P number of random test points hitting VOC, subscript C across whole cell). The distribution of gold particles across compartments is not random (P-value < 0.001). Compartments VOC1 and VOC2 are preferentially labelled (RLI > 1 and χ 2 accounts for > 10 % of total) The observed total of gold particles falling on all the FOVs from this cell type (ΣoNG) is 204 and the total number of random test points (ΣP) is 215. Totals of 74 gold particles and 12 test points are associated with VOC1. What is the expected number of gold particles (eNG) for VOC1? These totals suggest that, if 204 gold particles were randomly distributed on the ultrathin sections, eNG is expected to be 12 × 204/215 = 11.386. The RLI for VOC1 is estimated as oNG/eNG, which gives 74/11.386 = 6.50. This suggests that the labelling intensity of compartment VOC1 is almost seven times greater than expected for a random dispersal of gold particles. The partial χ 2 value for a compartment is obtained from observed and expected gold counts as
$$ {\left({\mathrm{oN}}_{\mathrm{G}}\hbox{--}\ {\mathrm{eN}}_{\mathrm{G}}\right)}^2/{\mathrm{eN}}_{\mathrm{G}}. $$For VOC1, the value is (74–11.386)2/11.386 = 344.33.
For the full dataset in Table 1, total χ 2 is 592.52 and, for 4 degrees of freedom (df, determined by 2–1 groups × 4–1 compartments), the probability level is P-value < 0.001. This means that we must reject the null hypothesis that the labelling pattern is random.
When observed and expected distributions are found to be significantly different, two criteria for deciding on the preferential labelling of a compartment must be satisfied: (1) the compartmental RLI must be > 1 and (2) the partial χ 2 value must account for a significant proportion (10 % or more) of total χ 2. With these criteria, the cells show preferential labelling of compartments VOC1 and VOC2 (Table 1).
For χ 2 analysis, a mixture of labelled and unlabelled compartments should be selected for inclusion in the study, especially if the labelled compartments have similar RLI values (Mayhew et al. 2002). The test also imposes conditions on the numbers of expected gold particles on individual compartments (Mayhew et al. 2004) and this will affect the choice of compartments and numbers of gold particles needed to be counted. If small, rare or poorly labelled compartments are included in the study, extra effort in terms of sampled FOVs will be required in order to count the gold particles associated with them. If the compartment is of minor interest only, then it can be treated as part of some larger compartment such as “residuum” or “rest of cell”.
To obtain indirect estimates of RLI, the LD of any VOC is first estimated simply and efficiently as
$$ {\mathrm{LD}}_{\mathrm{X}}={\mathrm{oN}}_{\mathrm{G}}/{\mathrm{P}}_{\mathrm{X}} $$where PX is the sum of test points falling on that VOC (Mayhew et al. 2003). The RLI value is then estimated as
$$ {\mathrm{RLI}}_{\mathrm{X}}={\mathrm{LD}}_{\mathrm{X}}/{\mathrm{LD}}_{\mathrm{C}}. $$Table 2 provides LD values for the same dataset as in Table 1. For example, the LD for VOC1 is 74/12 = 6.167 gold particles per test point and LDC is 204/215 = 0.949 gold particles per test point. Consequently, the RLI for VOC1 is 6.167/0.949 = 6.50 and the partial χ 2 is 344.33 (as in Table 1). Again, preferential labelling of VOC1 and VOC2 is apparent.
-
Scenario 2
Dealing with a set of SOCs. For this scenario, the example is a sample of labelled cells divided into a set of five SOCs and the simulated distribution is generated by counting intersections (I) with random test lines (see Table 3). The calculations are performed in a similar way to those described above for direct and indirect RLI estimates for VOCs. To avoid unnecessary duplication, the example is confined to direct estimation of RLI. For the full dataset, total χ 2 is 56.62 and P-value < 0.001. The null hypothesis that the labelling pattern is random is rejected. With the criteria for deciding on preferential labelling, these cells appear to show preferential labelling of compartments SOC1, SOC3 and SOC4.
-
Scenario 3
Dealing with a mixed set of VOCs and SOCs. For this scenario, we must first deal with two practical issues: (1) sectional images of VOCs and SOCs are not equivalent because SOCs appear on TEM ultrathin sections as membrane trace lengths, whereas VOCs occur as profile areas and (2) not all membrane traces are clearly visible on TEM ultrathin sections (some of them are vague or indistinct because their membranes were not cut orthogonally by the section plane and so are tilted away from the electron axis at various angles; Mayhew and Reith 1988).
Table 2 Testing the null hypothesis that the observed distribution of gold particles between VOCs in a single group of cells is random. RLI is estimated indirectly from LDX/LDC before undertaking a χ 2 analysis (oN G observed number of gold particles, P number of random test points hitting VOC, LD X labelling density of compartment, LD C labelling density of whole cell, RLI relative labelling index, subscript C across whole cell). The distribution of gold particles is not random (P-value < 0.001). Compartments VOC1 and VOC2 are preferentially labelled Table 3 Testing the null hypothesis that the observed distribution of gold particles between surface-occupying compartments (SOCs) in a single group of cells is random. RLI is estimated directly from the observed and expected numbers of gold particles before undertaking a χ 2 analysis (oN G observed number of gold particles, eN G expected number of gold particles, I number of intersections between SOC and random test lines, RLI relative labelling index, subscript C across whole cell). The distribution of gold particles across compartments is not random (P-value < 0.001). Compartments SOC1, SOC3 and SOC4 are preferentially labelled (RLI > 1 and χ 2 accounts for > 10 % of the total) To overcome non-equivalence, membrane trace lengths can be converted to profile areas by defining a “zone of acceptance” on both sides of each membrane trace (Mayhew and Lucocq 2008a). The width of this zone is taken to be twice the diameter of the gold particles. The profile area of the acceptance zone can be calculated by multiplying its overall width by its trace length (estimated by counting intersections with random test lines) or by counting the number of equivalent test points after randomly superimposing a lattice of test points (Mayhew and Lucocq 2008a, b).
To deal with membrane tilt in the section, observed numbers of gold particles falling on membranes can be obtained by restricting counts to membrane traces that are clearly visible because they are sectioned at favourable angles. Of course, these raw counts must be corrected for image loss in order to obtain corresponding eNG values. Correction factors can be derived by goniometry or from stereological estimates (Mayhew and Reith 1988; Mayhew and Lucocq 2008a, b).
Imagine a group of cells and a set of VOCs and SOCs labelled with 10-nm gold particles. The overall width (w) of the membrane acceptance zone from one side to the other would be 2(2 × 10) = 40 nm or 0.04 μm. Imagine that only clear or near-orthogonal membrane traces (tilted no more than 5° from the electron axis) were examined. For a critical angle of 5°, the correction factor for membrane image loss would be 9.03 (Mayhew and Reith 1988; Mayhew and Lucocq 2008a, b). Table 4 presents a possible dataset from such a study. A total of 13 gold particles were counted on clear membrane images of compartment SOC2 and so the corrected number would be 13 × 9.03 = 117.39. With 60 test line intersections (I) counted with the same clear images, the corrected total should be 60 × 9.03 = 541.80. By employing a lattice of “vertical” and “horizontal” test lines with a spacing of d = 0.5 μm on the scale of the specimen (equivalent to an area of 0.25 μm2 per test point), this equates to a total of (π/4 × I × d × w)/0.25 = (3.1416/4 × 541.80 × 0.5 × 0.04)/0.25 = 34.04 test points falling within the acceptance zone of SOC2 membranes.
Table 4 Testing the null hypothesis that the observed distribution of gold particles in cells with a mixture of VOCs and SOCs is random. Orthogonally sectioned membranes are treated as profiles and membrane image loss is corrected before estimating RLI from LDX/LDC and undertaking a χ 2 analysis. Here, the correction factor (based on examining clear membrane images only) is taken to be 9.03 (see values in parenthesis; oN G observed number of gold particles, eN G expected number of gold particles, P number of random test points hitting VOC, I number of intersections between SOC and random test lines, LD labelling density, RLI relative labelling index, subscript C across whole cell). The distribution of gold particles between compartments is not random (P-value < 0.001). Membrane compartments (SOC1 and SOC2) are preferentially labelled For each compartment, eNG values can be estimated from the corrected point totals and oNG counts and corresponding LD and RLI values can be calculated. For the complete dataset, total χ 2 is 123.28 and, for df = 4, P-value < 0.001. The RLIs (1.80 and 1.81) and partial χ 2 values (∼29 and 34 % of total) indicate that compartments SOC1 and SOC2, respectively, are preferentially labelled.
-
Scenario 1
-
[b]
Tackling Question 2. The aim here is to distinguish specific from non-specific labelling of target molecules. To achieve this, Lucocq and Gawden-Bone (2010) developed a method relying on the use of specimen-based control cells. One group of cells, here denoted by the symbol “+”, represents normal expression (e.g., wild-type) and another, symbol “−”, represents reduced expression (e.g., knockout or knockdown).
The virtual dataset in Table 5 uses data in Table 1 and treats the cells as belonging to the normal expression group. Observed numbers of gold particles associated with selected compartments (here, VOCs) are counted separately in the two group of cells to give the totals oNG+ and oNG-. By superimposing lattices of test points on randomly sampled FOVs, the observed gold counts are converted into labelling densities, LD + and LD-, expressed as gold particles per test point. The specific labelling density for each compartment, LDsp, is estimated as
$$ \mathrm{LDsp}=\mathrm{L}\mathrm{D}+-\mathrm{L}\mathrm{D}-. $$Table 5 Estimating specific labelling densities (LDsp) in two cell groups representing normal (+) and reduced (−) expression and testing the null hypothesis that the specific-labelling distributions of gold particles between VOCs is random. The normal expression cell is the same as that in Tables 1, 2 (oN G observed number of gold particles, eN G expected number of gold particles, P number of random test points hitting VOC, LD labelling density, RLI relative labelling index, subscript C across whole cell). The specific-labelling distribution of gold particles is not random (P-value < 0.001). Compartments VOC1 and VOC2 are preferentially labelled For example, LD+ for the compartment VOC1 is estimated to be 6.167 gold particles per test point in normal expression cells but its LD- after reduced expression is only 1.333 (Table 5). Therefore, LDsp for this VOC1 is equal to 4.834 gold particles per test point. This implies that about 78 % of the observed labelling of VOC1 is specific.
From the above, we can estimate the number of gold particles that represents specific labelling for each compartment (Table 5). Thus, LDsp for VOC1 is 4.834 gold particles per point but this compartment in the LD+ group contains 12 test points. Therefore, oNGsp for VOC1 is predicted to be 4.834 × 12 = 58.01. The total points falling on the cell (Σ = 215) and the total number of specific gold particles (Σ = 122.96) provide the expected number of specific gold particles for each compartment, eNGsp. Hence, the expected number falling on VOC1 is 12 × 122.96/215 = 6.86 gold particles and the specific RLI is given by 58.01/6.86 = 8.46. VOC1 appears to show at least eight times stronger labelling than that expected for a purely random spread of gold particles. The partial χ 2 for VOC1 is 381.39 and total χ 2 for the whole dataset is 609.31. For df = 4, P-value < 0.001. Calculations for the other VOCs indicate that only VOC1 and VOC2 are preferentially and specifically labelled.
-
[c]
Tackling Question 3. The aim here is to test whether the distribution of gold particles across compartments alters in two or more different groups of cells. The method might also be useful for examining patterns of labelling at various antibody dilutions. With this method, observed numerical frequency distributions of raw gold counts in various groups of cells are compared directly by contingency table analysis (Mayhew et al. 2002, 2003; Mayhew and Desoye 2004).
Consider the virtual dataset in Table 6. For a compartment in a given study group (here, control and treated A and treated B), the expected number of gold particles is calculated by multiplying the relevant column sum by the corresponding row sum and dividing by the grand row sum. For instance, the expected number of gold particles on membrane compartment SOC1 is given by 205 × 310/814 = 78.07. For the observed gold count of 99, partial χ 2 amounts to (99–78.07)2/78.07 = 5.61.
Table 6 Testing the null hypothesis that no difference exists between the observed distributions of SOC-associated gold particles in three groups of cells. Observed (expected) numbers of gold particles are estimated by contingency table analysis. The three distributions are different (P-value < 0.01). Control cells have greater-than-expected labelling of SOC1 membranes (χ 2 is ∼25 % of total) and less-than-expected labelling of SOC2 and SOC5 membranes (χ 2 ∼17 % and ∼22 % of total respectively). Cells in the treated B group have less-than-expected labelling of SOC1 membranes (χ 2 ∼11 % of total) but greater-than-expected labelling of SOC5 membranes (χ 2 ∼12 % of total) Total χ 2 for all three groups is 22.46 and, for df = 8 (3–1 groups × 5–1 compartments), P-value < 0.01. Therefore, the null hypothesis (no difference in distributions between groups) is rejected. Inspection of partial χ 2 values shows that control cells have greater-than-expected labelling of SOC1 but less-than-expected labelling of SOC2 and SOC5. In contrast, cells in the treated B group exhibit less-than-expected labelling of SOC1 but greater-than-expected labelling of SOC5.
With this method, magnification need not be known or standardised between groups. Statistical evaluation by contingency table analysis prefers that the expected numbers of gold particles should not be < 5. Aiming for similar column sums for total gold counts in each group of cells is prudent so that statistical handling is not distorted by large discrepancies between groups.
A constraint of this method is its limited ability to facilitate mechanistic interpretations of shifts in gold labelling distributions. In such situations, analysis can be advantageously supported by estimates of labelling intensities (for a real example, see Schmiedl et al. 2005).
-
[d]
Tackling Question 4. The aim here is to test whether, in a multi-labelling study, targets labelled with two or more different sizes of gold particle show evidence of colocalisation. Consider a triple-labelling study examining colocalisation in a set of SOCs by using gold particles of diameters 5 nm, 10 nm and 20 nm (Table 7). The method is essentially a variant of that described under Question 3.
Table 7 Testing the null hypothesis of identical membrane (SOC) colocalisation of three different antigens labelled using 5-, 10- and 20-nm gold particles. Observed (expected) numbers of gold particles are estimated by contingency table analysis. The three distributions of gold particles do not differ significantly (P-value = 0.99) giving evidence of colocalisation Using 5-nm gold particles, the following numbers of particles were associated with SOC1 (100), SOC2 (14), SOC3 (50), SOC4 (12) and SOC5 (26). Corresponding totals for 10-nm and 20-nm particles are summarised in Table 7. For these data, total χ 2 amounts to 1.82 and, for df = 8, P-value = 0.99. The distributions do not differ significantly and the data are consistent with colocalisation.
-
[e]
Tackling Question 5. The aim now is to test whether individual organelles that form a particular VOC exhibit colabelling (Mayhew and Lucocq 2011). The method presented here is limited to VOCs and to dual-labelling. Alternative approaches will be required to deal with large SOCs that are not associated with small vesicles or granules.
The analytical method involves assessment of whether a VOC profile (identified as containing a given target by labelling with a smaller gold particle) also labels for a second target (identified by a larger gold particle). To this end, individual VOC profiles are selected by using an SUR set of unbiased counting frames such as forbidden line frames (Gundersen 1977; Sterio 1984). Profiles are selected provided that they are entirely within the frame or touch its acceptable borders but do not touch its forbidden borders or their extensions. Profiles meeting these criteria are counted and assigned to one of four classes in a 2 × 2 table. The classes are: double-positive, positive only for one target, positive only for the other target and double-negative.
In the example provided in Table 8, VOC profiles were labelled by using 5-nm and 15-nm gold particles. Overall, 87 profiles were selected by the counting frames: 22 were found to be double-positive and 49 were double-negative. Only five profiles were labelled with 5-nm gold particles alone and 11- with 15-nm particles alone. Fisher’s exact probability test reveals that the labelling of profiles by the two sizes of gold particle is not independent (P-value < 0.001). Double-positive labelling differs from that which is to be expected for a random process. To help interpret the outcomes, an odds ratio is calculated. First, the ratio of labelled:unlabelled profiles for 15-nm gold particles is computed separately for each of the groups that are positive and negative for 5-nm gold. Then, an odds ratio is calculated from these initial ratios. The size of the odds ratio indicates whether a higher proportion of labelling by 15-nm particles occurs on profiles that are also labelled by 5-nm gold particles. For the data in Table 8, the odds ratio (4.400/0.224 = 19.60) indicates dual-labelling of these VOC profiles.
Table 8 Testing for the null hypothesis of colabelling independence of individual VOC profiles by antigens labelled by using 5-nm or 15-nm gold particles. Observed numbers are counted in variously labelled VOC profiles selected with unbiased counting frames and analysed by the Fisher exact test. The two labelling patterns are not independent (Fisher’s exact test yields P-value < 0.001). The odds ratio (4.400/0.224∼19.6) is consistent with dual-labelling -
[f]
Tackling Question 6. The aim now is to map labelling across cell compartments in 3D (Lucocq and Gawden-Bone 2009). An efficient sampling tool is the vertical rotator, which is used to provide random sections orthogonal to a convenient reference plane that might be intrinsic to the cells themselves (e.g., the basal membrane domain of a polarised epithelial cell or the metaphase plate of a cell undergoing mitosis) or external to them (e.g., the substratum of cultured cells).
Individual cells are selected by means of unbiased 3D counting tools (physical disectors) in order to identify those with a single point-like feature such as the nucleolus or centrosome. A short series of ultrathin sections spanning the feature is taken and immunolabelled. Cells displaying the feature are chosen if they appear on one section plane of the physical disector (the reference section) but not on a parallel section (the look-up section). These reference and look-up sections are separated by a known distance, t, equivalent to section thickness. FOVs are recorded and a vertical axis is identified on them that passes through the chosen feature of the selected cells.
On the sampled FOVs, gold particles associated with compartments (VOCs or SOCs) are identified and the distances (dv) from these gold particles to the vertical axis are classified with the help of a test lattice of systematic points (Nyengaard and Gundersen 2006) or lines running parallel to the vertical axis (Lucocq and Gawden-Bone 2010; Mayhew 2011). Alternatively, a ruler graduated into equidistant classes can be used. Numbers of gold particles falling into each compartment are summed for each class and then multiplied by the corresponding class mid-points. The sum of all these values (Σdv), multiplied by π/t, provides an estimator of the number of gold particles in the cell (Lucocq and Gawden-Bone 2010). The factor π/t is appropriate for post-embedding labelling because, clearly, the same gold particles cannot be present on both the reference and look-up sections.
A worked example is offered in Table 9 for a set of n = 15 cells sampled by using the physical disector and vertical rotator. The section thickness is 70 nm and the vertical axis passes through nucleoli appearing on the reference but not on the look-up section. A ruler with six size classes and a class interval of 0.8 μm on the specimen scale is employed and distances from gold particles to the vertical axis are classified for a set of three VOCs.
Table 9 Estimating the 3D spatial distribution of gold particles across three VOCs in n = 15 cells sampled by using the rotator. Distances in micrometers from a vertical axis (orthogonal to the cell substratum and passing through the nucleolus) are classified by using equidistant (0.8 μm) class intervals. Section thickness is 70 nm (equivalent to 0.07 μm) For VOC1, 17 gold particles were found within class 1, the mid-point for which was 0.4 μm. The sum of these distances was therefore 17 × 0.4 = 6.8 μm. For class 2, corresponding values were 44 gold particles, mid-point 1.2 μm and sum 52.8 μm. Summing across all classes, the cumulative distances for VOC1 amounted to Σdv = 267.6 μm. The number of gold particles in this set of cells was subsequently estimated as NG = Σdv × π/t = 267.6 × (3.1416/0.07) = 12009.9. Since this was obtained from n = 15 sampled cells, the number of gold particles labelling VOC1 was ∼801. Totals for VOC2 and VOC3 compartments were ∼1552 and 505 gold particles, respectively.
Discussion
The methods reviewed here comprise the first coherent set of methods for quantitative ICC at the ultrastructural level. All are based on sound principles of sampling, estimation and inferential statistics that are beneficial in terms of two cardinal statistical qualities, viz. precision and bias. The methods are also relatively simple and straightforward to apply. Although the presently worked examples are all synthetic (rather than based on real experimental data), the methods that they illustrate have been applied in a wide variety of studies involving bacteria, viruses and animal and plant cells and tissues (for a list of references, see Mayhew 2011). Moreover, they have been adopted in areas other than ICC in order to analyse, for instance, spatial distributions of nanoparticles within cells/tissues (Mühlfeld et al. 2007, 2008; Geiser et al. 2013) and the association of viruses with the symbiotic pair of Paramecium and Chlorella (Yashchenko et al. 2012). In principle, they could also be applied in immuno-quantum dot cytochemistry (e.g., Nisman et al. 2012; Killingsworth et al. 2012). Use of the disector and rotator tools offers the possibility for studying polarised, oriented or dividing cells and systems characterised by the movement of VOCs from one region to another (Lucocq and Gawden-Bone 2009). The combination of gold counting with other stereological tools opens up the further possibility of estimating LDs for VOCs as numbers of particles per cubic micrometer.
Apart from specificity, labelling efficiency (LE, a measure of the number of gold particles per target molecule) is another factor influencing the quantitation of gold labelling. Unfortunately, post-embedding labelling does not ensure that all of the target is labelled by gold particles and, sometimes, a target molecule might be associated with more than one particle. In practice, LE is influenced by various technical steps including fixation, embedding, labelling protocols, penetration of labelling reagents into the section and gold particle size. Methods for estimating LE have been discussed in greater detail elsewhere (Lucocq 1992; Griffiths 1993; Mayhew and Lucocq 2008b; Mayhew et al. 2009). Interestingly, a further potential development of the estimation procedure for labelling specificity (Lucocq and Gawden-Bone 2010) is to apply correction factors to apparent LE estimates. The method could be adapted also to incorporate an iterative approach to test the impact of various levels of knockdown between some arbitrary minimum level and 100 % knockout.
Notably, other quantitative methods and imaging modalities are available for immunogold quantification. In multiple-labelling studies, colocalisation has been analysed by other approaches including correlation functions (Philimonenko et al. 2000; Anderson et al. 2003; D’Amico and Skarmoutsou 2008b), which, for example, have been applied to HeLa cells to test for the colocalisation of nascent DNA and nuclear proteins. The correlation function approach tends to require more complex computations than those described here and the same is true of computer simulations for analysing spatial patterns of gold labelling on membranes (Nikonenko et al. 2000).
Several practical constraints ensue with regard to the use of various sizes of colloidal gold nanoparticles for multiple-labelling ICC. One factor is the ability to recognise unambiguously diverse sizes of particle and, in consequence, most studies have limited themselves to localising only two or three target molecules. Another factor is the observation that the efficiency of antibody binding is inversely related to particle size. For example, freeze-fracture immunolabelling of neutrophil leucocytes has shown that higher labelling intensities are obtained with ultrasmall (0.8–1.5 nm) gold particles in comparison to 5–15 nm colloidal gold particles (Robinson et al. 2000). Alternative approaches have been developed in attempts to deal with this issue.
One approach has involved the use of equally-sized nanoparticles made of different metals, e.g., gold, platinum and palladium particles of about 6 nm in diameter, bound to different primary antibodies (Bleher et al. 2008). To this end, multiply-labelled cryosections of skeletal muscle have been analysed for elemental composition by electron spectroscopic imaging via energy-filtering TEM. A variant on this approach has been the use of colloidal gold and quantum dot particles in combination with scanning transmission electron microscopy (STEM) and elemental composition analysis (Loukanov et al. 2010). An alternative approach (Philimonenko et al. 2014) has utilised other electron-dense nanoparticles identifiable on the basis of their differences in shape rather than size. With this approach, five different targets have been localised simultaneously in the nuclei of Hela cells by using two different sizes of colloidal gold (6 nm and 12 nm) together with gold-silver core-shell nanoparticles (doughnut-shaped, ∼13 nm), palladium nanoparticles (cubic, ∼15 nm) and gold nanoparticles (rod-shaped, ∼16 nm × 6 nm). Clearly, this approach has the potential to extend the range of multiple-labelling and the investigation of interactions between three or more targets.
As alluded to above, limitations of TEM include the tiny volume of specimen that can be processed and the thickness of section (50–90 nm) that needs to be cut to ensure good lateral resolution. In order to image larger volumes of the specimen, 3D reconstructions can be undertaken by serial physical sectioning or some form of tomography. Technical issues are associated with serial sectioning including section compression and damage/loss, knife score marks, specimen hardness and the maintenance of the faithful registration of reconstructed sections. At the TEM level, these problems are particularly acute and the production of ultrathin sections is a time-consuming and less efficient procedure. However, physical ultrathin sectioning underlies various forms of array tomography: for example, ribbons of sections can be deposited on suitable substrates and viewed by SEM (Micheva and Smith 2007; Wacker and Schroeder 2013) or uncoated block-faces of TEM sections can be viewed by using SEM in backscattering mode (Denk and Horstmann 2004). Both SEM and TEM can be used in conjunction with array tomography and ICC (Micheva et al. 2010; Kay et al. 2013).
Reconstruction in 3D by physical slicing can be avoided by electron tomography (Vanhecke et al. 2007) and STEM tomography (Baudoin et al. 2013). The techniques can be combined with pre-embedding immunolabelling. Interestingly, stereological tools can be combined with electron tomography (Vanhecke et al. 2007) to estimate compartment volumes, surface areas and numbers. Indeed, the smaller section thicknesses (down to a few nanometers) serve to reduce sources of bias that are difficult to correct in other ways. Application to pre-embedded immunogold-labelled sections (Griffiths 1993) would be required to determine absolute numbers of gold particles and to express LDs per cubic micrometer of VOC or per square mucrometer of SOC. Again, these 3D reconstruction methods are more demanding in terms of computing power and time. Moreover, they generate a large amount of information about a small sampling volume. This re-emphasises the need to embark on rigorous sampling protocols that balance noise (sampling variation + estimation precision) against the signal represented by the biological variation between independent items (e.g., cell groups, organisms) selected at the highest level of the multistage sampling cascade (Gundersen and Østerby 1981; Gupta et al. 1983; Mühlfeld et al. 2010).
Finally, although the stereological methods described here are performed manually, they offer the advantage of low cost and the twin statistical benefits of reasonable precision and minimal or no bias. Steps towards automatic or semi-automatic counting of immunogold particles on TEM ultrathin sections have been taken (see Lebonvallet et al. 1991; Brandt et al. 2001; Monteiro-Leal et al. 2003; D’Amico and Skarmoutsou 2008a, b; Wang et al. 2011) but these do not form a coherent and comprehensive set in the sense of the present methods. Although not bias-free, the approach described by Wang et al. (2011) can detect particles on single- and dual-labelled sections with low incidences of false-negative and false-positive recognition and of particle size misclassification. The approach can also be applied to pre-embedding labelling studies. Further developments to include efficient and minimally biased estimates for statistical analysis of spatial distributions of gold labelling are awaited.
References
Amiry-Moghaddam M, Ottersen OP (2013) Immunogold cytochemistry in neuroscience. Nat Neurosci 16:798–804
Anderson JB, Carol AA, Brown VK, Anderson LE (2003) A quantitative method for assessing co-localization in immunolabeled thin section electron micrographs. J Struct Biol 143:95–106
Baddeley AJ, Gundersen HJG, Cruz-Orive LM (1986) Estimation of surface area from vertical sections. J Microsc 142:259–276
Baudoin J-P, Jerome WG, Kübel C, Jonge N de (2013) Whole-cell analysis of low-density lipoprotein uptake by macrophages using STEM tomography. PLoS One 8:e55022
Bendayan M (1982) Double immunocytochemical labelling applying the protein A gold technique. J Histochem Cytochem 30:81–85
Bleher R, Kandela I, Meyer DA, Albrecht RM (2008) Immuno-EM using colloidal metal nanoparticles and electron spectroscopic imaging for co-localization at high spatial resolution. J Microsc 230:388–395
Brandt S, Heikkonen J, Engelhardt P (2001) Multiphase method for automatic alignment of transmission electron microscope images using markers. J Struct Biol 133:10–22
D’Amico F, Skarmoutsou E (2008a) Quantifying immunogold labelling in transmission immunoelectron microscopy. J Microsc 230:9–15
D’Amico F, Skarmoutsou E (2008b) Clustering and colocalization in transmission immunoelectron microscopy: a brief review. Micron 39:1–6
Daly LE, Bourke GJ (2000) Interpretation and uses of medical statistics. Blackwell Science, Oxford
Denk W, Horstmann H (2004) Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS Biol 2:e329
Geiser M, Quaile O, Wenk A, Wigge C, Eigeldinger-Berthou S, Hirn S, Schäffler M, Schleh C, Mőller W, Mall MA, Kreyling WG (2013) Cellular uptake and localization of inhaled gold nanoparticles in lungs of mice with chronic obstructive pulmonary disease. Part Fibre Toxicol 10:19
Geuze HJ, Slot JW, Ley P van der, Scheffer RCT (1981) Use of colloidal gold particles in double-labelling immunoelectron microscopy of ultrathin tissue sections. J Cell Biol 89:653–659
Griffiths G (1993) Fine structure immunocytochemistry. Springer, Heidelberg
Griffiths G, Lucocq JM (2014) Antibodies for immunolabeling by light and electron microscopy: not for the faint hearted. Histochem Cell Biol 142:347–360
Gundersen HJG (1977) Notes on the estimation of the numerical density of arbitrary profiles: the edge effect. J Microsc 111:219–223
Gundersen HJG, Jensen EB (1987) The efficiency of systematic sampling in stereology and its prediction. J Microsc 147:229–263
Gundersen HJG, Østerby R (1981) Optimizing sampling efficiency of stereological studies in biology: or “do more less well!”. J Microsc 121:65–73
Gundersen HJG, Jensen EBV, Kiêu K, Nielsen J (1999) The efficiency of systematic sampling in stereology: reconsidered. J Microsc 193:199–211
Gupta M, Mayhew TM, Bedi KS, Sharma AK, White FH (1983) Inter-animal variation and its influence on the overall precision of morphometric estimates based on nested sampling designs. J Microsc 131:147–154
Howard CV, Reed MG (2005) Unbiased stereology. Three-dimensional measurement in microscopy, 2nd edn. Garland Science/BIOS Scientific, Abingdon
Kay KR, Smith C, Wright AK, Serrano-Pozo A, Pooler AM, Koffie R, Bastin ME, Bak TH, Abrahams S, Kopeikina KJ, McGuone D, Frosch MP, Gillingwater TH, Hyman BT, Spires-Jones TL (2013) Studying synapses in human brain with array tomography and electron microscopy. Nat Protoc 8:1366–1380
Killingsworth MC, Lai K, Wu X, Yong JLC, Lee CS (2012) Quantum dot immunocytochemical localization of somatostatin in somatostatinoma by widefield epifluorescence, super-resolution light, and immunoelectron microscopy. J Histochem Cytochem 60:832–843
Klein T, Proppert S, Sauer M (2014) Eight years of single-molecule localization microscopy. Histochem Cell Biol 141:561–575
Koster AJ, Klumperman J (2003) Electron microscopy in cell biology: integrating structure and function. Nat Rev Mol Cell Biol 4 (suppl):SS6–SS10
Lebonvallet S, Mennesson T, Bonnet N, Girod S, Plotkowski C, Hinnrasky J, Puchelle E (1991) Semi-automatic quantitation of dense markers in cytochemistry. Histochem Cell Biol 96:245–250
Loukanov A, Karnasawa N, Danev R, Shigemoto R, Nagayama K (2010) Immunolocalization of multiple membrane proteins on a carbon replica with STEM and EDX. Ultramicroscopy 110:366–374
Lucocq J (1992) Quantitation of gold labelling and estimation of labelling efficiency with a stereological counting method. J Histochem Cytochem 40:1929–1936
Lucocq J (1994) Quantitation of gold labelling and antigens in immunolabelled ultrathin sections. J Anat 184:1–13
Lucocq J (2012) Can data provenance go the full monty? Trends Cell Biol 22:229–230
Lucocq JM, Gawden-Bone C (2009) A stereological approach for estimation of cellular immunolabeling and its spatial distribution in oriented sections using the rotator. J Histochem Cytochem 57:709–719
Lucocq JM, Gawden-Bone C (2010) Quantitative assessment of specificity in immunoelectron microscopy. J Histochem Cytochem 58:917–927
Lucocq JM, Hacker C (2013) Cutting a fine figure. On the use of thin sections in electron microscopy to quantify autophagy. Autophagy 9:1–6
Lucocq JM, Habermann A, Watt S, Backer JM, Mayhew TM, Griffiths G (2004) A rapid method for assessing the distribution of gold labelling on thin sections. J Histochem Cytochem 52:991–1000
Lucocq JM, Mayhew TM, Schwab Y, Steyer AM, Hacker C (2014) Systems biology in 3D space—enter the morphome. Trends Cell Biol. doi:10.1016/j.tcb.2014.09.008
Mattfeldt T, Mall G, Gharehbaghi H, Möller P (1990) Estimation of surface area and length with the orientator. J Microsc 159:301–317
Mayhew TM (1991) The new stereological methods for interpreting functional morphology from slices of cells and organs. Exp Physiol 76:639–665
Mayhew TM (2008) Taking tissue samples from the placenta: an illustration of principles and strategies. Placenta 29:1–14
Mayhew TM (2011) Mapping the distributions and quantifying the labelling intensities of cell compartments by immunoelectron microscopy: progress towards a coherent set of methods. J Anat 219:647–660
Mayhew TM, Desoye G (2004) A simple method for comparing immunogold distributions in two or more experimental groups illustrated using GLUT1 labelling of isolated trophoblast cells. Placenta 25:580–584
Mayhew TM, Lucocq JM (2008a) Quantifying immunogold labelling patterns of cellular compartments when they comprise mixtures of membranes (surface-occupying) and organelles (volume-occupying). Histochem Cell Biol 129:367–378
Mayhew TM, Lucocq JM (2008b) Developments in cell biology for quantitative immunoelectron microscopy based on thin sections—a review. Histochem Cell Biol 130:299–313
Mayhew TM, Lucocq JM (2011) Multiple-labelling immunoEM using different sizes of colloidal gold: alternative approaches to test for differential distribution and colocalization in subcellular structures. Histochem Cell Biol 135:317–326
Mayhew TM, Reith A (1988) Practical ways to correct cytomembrane surface densities for the loss of membrane images that results from oblique sectioning. In: Reith A, Mayhew TM (eds) Stereology and morphometry in electron microscopy. Problems and solutions. Hemisphere, New York
Mayhew TM, Lucocq JM, Griffiths G (2002) Relative labelling index: a novel stereological approach to test for non-random immunogold labelling of organelles and membranes on transmission electron microscopy thin sections. J Microsc 205:153–164
Mayhew T, Griffiths G, Habermann A, Lucocq J, Emre N, Webster P (2003) A simpler way of comparing the labelling densities of cellular compartments illustrated using data from VPARP and LAMP-1 immunogold labelling experiments. Histochem Cell Biol 119:333–341
Mayhew TM, Griffiths G, Lucocq JM (2004) Applications of an efficient method for comparing immunogold labelling patterns in the same sets of compartments in different groups of cells. Histochem Cell Biol 122:171–177
Mayhew TM, Mühlfeld C, Vanhecke D, Ochs M (2009) A review of recent methods for efficiently quantifying immunogold and other nanoparticles using TEM sections through cells, tissues and organs. Ann Anat 191:153–170
Micheva KD, Smith SJ (2007) Array tomography: a new tool for imaging the molecular architecture and ultrastructure of neural circuits. Neuron 55:25–36
Micheva KD, Busse B, Weller NC, O’Rourke N, Stephen J, Smith SJ (2010) Single synapse analysis of a diverse synapse population: proteomic imaging methods and markers. Neuron 68:639–653
Mironov AA Jr, Mironov AA (1998) Estimation of subcellular organelle volume from ultrathin sections through centrioles with a discretized version of the vertical rotator. J Microsc 192:29–36
Monteiro-Leal LH, Troster H, Campanati L, Spring H, Trendelenburg MF (2003) Gold finder: a computer method for fast automatic double gold labelling detection, counting, and color overlay in electron microscopic images. J Struct Biol 141:228–239
Mühlfeld C, Mayhew TM, Gehr P, Rothen-Rutihauser B (2007) A novel quantitative method for analysing the distribution of nanoparticles between different tissue and intracellular compartments. J Aerosol Med 20:395–407
Mühlfeld C, Rothen-Rutishauser B, Blank F, Vanhecke D, Ochs M, Gehr P (2008) Interactions of nanoparticles with pulmonary structures and cellular responses. Am J Physiol Lung Cell Mol Physiol 294:L817–L829
Mühlfeld C, Nyengaard JR, Mayhew TM (2010) A review of state-of-the-art stereology for better quantitative 3D morphology in cardiac research. Cardiovasc Pathol 19:65–82
Nickel BM, DeFranco BH, Gay VL, Murray SA (2008) Clathrin and Cx43 gap junction plaque endoexocytosis. Biochem Biophys Res Commun 374:679–682
Nikonenko AG, Nikonenko IR, Skibo GG (2000) Computer simulation approach to the quantification of immunogold labelling on plasma membrane of cultured neurons. J Neurosci Meth 96:11–17
Nisman R, Dellaire G, Ren Y, Li R, Bazett-Jones DP (2012) Application of quantum dots as probes for correlative fluorescence, conventional, and energy-filtered transmission electron microscopy. J Histochem Cytochem 52:13–18
Nyengaard JR, Gundersen HJG (1992) The isector: a simple and direct method for generating isotropic, uniform random sections from small specimens. J Microsc 165:427–431
Nyengaard JR, Gundersen HJG (2006) Direct and efficient stereological estimation of total cell quantities using electron microscopy. J Microsc 222:182–187
Oddone A, Vilanova IV, Tam J, Lakadamyali M (2014) Super-resolution imaging with stochastic single-molecule localization: concepts, technical developments, and biological applications. Microsc Res Tech 77:502–509
Owen DM, Magenau A, Williamson DJ, Gaus K (2013) Super-resolution imaging by localization microscopy. Meth Mol Biol 950:81–93
Petrie A, Sabin C (2000) Medical statistics at a glance. Blackwell Science, Oxford
Philimonenko AA, Janáček J, Hozák P (2000) Statistical evaluation of colocalization patterns in immunogold labelling experiments. J Struct Biol 132:201–210
Philimonenko VV, Philimonenko AA, Šloufová I, Hrubý M, Novotný F, Halbhuber Z, Krivjanská M, Nebesářová J, Šlouf M, Hozák P (2014) Simultaneous detection of multiple targets for ultrastructural immunocytochemistry. Histochem Cell Biol 141:229–329
Robinson JM, Takizawa T, Vandré DD (2000) Enhanced labelling efficiency using ultrasmall immunogold probes: immunocytochemistry. J Histochem Cytochem 48:487–492
Roth J (1996) The silver anniversary of gold: 25 years of the colloidal gold marker system for immunocytochemistry and histochemistry. Histochem Cell Biol 106:1–8
Schmiedl A, Ochs M, Mühlfeld C, Johnen G, Brasch F (2005) Distribution of surfactant proteins in type II pneumocytes of newborn, 14-day old, and adult rats: an immunoelectron microscopic and stereological study. Histochem Cell Biol 124:465–476
Sibarita J-B (2014) High-density single-particle tracking: quantifying molecule organization and dynamics at the nanoscale. Histochem Cell Biol 141:587–595
Slot JW, Geuze HJ (1985) A new method for preparing gold probes for multiple-labelling cytochemistry. Eur J Cell Biol 38:87–93
Sterio DC (1984) The unbiased estimation of number and sizes of arbitrary particles using the disector. J Microsc 134:127–136
Vanhecke D, Studer D, Ochs M (2007) Stereology meets electron tomography: towards quantitative 3D electron microscopy. J Struct Biol 159:443–450
Vedel-Jensen EB, Gundersen HJG (1993) The rotator. J Microsc 171:35–44
Wacker I, Schroeder RR (2013) Array tomography. J Microsc 252:93–99
Wang R, Pokhariya H, McKenna SJ, Lucocq J (2011) Recognition of immunogold markers in electron micrographs. J Struct Biol 176:151–158
Webster P, Schwarz H, Griffiths G (2008) Preparation of cells and tissues for immuno EM. Meth Cell Biol 88:45–58
Yashchenko VV, Gavrilova OV, Rautian MS, Jakobsen KS (2012) Association of Paramecium bursaria Chlorella viruses with Paramecium bursaria cells: ultrastructural studies. Eur J Protistol 48:149–159
Acknowledgments
For over 40 years, I have greatly enjoyed fruitful interactions and collaborations with many colleagues, students and fellow teachers on international courses of stereology, electron microscopy and cell and molecular biology.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mayhew, T.M. Quantitative immunocytochemistry at the ultrastructural level: a stereology-based approach to molecular nanomorphomics. Cell Tissue Res 360, 43–59 (2015). https://doi.org/10.1007/s00441-014-2038-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00441-014-2038-y