Abstract
The ion regulation mechanisms of fishes have been recently studied in zebrafish (Danio rerio), a stenohaline species. However, recent advances using this organism are not necessarily applicable to euryhaline fishes. The euryhaline species medaka (Oryzias latipes), which, like zebrafish, is genetically well categorized and amenable to molecular manipulation, was proposed as an alternative model for studying osmoregulation during acclimation to different salinities. To establish its suitability as an alternative, the present study was conducted to (1) identify different types of ionocytes in the embryonic skin and (2) analyze gene expressions of the transporters during seawater acclimation. Double/triple in situ hybridization and/or immunocytochemistry revealed that freshwater (FW) medaka contain three types of ionocyte: (1) Na+/H+ exchanger 3 (NHE3) cells with apical NHE3 and basolateral Na+-K+-2Cl− cotransporter (NKCC), Na+-K+-ATPase (NKA) and anion exchanger (AE); (2) Na+-Cl− cotransporter (NCC) cells with apical NCC and basolateral H+-ATPase; and (3) epithelial Ca2+ channel (ECaC) cells [presumed accessory (AC) cells] with apical ECaC. On the other hand, seawater (SW) medaka has a single predominant ionocyte type, which possesses apical cystic fibrosis transmembrane conductance regulator (CFTR) and NHE3 and basolateral NKCC and NKA and is accompanied by smaller AC cells that express lower levels of basolateral NKA. Reciprocal gene expressions of decreased NHE3, AE, NCC and ECaC and increased CFTR and NKCC in medaka gills during SW were revealed by quantative PCR analysis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Euryhaline fishes are able to survive within a wide range of salinities. To adapt to dramatic fluctuations of osmotic gradients in aquatic environments, fish gills contain ionocytes (also called chloride cells), which are specialized mitochondrion-rich cells critical for maintaining the ionic compositions of body fluids. In freshwater (FW), ionocytes actively absorb Na+ and Cl− from external FW to compensate for the passive loss of ions in the urine and diffusion through the body and gill surfaces. In contrast, fishes in seawater (SW) must excrete excess Na+ and Cl− through gill ionocytes to maintain the osmotic pressure of blood substantially below that of the external media (Hirose et al. 2003; Evans et al. 2005; Hwang and Lee 2007). The mechanism by which SW-type ionocytes secrete NaCl has been studied for several decades. In a well-accepted model, the basolateral Na+-K+-2Cl− cotransporter (NKCC) carries one sodium, one potassium and two chloride ions into the cell down their electrochemical gradients, which are generated by the action of Na+-K+-ATPase (NKA). Accumulated intracellular Cl− is then extruded across the apical membrane via the apical cystic fibrosis transmembrane conductance regulator (CFTR) Cl− channel (Hirose et al. 2003; Evans et al. 2005; Hwang and Lee 2007; Evans 2008). Moreover, the transepithelial electrical potential across the gill epithelium drives Na+ across the leaky junction between the ionocytes and the accessory (AC) cells (Hwang et al. 2011). Some studies have demonstrated K+ channels to be involved in the recycling or secretion of K+ in SW ionocytes (Suzuki et al. 1999; Tse et al. 2006; Furukawa et al. 2012 ) and identification of K+ channels in medaka remains to be done in the future. However, the ion-uptake mechanism and ionocyte subtypes in FW fishes are more complicated and remain a source of contention (Evans 2011; Hwang et al. 2011; Dymowska et al. 2012; Hiroi and McCormick 2012; Hwang and Lin 2013).
Recent studies of the skin and gills of zebrafish (Danio rerio) have demonstrated that at least four ionocyte subtypes with distinct functions and mechanisms are involved in ion uptake (Chang and Hwang 2011; Dymowska et al. 2012; Hwang and Lin 2013; Hwang and Chou 2013); these ionocytes are as follows: (1) H+-ATPase-rich (HR) cells with apical H+-ATPase (HA), Na+/H+ exchanger (NHE3b) and Rhesus glycoproteins (Rhcg1), which are responsible for Na+ uptake/acid secretion/ammonia excretion; (2) NKA-rich (NaR) cells with apical epithelial Ca2+ channel (ECaC), which function primarily in Ca2+ uptake; (3) Na+-Cl− cotransporter-expressing (NCC) cells, which are involved in Cl− and/or Na+ uptake; and (4) K+ secretion (KS) cells, which contain mRNA encoding the K+ channel Kir1.1 (ROMK) and have been proposed to be involved in K+ secretion. The embryonic skin and adult gills of euryhaline tilapia (Oreochromis mossambicus) contain three FW-types of ionocyte: (1) basolateral-NKA-expressing type I ionocytes; (2) basolateral-NKA- and apical-NCC-expressing type II ionocytes; and (3) basolateral-NKA/NKCC- and apical-NHE3-expressing type III ionocytes (Hiroi et al. 2008; Inokuchi et al. 2009). The findings of acclimation experiments with different salinities or ionic compositions suggest that type II ionocytes are involved mainly in Cl−/Na+ uptake and type III cells in Na+ uptake/acid secretion. During the transfer from FW to SW, type III cells transform into type IV (SW-type) cells, which secrete NaCl via apical CFTR and basolateral NKCC and NKA (Hiroi et al. 2008; Inokuchi et al. 2008, 2009). However, recent studies have only identified two subtypes of ionocyte in the gills of FW-acclimated euryhaline rainbow trout (Oncorhynchus mykiss); these ionocytes differed as to whether they expressed peanut lectin agglutinin (PNA) or not (PNA+ and PNA−) (Galvez et al. 2002). Isolated PNA− cells exhibited higher expression of HA and engaged in bafilomycin-sensitive, acid-activated Na+ uptake; PNA+ cells, on the other hand, were proposed to be involved in Cl− uptake/base secretion and Ca2+ uptake (Galvez et al. 2002). Apparently, FW-type ionocyte subtypes are highly variable between fishes. Such diversity has been ascribed to differences in the species studied, as well as the criteria for identification and/or the methods used to functionally analyze the ionocytes (or transporters); therefore, many issues surrounding FW ionocytes remain the subject of debate (Hwang et al. 2011; Hwang and Lin 2013). As a consequence, additional work is required to generate a comprehensive working model of the mechanisms of fish ion regulation.
Zebrafish, a stenohaline FW fish, is a commonly used animal model for human disease and drug screening (Ali et al. 2011) and is also one of the most extensively studied species for determining the functions and regulatory pathways of ionocytes in fishes (Hwang and Chou 2013). Utilization of genome databases and advanced cellular/molecular physiological approaches has enabled ion uptake mechanisms to be more precisely examined in zebrafish over the last decade; however, the knowledge gained from zebrafish cannot be directly applied to many euryhaline and marine species (Hwang 2009; Hwang and Perry 2010; Evans 2011; Hwang et al. 2011; Dymowska et al. 2012; Hwang and Chou 2013). Thus, there is a growing need for an alternative model; one possibility is medaka (Oryzias latipes), a euryhaline species that is highly adaptable to different salinities and, like zebrafish, is amenable to genetic manipulation and suitable for studies of organogenesis (Wittbrodt et al. 2002; Takeda and Shimada 2010). Previous studies have identified only two types of ionocyte in FW-acclimated medaka and one type in SW-acclimated medaka (Kang et al. 2008, 2010; Wu et al. 2010; Lin et al. 2012). The expression and function of ion transporters in such ionocytes have not been well characterized and it is possible that additional types of ionocyte in the skin or gills of medaka remain to be identified.
To establish medaka as a new model for fish ion regulation, the present study was designed to identify: (1) whether other ionocyte types exist in medaka, in addition to those previously described (Lin et al. 2012); (2) the ion transporters specifically expressed in medaka ionocytes; and (3) the gene expressions of ion transporters after SW transfer.
Materials and methods
Experimental animals
Mature Japanese medaka (Oryzias latipes) were reared in tanks with circulating tap water at 27 °C, with a photoperiod of 14 h light/10 h dark. Females spawned every day and fertilized egg clusters were collected from the belly of females and rinsed with running tap water, in order to remove any mucus and debris and separate the clusters into single eggs. The eggs were incubated in different media depending on experiments (see below). Embryos usually hatched at 7 days post-fertilization (dpf) and newly hatched embryos were used for the following experiments. The experimental protocols were approved by the Academia Sinica Institutional Animal Care and Utilization Committee (approval no.: RFiZOOHP2009060).
Acclimation experiment
Seawater (SW) of 30 ‰ was prepared by adding the appropriate amounts of sea salt (Instant Ocean; Aquarium Systems, Mentor, OH, USA) to the local tap water (FW). High-Na+ (9.2–10.5 mM), low-Na+ (0.03–0.05 mM), high-Cl− (9.5–10.3 mM) and/or low-Cl− (0.03–0.04 mM) artificial FWs were prepared by adding adequate CaSO4.2H2O, MgSO4.7H2O, NaCl, Na2SO4, MgCl2.6H2O, K2HPO4 and KH2PO4 to double-deionized water (Milli-RO60; Millipore, Temecula, CA, USA) (Wang et al. 2009). The concentrations of other major ions (Ca2+, 0.16–0.23 mM; Mg2+, 0.18–0.24 mM; K+, 0.15–0.22 mM; pH, 6.7–6.9) in the artificial FWs mimicked the levels of the local tap water (Wang et al. 2009). Adult medaka were acclimated to different artificial FWs or SW for 2 weeks to collect gills for qRT-PCR and the waters were replaced every 2 days. For SW larvae, fertilized eggs were directly transferred from FW to SW until hatched (7 dpf) and the waters were replaced every day. In SW transfer experiments, gills of adult medaka were collected at 3, 6, 12 and 24 h after transfer of the fish from FW to SW.
Molecular cloning and phylogenetic analysis
Partial open reading frames of medaka slc4a1a (AE1a, Ensembl gene ID: ENSORLT00000006918), slc4a1b (AE1b, ENSORLT00000003183), slc12a2a (NKCC, ENSORLT00000024013) and trpv6 (ECaC; ENSORLT00000014936) homologs obtained from the genome were confirmed with reference to the expressed sequence tag database. For NCCs, peptide sequences from other species (teleosts were given the highest priority) were used to BLAST genome databases (the Ensembl Genome Browser system) for medaka homologs. In silico predictions of full-length medaka NCC homologs obtained from the genome were confirmed with reference to the GenBank database. Specific primers (Table 2) were designed for reverse-transcriptase polymerase chain reaction (RT-PCR) analysis. PCR products thus obtained were subcloned into a pGEM-T Easy vector (Promega, Madison, WI, USA) and the nucleotide sequences were determined with an ABI 377 sequencer (Applied Biosystems, Warrington, UK). Sequence analysis was conducted using BLASTx (NCBI). Identified candidates were confirmed to be members of the core NCC protein family, through alignment of the deduced amino-acid sequences of cloned medaka NCCs with all known NCC protein sequences available in public databases using ClustalX (Table 1); candidates were then subjected to phylogenetic analysis using the neighbor-joining (NJ) method. Ten thousand bootstrap replicate analyses were carried out with MEGA 5.
RT-PCR
Medaka embryos, gills, or organs were collected and homogenized in Trizol reagent (Ambion, Woodward, TX, USA). Total RNA was purified following the manufacturer’s protocol. Genomic DNA was removed by treating total RNA with DNase1 (Qiagen, Hilden, Germany) at 37 °C for 15 min. The total amount and quality of RNA was determined at absorbances of 260 and 280 nm by spectrophotometry (ND-1000; NanoDrop Technol, Wilmington, DE, USA). All RNA pellets were stored at −20 °C. For cDNA synthesis, 5 μg of total RNA was reverse-transcribed in a final volume of 20 μl containing 0.5 mM dNTPs, 2.5 μM oligo(dT)20, 5 mM dithiothreitol, 40 units of an RNase inhibitor and 200 units of SuperScript III RT (Invitrogen, Carlsbad, CA, USA) for 1.5 h at 55 °C, followed by incubation for 15 min at 70 °C. Thereafter, remnant RNA was removed by incubation with 20 units of Escherichia coli RNase H (Invitrogen) for 20 min at 37 °C. For PCR amplification, 1 μl of cDNA (<500 ng) was used as template in a 25-μl final reaction volume containing 0.25 μM dNTP, 1.25 units of Gen-Tag polymerase (Genemark, Taipei, Taiwan) and 0.2 μM of each primer. Thirty cycles were performed for each reaction.
Expression of medaka ncc gene mRNA in different tissues
The expression of the medaka ncc, ncc-like 1 and ncc-like 2 genes were examined in various organs using RT-PCR. Total RNA samples were extracted from the brain, gills, eyes, heart, liver, intestines, kidneys, muscles and fins of medaka. Samples were subjected to RT-PCR analysis with the primer sets shown in Table 2.
Quantitative (q)RT-PCR
Expression levels of ae1b, ecac,slc9a3 (ENSORLG00000009128), ncc-like2, abcc7 (ENSORLG00000019555) and slc12a2a mRNAs were measured by qRT-PCR with a Roche Lightcycler 480 (Roche, Penzberg, Germany). The final reaction volume in each well was 10 μl, which consisted of 5 μl of 2× SYBR green master mix (Roche), 3.2 ng of cDNA and 50 nM of primers. Standard curves for each gene were generated in the linear range and the gene encoding ribosomal protein (RP)L7 (ENSORLT00000009998) was used as an internal control. The primer sets used for qRT-PCR are shown in Table 2. The specificity of the primer sets was confirmed by the presence of a single peak in the dissociation curve analysis and by the detection of a single band of the correct size by gel electrophoresis.
Whole-mount in situ hybridization
For in situ hybridization, primers (Table 2) were designed against the conserved regions of ncc-like 1, ncc-like 2, ae1b, ecac and nkcc and used to obtain DNA fragments by PCR; these were individually inserted into the pGEM-T Easy vector (Promega). The inserted fragments were amplified with the T7 and SP6 primers by PCR and the respective products were used as templates for in vitro transcription with T7 or SP6 RNA polymerase (Roche) in the presence of digoxigenin (DIG)-UTP (Roche), to synthesize sense and anti-sense probes, respectively. DIG-labeled RNA probes were examined using RNA gels and their quality and concentrations were determined using dot blot assays. Medaka embryos were anesthetized on ice and fixed with 4 % paraformaldehyde in phosphate-buffered saline (PBS; 1.4 mM NaCl, 0.2 mM KCl, 0.1 mM Na2HPO4 and 0.002 mM KH2PO4; pH 7.4) solution at 4 °C overnight. Afterward, samples were washed with diethylpyrocarbonate (DEPC)-PBST (PBS with 0.1 % Tween-20) several times (10 min/wash). Samples were subsequently incubated with hybridization buffer (HyB; 50 % formamide, 5× SSC, and 0.1 % Tween 20) at 65 °C for 5 min and then with HyB containing 500 μg/ml yeast tRNA at 65 °C for 4 h. Following overnight hybridization with 100 ng/ml DIG-labeled antisense or sense RNA probe, embryos were serially washed with 50 % formamide-2× SSC (65 °C for 20 min), 2× SSC (65 °C for 10 min), 2× SSC (65 °C for 10 min), 0.2× SSC (65 °C for 30 min, twice) and PBST (room temperature for 10 min). Embryos were then immunoreacted with an alkaline phosphatase-coupled anti-DIG antibody (1:8,000) and stained with nitro blue tetrazolium (NBT) (Roche) and 5-bromo-4-chloro-3-indolyl phosphate (NCIP) (Roche). Fluorescence staining was conducted with a commercial kit, the TSA Plus Fluorescence System (Perkin-Elmer, Boston, MA, USA). Fluorescence signals were amplified through fluorescein-tyramide signal amplification (TSA).
Whole-mount immunocytochemistry
For double immunocytochemistry, medaka samples were fixed with 4 % paraformaldehyde in a PBS solution at 4 °C for 2 h. After being washed with PBS, the samples were incubated with 3 % bovine serum albumin (BSA) for 1 h to block nonspecific binding. Samples were then incubated overnight at 4 °C with the primary antibodies. After washing with PBS for 20 min, samples were further incubated in Alexa Fluor 568 goat anti-rabbit (Molecular Probes; diluted 1: 200 with PBS) and Alexa Fluor 488 goat anti-mouse antibodies (Molecular Probes; diluted 1:200 with PBS) for 2 h at room temperature and then observed under a microscope. The primary antibodies used were as follows: anti-medaka NHE3 polyclonal (diluted 1:100) (Lin et al. 2012), anti-human NKCC (T4) monoclonal (diluted 1:100) [Developmental Studies Hybridoma Bank (DSHB), University of Iowa, Ames, IA, USA], anti-avian NKA (α5) monoclonal (diluted 1: 200) (DSHB), anti-killifish (Fundulus heteroclitus) HA polyclonal (diluted 1:100) (Katoh et al. 2003) and anti-human CFTR monoclonal (diluted 1:100) (R&D Systems, Boston, MA, USA). The anti-NCC antibody was generated in rabbits injected with a 19-residue synthetic peptide (52-SEPQIEGEQPPTPKGKTPI-70) of medaka NCC-like 2, used at a dilution of 1:100. For triple in situ hybridization and immunocytochemistry, medaka samples were first hybridized in situ and subsequently subjected to immunohistochemistry. After in situ hybridization, the ae1b, ncc-like 2, slc12a2a and trpv6 genes were washed with PBS and incubated with 3 % BSA for 2 h, before being incubated overnight at 4 °C with the indicated primary antibody. The samples were subsequently incubated with Alexa Fluor 488 (or 568) goat anti-rabbit and Alexa Fluor 568 (or 488) goat anti-mouse antibodies for 2 h at room temperature. Images were obtained with an upright microscope (Imager M1; Carl Zeiss, Oberkochen, Germany) or a Leica TCS-SP5 confocal laser scanning microscope (Leica Lasertechnik, Heidelberg, Germany). To generate z-plan images, 30 serial sections (0.5 μm/section; at a total thickness of 15 μm) of confocal microscopic images were acquired and subjected to image reconstruction and analysis.
Western blot analysis
Western blotting was performed to characterize the anti-NCC antiserum. Gill protein samples (50 mg/well) were subjected to 10 % sodium dodecyl sulfate polyacrylamide gel electrophoresis at 100 V for 2 h. After separation, proteins were transferred to polyvinylidene difluoride membranes (Millipore) at 100 V for 2 h. After being blocked for 1.5 h in 5 % nonfat milk, blots were incubated first with anti-NCC antiserum (overnight at 4 °C, diluted 1:500) and then with an AP-conjugated goat anti-rabbit IgG antibody (Jackson Laboratories, West Grove, PA, USA; 2 h at room temperature, diluted 1:5,000). Blots were developed with BCIP/NBT.
Statistical analysis
Values are presented as the means ± standard deviation (SD). Values from each condition were analyzed using a one-way analysis of variance (ANOVA) followed by Tukey’s pairwise comparisons. Student’s unpaired t test (two-tailed) was used for simple comparisons of 2 means. In all cases, significance was accepted at a level of 0.05.
Results
Genomic prediction, identification and annotation of medaka NCCs
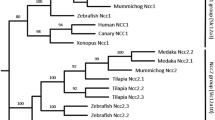
Based on sequences from the Ensembl genome database, the following three NCC homologs in medaka were predicted, cloned and annotated: olslc12a3 (olNCC; NP_001038545.1), olslc12a10a (olNCC-like 1; KJ489428) and olslc12a10b (olNCC-like 2; KJ489429) (Fig. 1). The coding regions of the olslc12a3, olslc12a10a and olslc12a10b cDNAs are 3084, 3006 and 3015 bp in length, respectively (Supporting Information). The genomic locations of these genes were determined, to confirm their identities as medaka NCC isoforms ( Table 1). In the medaka genome, the three NCC homologs are located on different chromosomes. The olNCC gene, which is clustered with conventional NCC (SLC12A3), is composed of 26 exons, as also observed in human (Homo sapiens) and dog (Canis lupus familiaris) (data not shown). The teleost-specific olNCC-like 1 and olNCC-like 2 (SLC12A10) genes consist in 27 exons each (data not shown). Comparisons of the genomic exon-intron structure revealed that olNCC-like 1 and olNCC-like 2 are most similar to zebrafish NCC-like 2 (Fig. 1), which is expressed in the gills (Wang et al. 2009); we therefore chose to focus on these two genes in the following experiments.
Routed phylogenetic analysis of the SLC12A3 and SLC12A10 amino acid sequences. The putative NCC sequences of other species were obtained from the Ensembl database, as shown in Table 1. Consensus trees were generated using the neighbor-joining method with the pairwise deletion gap calculating option. The results were confirmed by 10,000 bootstraps. Numbers indicate bootstrap values and the scale bar units are the number of amino acid substitutions per site. dr Danio rerio; ol Oryzias latipes; xm Xiphophorus maculates; on Oreochromis niloticus ; ga Gasterosteus aculeatus; hs Homo saplens; cf Canis lupus familiaris ; mm Mus musculus; ac Anolis carolinensis; gg Gallus gallus; xt Xenopus laevis; lc Latimeria chalumnae; tn Tetraodon nigroviridis
Expressions of ncc genes in different tissues of FW medaka
Medaka slc12a3, slc12a10a and slc12a10b exhibit tissue-specific patterns of expression, as shown by RT-PCR (Fig. 2). The slc12a3 gene is mainly expressed in the heart, intestine and kidney, while slc12a10b is expressed in the gills, liver and spleen; on the other hand, the expression of slc12a10a appears to be more universal, with mRNA transcripts in the brain, gills, eye, intestine, spleen, kidney and muscle (Fig. 2). Notably, both slc12a10a and slc12a10b were strongly expressed in the gills (Fig. 2).
NCC-expressing ionocytes in FW medaka
This study has thus far demonstrated that two paralogs, slc12a10a and slc12a10b, encoding teleost-specific NCC-like proteins, are expressed in the gills of medaka. In situ hybridization with specific probes was subsequently performed to determine the expression patterns of these two genes. Hybridization with probes against slc12a10b (ncc-like 2) revealed the typical salt-and-pepper pattern of ionocytes (Fig. 3a), while slc12a10a (ncc-like 1) signals were mainly distributed in the branchial and nearby yolk-sac areas, with a pattern distinct from that of ionocytes (Fig. 3b). The slc12a10b (ncc-like 2) gene was therefore the target for the subsequent experiments. Triple labeling of ncc-like 2 mRNA, NKA and NHE3 demonstrated that ncc-like 2 is expressed in a subset of NKA-labeled ionocytes (Fig. 3c, d, f–i) but not in NHE-labeled ones (Fig. 3e, f, h, i). For further confirmation, double immunocytochemical analysis was performed with anti-medaka NCC-like 2 antisera and two heterologous antibodies (T4 and α5) (Fig. 4a–g). T4 antibody can be used to detect both apical NCC and basolateral NKCC in tilapia ionocytes (Hiroi et al. 2008; Hiroi and McCormick 2012). Similarly, T4 was able to label the apical NCC and basolateral NKCC in two distinct groups of ionocytes in medaka (Fig. 4a). Medaka NCC antisera only labeled apical ionocytes detected by T4 (Fig. 4b, c). Furthermore, NCC signals were located at the apical membrane of the NKA-labeled ionocytes, as demonstrated by observation of double labeling with the anti-medaka NCC (Fig. 4d, e) and α5 (anti-NKA) (Fig. 4e) antibodies under high magnification (Fig. 4f) and z-plan image (Fig. 4g). Western blot using NCC-like 2 antisera revealed a single immunoreactive band of 113 kDa (the predicted size) in medaka gill samples (Fig. 4h), confirming antibody specificity.
In situ hybridization against ncc-like 1 and -like 2 mRNAs and triple labeling together with NKA and NHE3 proteins in FW medaka at 7 dpf. Hybridization against ncc-like 2 (a, arrows) but not ncc-like 1 mRNA (b, asterisks), revealed the salt-and-pepper pattern of ionocytes. The same embryo was triple labeled against ncc-like 2 mRNA (c), NKA (d) and NHE3 (e), as shown as a merged image in (f). Ionocytes from the same embryo were triple labeled with ncc-like 2 mRNA (g) and NKA and NHE3 (h), as shown as a merged image in (i). Arrows indicate cells expressing ncc-like 2 mRNA (dashed line) and arrowheads indicate cells with NHE3 signals (dotted line; c–i). Scale: (a, b) 100 μm, (c–f) 50 μm, (g–i) 10 μm
Double immunocytochemical labeling of NCC and NKA in FW medaka at 7 dpf. Staining of the same embryos with T4 antibody (T4) (a) and anti-medaka NCC-like 2 antibody (NCC) (b); the merged image is shown in (c). Arrows indicate ionocytes with both T4 and NCC signals (dashed line) and arrowheads indicate ionocytes with only T4 signals (dotted line). Staining of the same embryos with antibodies against medaka NCC-like 2 (NCC) (d, e) and NKA (e). Arrows indicate ionocytes with both NCC and NKA signals (dashed line). Magnified (f) and z-plan (g) images of ionocytes with both NKA and NCC signals. Scale (a–c) 50 μm, (d, e) 100 μm, (f, g) 10 μm. Western blot of FW medaka gill tissues using an anti-medaka NCC-like 2 antibody (h) revealed a single immunoreactive band (arrow)
Subsequent triple in situ (ncc-like 2 mRNA) and immunocytochemical experiments using anti-killifish HA and α5 confirmed the NCC-expressing ionocytes to be the HA cells identified in our previous study (Lin et al. 2012). The ncc-like 2 mRNA signals (Fig. 5a) specifically colocalized with HA (i.e., the HA cells) (Fig. 5b) in a portion of the NKA-labeled ionocytes (Fig. 5c, d). qRT-PCR was further conducted to examine ncc-like 2 mRNA expressions in gills of medaka acclimated to different Na+ and Cl− environments (Fig. 5e). Comparing high-Na/high-Cl and high-Na/low-Cl, the low-Cl environment increased ncc-like 2 mRNA expressions in medaka gills (Fig. 5e). This result showed the role of NCC in Cl− uptake mechanism of ionocytes, which is also demonstrated in zebrafish (Wang et al. 2009). On the other hand, comparing high-Na/low-Cl and low-Na/low-Cl, the high-Na environment also increased ncc-like 2 mRNA expressions in medaka gills (Fig. 5e)
Triple labeling of ncc-like mRNA (a, d) and HA (b, d) and NKA (c, d) proteins in FW medaka at 7 dpf. The three images are merged in (d). a–d Arrows indicate colocalization of ncc-like mRNA and HA. Arrowheads indicate cells without ncc-like 2 mRNA signals. Scale 10 μm. (e) qRT-PCR analysis of ncc-like 2 mRNA in the gills of madaka adults acclimated to high-Na+/high-Cl− (H-Na/H-Cl), high-Na+/low-Cl− (H-Na/L-Cl) and low-Na+/low-Cl− (L-Na/L-Cl) artificial FWs for 2 weeks. Values were normalized to rpl7 and are expressed as means ± SD (n = 9). Different letters above columns indicate significant difference by one-way ANOVA (Tukey’s pairwise comparisons)
NHE-expressing ionocytes in FW medaka
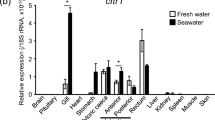
The previously identified NHE-expressing ionocytes (Lin et al. 2012) were further characterized based on the expression of other transporters. Similar to zebrafish (Lee et al. 2011), medaka has two orthologs of slc4a, slc4a1a (ae1a) and slc4a1b (ae1b), while ae1b mRNA was expressed in the salt-and-pepper pattern of ionocytes (Fig. 6a) and ae1a mRNA was mainly distributed in the branchial area, with a pattern unlike that of ionocytes (Fig. 6b). Triple in situ hybridization and immunocytochemistry demonstrated that ae1b mRNA localized with apical NHE3 and basolateral NKA proteins in the same ionocytes, at a 1:1 ratio (Fig. 6c-h). On the other hand, the expression of ae1b mRNA in the adult gills of SW medaka was too low to be detected by in situ hybridization (data not shown). In support of this observation, expression of ae1b mRNA was significantly lower in SW gills as compared to FW gills (Fig. 6i). Subsequent experiments involving double immunocytochemistry with the anti-medaka NHE3 and T4 antibodies demonstrated colocalization of apical NHE3 and basolateral NKCC in the same ionocytes, also at a one-to-one ratio, in FW medaka (Fig. 7a-c).
In situ hybridization against ae1b (slc4a1b) and ae1a (slc4a1a) mRNA and triple labeling together with NKA and NHE3 proteins in FW medaka at 7 dpf and salinity effect on ae1b mRNA expression in adult gills (I). Expression of ae1b mRNA (a, arrows) but not ae1a mRNA (b, asterisks), resembled the salt-and-pepper pattern of ionocytes. The same embryo triple labeled with ae1b mRNA (c) and NKA and NHE3 (d). The merged image is shown in (e). Arrows indicate cells exhibiting colocalization signals. The same ionocytes labeled with ae1b mRNA (f) and NKA and NHE3 (g) are shown at higher magnification. The merged and magnified image is shown in (h). Scale (a, b) 100 μm, (c–e) 50 μm, (f–h) 10 μm. i qRT-PCR analysis of ae1b mRNA in the gills of FW- and SW-acclimated medaka adults. Values were normalized to rpl7 and are expressed as means ± SD (n = 6). ***Significant difference (Student’s t test, p < 0.001)
ECaC-expressing ionocytes in FW medaka
In addition to NCC- and NHE-expressing ionocytes, another type of ionocyte was identified in medaka. Triple in situ and immunocytochemistry experiments enabled the detection of trpv6 (ecac) mRNA in a specific group of NKA-labeled ionocytes adjacent to NHE-type ionocytes (Fig. 8). About 20 % of the NHE cells were accompanied by ECaC-expressing ionocytes (Fig. 8a). Enhanced magnification and z-plan image revealed that the ECaC ionocytes were smaller and exhibited weaker NKA signals as compared to NHE cells (Fig. 8b–g). Such properties suggest that the ECaC-expressing ionocytes may be accessory (AC) cells, which were initially identified in SW teleosts (Hootman and Philpott 1980).
Triple labeling of trpv6 (ecac) mRNA (green) and NHE3 (blue) and NKA (red) proteins in FW-acclimated 7-dpf embryo (a–g) and salinity effect on ecac mRNA expression in adult gills (h). Arrows in (a) indicate NHE cells accompanied by ecac mRNA-expressing cells. Magnified images of cells stained with NKA (b), NHE3 (c), or ecac mRNA (d) and the merged image (e) are shown. Z-plan images of the same cells are shown in (f–g). (h) qRT-PCR analysis of ecac mRNA in the gills of FW- and SW-acclimated medaka adults. Values were normalized to rpl7 and are expressed as means ± SD (n = 6). ***Significant difference (Student’s t test, p < 0.001)
SW AC cells were subsequently compared to FW ECaC ionocytes. SW AC cells exhibited NKA signals (Shen et al. 2011) but expression of ecac mRNA was too low to be detected by in situ hybridization (data not shown). In support of this observation, expression of ecac mRNA was significantly lower in SW gills as compared to FW gills (Fig. 8h).
Ionocytes in SW medaka
Triple in situ hybridization and immunocytochemistry were also used to identify co-expressed ion transporters in the ionocytes of SW medaka. The mRNA encoding slc12a2a (nkcc) was colocalized with CFTR and NKA (in a one-to-one manner) in the predominant type of SW ionocyte (Fig. 9a–d). High-magnification images revealed that CFTR expression was confined to the apical region of ionocytes (Fig. 9e–h). Furthermore, CFTR signals were not observed in SW ACs, which were characterized by their small size and relatively low expression of NKA (Fig. 9i–k) (as for FW AC cells; Fig. 8). A previous study showed that around 60 % of SW ionocytes are accompanied with ACs and the functions of SW ionocyte were developed within 5 h after transferred from FW to SW (Shen et al. 2011). In this study, gene expression of slc9a3 (NHE3), ncc-like2, abcc7 (CFTR) and slc12a2a were examined by qRT-PCR after SW transfer at 3, 6, 12 and 24 h (Fig. 10). Following SW acclimated duration, expression of slc9a3 and ncc-like2 were decreased and had significant difference after 6 h (Fig. 10a, b). On the other side, expression of abcc7, slc12a2a was progressively increased and had significant difference after 12 h (Fig. 10c, d).
Triple and double labeling of slc12a2a (nkcc) mRNA (blue; a–e, h) and CFTR (green; b, d, f, h, i, k) and NKA (red; c, d, g, h, j, k) proteins in SW medaka at 7 dpf. (a–c) The same embryo was stained with the indicated marker. (d) The merged image. Arrows indicate ionocytes with colocalized signals (dotted line). (e–g) Magnified images of the above. (h) The merged image. (i – k) Arrows indicate colocalization signals in SW-type ionocytes and arrowheads indicate accompanying accessory cells without CFTR, in the same embryo. Scale (a–d) 50 μm, (e–h) 10 μm, (i–k) 50 μm
qRT-PCR analysis of slc9a3 (a), ncc-like2 (b), abcc7 (c) and slc12a2a (D) mRNAs in the gills of madaka adults transferred from FW to SW. Control (FW) was sampled at 0 and 24 h and the SW group was sampled at 3, 6, 12 and 24 h after transfer. Values were normalized by rpl7. Values are expressed as means ± SD (n = 6). Different letters above columns indicate significant difference by one-way ANOVA (Tukey’s comparison, P < 0.05)
Discussion
The findings of the current and earlier studies indicate that the embryonic skin of SW- and FW-acclimated medaka contain two and three types of ionocyte, respectively (Wu et al. 2010; Lin et al. 2012; Liu et al. 2013). The three ionocyte types in FW medaka are as follows: (1) NHE cells with apical NHE3 and Rhcg1 and basolateral NKCC, NKA, Rhbg and AE1, (2) ECaC cells (presumably AC cells) with apical ECaC and basolateral NKA and (3) HA cells with apical NCC and basolateral HA and NKA (Fig. 11). On the other hand, SW medaka have one predominant type of SW ionocyte with apical CFTR, NHE3, NHE2 and Rhcg1 and basolateral NKCC, NKA and Rhbg, which are accompanied by smaller AC cells that express less basolateral NKA. This study showed that the gene expressions of transporters (NHE3, AE1, ECaC, NCC) in FW ionocytes were down-regulated after SW transfer. In contrast, the gene expressions of transporters (CFTR, NKCC) in SW ionocytes were up-regulated.
The proposed model of ionocyte subtype function in FW and SW medaka. For details, please refer to the text. AC accessory cell; AE1 Cl−/HCO3 − exchanger; CFTR cystic fibrosis transmembrane conductance regulator; ECaC epithelial Ca2+ channel; HA H+-ATPase; NCC Na+-Cl− cotransporter; NHE2/3 Na+/H+ exchangers; NKA Na+-K+-ATPase; NKCC Na+-K+-2Cl− cotransporter; Rhgb and Rhbg Rhesus glycoproteins
The fish-specific NCC-like protein (slc12a10), initially identified in tilapia (Hiroi et al. 2008) and subsequently in zebrafish (Wang et al. 2009), has now been characterized in medaka. Similar to the situation in zebrafish (Wang et al. 2009), only one of the paralogs, slc12a10.b (ncc-like 2), was found to be predominantly expressed in the gills of medaka (Fig. 2), in the salt-and-pepper pattern of ionocytes (Fig. 3a). In zebrafish, NCC-type ionocytes (which are distinct from the two other major types of HR- and NaR-ionocyte) are restricted to the skin near the head, the branchial arch and the upper part of the yolk sac (Pan et al. 2005; Lin et al. 2006; Wang et al. 2009). Similarly in medaka, ionocytes expressing ncc-like 2 mRNA signals were found to be restricted to areas of skin near the branchial arch (Fig. 3a), unlike previously identified NHE3-expressing ionocytes (Lin et al. 2012). In support of these cells being distinct populations, ncc-like 2-expressing ionocytes with basolateral Na+-K+-ATPase were distinguished from NHE3 cells by triple labeling (Fig. 3c–i). T4 antibody (previously used to detect both apical NCC and basolateral NKCC in fish gills (Hiroi et al. 2005; Hiroi and McCormick 2012)) and our newly established anti-medaka NCC-like 2 antibody were used to demonstrate apical localization of NCC-like 2 in NCC type ionocytes in medaka (Fig. 4). In tilapia, zebrafish and medaka, NCC-expressing ionocytes are distinct from NHE3-expressing ionocytes (Hiroi et al. 2005; Wang et al. 2009; the present study). NHE3 in the apical membrane of zebrafish HR-type ionocytes and medaka NHE-expressing ionocytes are involved in Na+ uptake, acid secretion and NH4 + excretion (Yan et al. 2007; Esaki et al. 2007; Shih et al. 2012; Wu et al. 2010; Liu et al. 2013; Hwang and Chou 2013), while the NCC-like protein in zebrafish NCC ionocytes plays a major role in Cl− uptake and a minor or supplementary role in Na+ uptake (Wang et al. 2009; Chang et al. 2013). Furthermore NHE and NCC exhibit functional redundancy in zebrafish ionocytes (Chang et al. 2013). Down-regulation of NHE expression in acid acclimated zebrafish gills is concomitant up-regulation of NCC cell density (Yan et al. 2007; Chang et al. 2013). In this present study, we showed up-regulated NCC expression in high-Na acclimated gills (Fig. 5e), which may be a response of functional redundancy for the down-regulated NHE expression in high-Na acclimated gills that was demonstrated in zebrafish (Yan et al. 2007). However, further experiments should be conducted in medaka. Unlike tilapia, zebrafish and medaka, FW killifish contain a single type of gill ionocyte, which was recently proposed to express both NHE and NCC (Dymowska et al. 2012); this proposal was based on the immunolabeling of apical NCC in gill ionocytes (identified by the presence of Na+-K+-ATPase) with the T4 antibody (Katoh et al. 2008) and the localization of NHE2 mRNA in a subset of gill cells (Edwards et al. 2010). This difference is interesting from an evolutionary physiology perspective. However, to date, no evidence of colocalization of NHE and NCC in the same ionocytes is available and the function of apical NCC has been questioned due to the lack of Cl− uptake in this species (Dymowska et al. 2012).
Previous functional analysis studies have demonstrated that NCC-expressing ionocytes in tilapia (Type-II ionocytes) and zebrafish (NCC ionocytes) are involved in the uptake of Cl− and Na+ (Hiroi et al. 2008; Inokuchi et al. 2009; Wang et al. 2009; Chang et al. 2013) and these functions are presumably conserved in medaka (Fig. 5e). Expression of basolateral HA in the NCC type ionocytes in medaka suggests they may also have other roles. HA is usually expressed at the apical membrane of cells but HA-expressing cell types exhibit considerable diversity between fishes. HA is apically expressed in the NHE3-expressing HR cells of zebrafish, non-NHE3-expressing PNA-cells of trout and in the pavement cells of tilapia (Hiroi et al. 1998; Wilson et al. 2000; Galvez et al. 2002; Yan et al. 2007). Unlike HA in these cells, HA is basolaterally localized in the gill ionocytes of killifish (Katoh et al. 2003). HA is also localized in the basolateral membrane of NCC-expressing ionocytes in medaka (Fig. 5), indicating that the NCC-expressing and the previously-identified HA-expressing ionocytes (Lin et al. 2012) are the same. HA expressed in the apical membrane is thought to excrete acid and maintain the acid–base balance of the fish body (Galvez et al. 2002; Lin et al. 2006; Horng et al. 2007). Basolateral HA in the Type B intercalated cells of the mammalian nephron is also involved in acid–base regulation, through cooperating with apical pendrin to secrete base (which is equivalent to absorbing acid) (Wagner et al. 2011). In marine teleost, ionocyte with basolateral HA is postulated as base-secreting cells (Claiborne et al. 2002, 2008). However, the roles of basolaterally-expressed HA in FW fish gill ionocytes are uncertain. Killifish acclimated to low-NaCl FW exhibited higher immunofluorescent staining of basolateral HA (and Na+-K+-ATPase) in the gill ionocytes than equivalents acclimated to high-NaCl FW (Katoh et al. 2003). In medaka, HA mRNA and HA ionocyte density were enhanced by acclimation to acidified FW (Lin et al. 2012). NCC type ionocytes in tilapia (Furukawa et al. 2011) and zebrafish (Lee et al. 2011) express basolateral Na+/HCO3 − co-transporter (NBCe1), which has long been associated with Na+ uptake and acid–base regulation mechanisms in FW fish gills (Evans et al. 2005; Evans 2011; Hwang et al. 2011; Dymowska et al. 2012: Hwang and Chou 2013). Taken together, the NCC type ionocytes (i.e., those previously reported to express HA; Lin et al. 2012), which express apical NCC-like 2 and basolateral HA, may be involved in NaCl uptake and acid–base regulation; however, this hypothesis requires additional supporting evidence, such as identification of other transporters (e.g., NBCe1, pendrin, carbonic anhydrase) and functional analyses of those transporters with molecular physiological approaches.
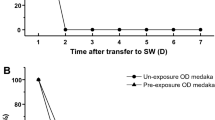
In FW medaka, over 90 % of the ionocytes in the embryonic skin are NHE3-expressing cells (the predominant ionocyte type) (Lin et al. 2012). Medaka NHE3-expressing ionocytes, like zebrafish HR cells (Lee et al. 2011), co-express AE1b (Fig. 6); AE1b’s role in basolateral HCO3 − transport is required for transepithelial acid secretion (Lee et al. 2011) and this may hold true for medaka NHE3 cells. Medaka NHE3-expressing ionocytes are also similar to Type-III ionocytes in tilapia (Hiroi et al. 2005), as they both express NKCC at the basolateral membrane (Fig. 7). Moreover, the NHE-expressing ionocytes of FW medaka also express apical Rhcg1 and basolateral Rhbg (Wu et al. 2010). Comparing SW and FW ionocytes in medaka, the predominant SW-type express additional CFTR and NHE2 compared to the FW-type NHE-expressing ionocytes (Fig. 11). Likewise, in tilapia, SW Type-IV ionocytes express additional CFTR in the apical membrane compared with FW Type-III cells (Hiroi et al. 2005). Sequential observations of individual ionocytes in vivo indicated that 75 % of the ionocytes survived 96 h after transfer of tilapia embryo from FW to SW, implying a possibility of transformation from the pre-exiting FW ionocytes into the SW ones (Hiroi et al. 1999). Subsequently, studies showed that decrease of the type-III cells and increase of the type-IV cells in the cell density occurred within 6–24 h after transfer from FW to SW, suggesting that the FW-type ionocytes (Type-III cells) directly transform into the SW-type ones (Type-IV cells) in tilapia (Hiroi et al. 2005; Choi et al. 2011). Direct transformation between FW NHE3-expressing ionocytes and SW-type ionocytes is also proposed to occur in medaka. We previously used SIET sequential detection to show that the Na+/Cl− secretion ability of skin ionocytes develops within 5 h of transfer from FW to SW (Shen et al. 2011). Nevertheless, the gene expression of CFTR and NKCC were significantly increased only at 12 and 24 h after transfer, which was slower than the up-regulation of Na+/Cl− secreting function (Fig. 10). Therefore, the acute functional change from salt uptake to secretion is likely achieved by rapid induction of CFTR and NKCC expression in pre-existing ionocytes (Hwang et al. 2011; Hwang and Lin 2013) Moreover, NKCC signal was detected in NHE3-expressing cells in FW medaka, suggesting a preparation for rapid functioning after SW transfer (Fig. 7; Kang et al. 2010). The expression of NKCC in FW ionocytes has also been reported in other fishes including tilapia, milkfish and pufferfish (Hiroi et al. 2008; Tang et al. 2011).
AC cells are usually found accompanied by ionocytes in SW teleosts; however, AC cells are not common in FW fishes (Evans et al. 2005). Generally, AC cells contain fewer mitochondria and express lower levels of NKA (Hootman and Philpott 1980); however, the identity and function of ACs remain a puzzle (Evans et al. 2005). It was thought that AC cells and the accompanying ionocytes share the same apical crypt in SW teleosts and form leaky junctions for paracellular Na+ extrusion (Sardet et al. 1979; Hootman and Philpott 1980; Hwang and Hirano 1985). Our previous studies in medaka demonstrated that the AC cell–ionocyte complex secretes Na+ and Cl−, whereas single ionocytes secrete only Cl− in SW embryos; these data provide functional evidence for a role for AC cells in Na+ excretion (Shen et al. 2011). The present study indicates that trpv6 (ecac) mRNA is expressed in the presumed AC cells (i.e., ECaC-expressing ionocytes) of FW medaka (Fig. 8a–g) and this may provide a new line of inquiry for the future investigation of AC cell function. TRPV6 has recently been reported to be important for Ca2+ absorption in the ionocytes of zebrafish and trout (Pan et al. 2005; Shahsavarani et al. 2006; Liao et al. 2007). Expression of ecac in AC cells suggests a role in Ca2+ absorption; however, such a role cannot be confirmed without the collection of additional molecular and physiological evidence. Decreased expression of ecac mRNA was also observed in SW medaka, as compared to their FW equivalents (Fig. 8h). Previous studies have demonstrated that Ca2+ is taken up by gills in both SW and FW fishes (Van der Heijden et al. 1999; Marshall and Bryson 1998). In SW with 8–10 mM Ca2+ (a much higher concentration of calcium than that in the plasma), teleosts must reduce gill Ca2+ uptake or increase the metabolic clearance rate of Ca2+ (Flik and Verbost 1993). The current finding that ecac mRNA is significantly decreased (confirmed by both qPCR and in situ hybridization; data not shown) in SW medaka suggests that expression of this gene is regulated during FW/SW adaptation. The function of AC cells in Ca2+ handling during FW/SW adaptation is a subject for future investigation.
In summary, 3 types of ionocytes: NHE3 cells, NCC cells (previously identified as HA cells; Lin et al. 2012) and ECaC cells (presumed AC cells) in FW and two types of ionocytes: SW-type ionocytes and AC cells in SW medaka were identified and characterized, based on specific expression of distinctive ion transporters (Fig. 11). The NHE3 cells are the predominant type of ionocytes in FW and are responsible for acid secretion, Na+ uptake and NH4 + excretion. Although further functional analyses are necessary, NCC cells and ECaC cells are proposed to be involved in NaCl uptake (or acid–base regulation) and Ca2+ uptake, respectively. On the other hand, SW-type ionocytes are the major player in acid secretion, NaCl secretion and NH4 + excretion. The present findings in medaka promise to reveal further similarities and differences in the development and function of ionocyte subtypes between fishes from a comparative point of view. However, continued identification and functional analysis of other co-expressed ion transporters in each ionocyte type are required to formulate a comprehensive model of the ionic and acid–base regulation mechanisms in medaka. As a model organism, medaka has similar advantages to zebrafish, including its suitability for loss- (and gain-) of function studies and the availability of cellular/molecular approaches and genetic databases. However, unlike the stenohaline zebrafish, the euryhaline medaka is a more suitable model for research into ionic and acid–base regulation during acclimation to a wide range of salinities.
References
Ali S, Champagne DL, Spaink HP, Richardson MK (2011) Zebrafish embryos and larvae: a new generation of disease models and drug screens. Birth Defects Res C 93:115–133
Chang WJ, Hwang PP (2011) Development of zebrafish epidermis. Birth Defects Res C 93:205–214
Chang WJ, Wang YF, Hu HJ, Wang JH, Lee TH, Hwang PP (2013) Compensatory regulation of Na+ absorption by Na+/H+ exchanger and Na+-Cl− cotransporter in zebrafish (Danio rerio). Front Zool 10:46
Choi JH, Lee KM, Inokuchi M, Kaneko T (2011) Morphofunctional modifications in gill mitochondria-rich cells of Mozambique tilapia transferred from freshwater to 70 % seawater, detected by dual observations of whole-mount immunocytochemistry and scanning electron microscopy. Comp Biochem Physiol 158A:132–142
Claiborne JB, Edwards SL, Morrison-Shetlar AI (2002) Acid–base regulation in fishes: cellular and molecular mechanisms. J Exp Zool 293:302–319
Claiborne JB, Choe KP, Morrison-Shetlar AI, Weakley JC, Havird J, Freiji A, Evans DH, Edwards SL (2008) Molecular detection and immunological localization of gill Na+/H+ exchanger in the dogfish (Squalus acanthias). Am J Physiol Regul Integr Comp Physiol 294:R1092–R1102
Dymowska A, Hwang PP, Goss GG (2012) Structure and function of ionocytes in the freshwater fish gill. Respir Physiol Neurobiol 184:282–292
Edwards SL, Weakley JC, Diamanduros AW, Claiborne JB (2010) Molecular identification of Na+-H+ exchanger isoforms (NHE2) in the gills of the euryhaline teleost Fundulus heteroclitus. J Fish Biol 76:415–426
Esaki M, Hoshijima K, Kobayashi S, Fukuda H, Kawakami K, Hirose S (2007) Visualization in zebrafish larvae of Na+ uptake in mitochondria-rich cells whose differentiation is dependent on foxi3a. Am J Physiol Regul Integr Comp Physiol 292:R470–R480
Evans DH (2008) Teleost fish osmoregulation: what have we learned since August Krogh, Homer Smith, and Ancel Keys. Am J Physiol Regul Integr Comp Physiol 295:R704–R713
Evans DH (2011) Freshwater fish gill ion transport: August Krogh to morpholinos and microprobes. Acta Physiol (Oxf) 202:349–359
Evans DH, Piermarini PM, Choe KP (2005) The multifunctional fish gill: dominant site of gas exchange, osmoregulation, acid–base regulation, and excretion of nitrogenous waste. Physiol Rev 85:97–177
Flik G, Verbost PM (1993) Calcium transport in fish gills and intestine. J Exp Biol 184:17–29
Furukawa F, Watanabe S, Inokuchi M, Kaneko T (2011) Responses of gill mitochondria-rich cells in mozambique tilapia exposed to acidic environments (pH 4.0) in combination with different salinities. Comp Biochem Physiol 158A:468–476
Furukawa F, Watanabe S, Kimura S, Kaneko T (2012) Potassium excretion through ROMK potassium channel expressed in gill mitochondrion-rich cells of Mozambique tilapia. Am J Physiol Regul Integr Comp Physiol 302:R568–R576
Galvez F, Reid SD, Hawkings G, Goss GG (2002) Isolation and characterization of mitochondria-rich cell types from the gill of freshwater rainbow trout. Am J Physiol Regul Integr Comp Physiol 282:R658–R668
Hiroi J, McCormick SD (2012) New insights into gill ionocyte and ion transporter function in euryhaline and diadromous fish. Respir Physiol Neurobiol 184:257–268
Hiroi J, Kaneko T, Uchida K, Hasegawa S, Tanaka M (1998) Immunolocalization of Vacuolar-Type H+-ATPase in the Yolk-Sac Membrane of Tilapia (Oreochromis mossambicus) Larvae. Zool Sci 15:447–453
Hiroi J, Kaneko T, Tanaka M (1999) In vivo sequential changes in chloride cell morphology in the yolk-sac membrane of Mozambique tilapia (Oreochromis mossambicus) embryos and larvae during seawater adaptation. J Exp Biol 202:3485–3495
Hiroi J, McCormick SD, Ohtani-Kaneko R, Kaneko T (2005) Functional classification of mitochondrion-rich cells in euryhaline Mozambique tilapia (Oreochromis mossambicus) embryos, by means of triple immunofluorescence staining for Na+/K+-ATPase, Na+/K+/2Cl− cotransporter and CFTR anion channel. J Exp Biol 208:2023–2036
Hiroi J, Yasumasu S, McCormick SD, Hwang PP, Kaneko T (2008) Evidences for an apical Na+-Cl− cotransporter involved in ion uptake in a teleost fish. J Exp Biol 211:2584–2599
Hirose S, Kaneko T, Naito N, Takei Y (2003) Molecular biology of major components of chloride cells. Comp Biochem Physiol 136B:593–620
Hootman SR, Philpott CW (1980) Accessory cells in teleost branchial epithelium. Am J Physiol Regul Integr Comp Physiol 238:R199–R206
Horng JL, Lin LY, Huang CJ, Katoh F, Kaneko T, Hwang PP (2007) Knockdown of V-ATPase subunit A (atp6v1a) impairs acid secretion and ion balance in zebrafish (Danio rerio). Am J Physiol Regul Integr Comp Physiol 292:R2068–R2076
Hwang PP (2009) Ion uptake and acid secretion in zebrafish (Danio rerio). J Exp Biol 212:1745–1752
Hwang PP, Chou MY (2013) Zebrafish as an animal model to study ion homeostasis. Pflugers Arch 465:1233–1247
Hwang PP, Hirano R (1985) Effects of environmental salinity on intercellular organization and junctional structure of chlorid cells in early stages of teleost development. J Exp Zool 236:115–126
Hwang PP, Lee TH (2007) New insights into fish ion regulation and mitochondrion-rich cells. Comp Biochem Physiol A 148:479–497
Hwang PP, Lin LY (2013) Gill ionic transport, acid–base regulation, and nitrogen excretion. In: Evans DH, Claiborne JB, Currie S (ed) The Physiology of Fishes, 4th eEon. CRC Press, Boca Raton
Hwang PP, Perry S (2010) Ionic and acid–base regulation. In: Perry S, Ekker, M, Farrell, AP , Brauner, CJ (ed) Fish Physiology, vol. 29. Academic, New York, pp 311–314
Hwang PP, Lee TH, Lin LY (2011) Ion regulation in fish gills: recent progress in the cellular and molecular mechanisms. Am J Physiol Regul Integr Comp Physiol 301:R28–R47
Inokuchi M, Hiroi J, Watanabe S, Lee KM, Kaneko T (2008) Gene expression and morphological localization of NHE3, NCC and NKCC1a in branchial mitochondria-rich cells of Mozambique tilapia (Oreochromis mossambicus) acclimated to a wide range of salinities. Comp Biochem Physiol A 151:151–158
Inokuchi M, Hiroi J, Watanabe S, Hwang PP, Kaneko T (2009) Morphological and functional classification of ion-absorbing mitochondria-rich cells in the gills of Mozambique tilapia. J Exp Biol 212:1003–1010
Kang CK, Tsai SC, Lee TH, Hwang PP (2008) Differential expression of branchial Na+/K+-ATPase of two medaka species, Oryzias latipes and Oryzias dancena, with different salinity tolerances acclimated to fresh water, brackish water and seawater. Comp Biochem Physiol A 151:566–575
Kang CK, Tsai HJ, Liu CC, Lee TH, Hwang PP (2010) Salinity-dependent expression of a Na+, K+, 2Cl− cotransporter in gills of the brackish medaka Oryzias dancena: a molecular correlate for hyposmoregulatory endurance. Comp Biochem Physiol A 157:7–18
Katoh F, Hyodo S, Kaneko T (2003) Vacuolar-type proton pump in the basolateral plasma membrane energizes ion uptake in branchial mitochondria-rich cells of killifish Fundulus heteroclitus, adapted to a low ion environment. J Exp Biol 206:793–803
Katoh F, Cozzi RRF, Marshall WS, Goss GG (2008) Distinct Na+/K+/2Cl− cotransporter localization in kidneys and gills of two euryhaline species, rainbow trout and killifish. Cell Tissue Res 334:265–281
Lee YC, Yan JJ, Cruz SA, Horng JL, Hwang PP (2011) Anion exchanger 1b, but not sodium-bicarbonate cotransporter 1b, plays a role in transport functions of zebrafish H+-ATPase-rich cells. Am J Physiol Cell Physiol 300:C295–C307
Liao BK, Deng AN, Chen SC, Chou MY, Hwang PP (2007) Expression and water calcium dependence of calcium transporter isoforms in zebrafish gill mitochondrion-rich cells. BMC Genomics 8:354
Lin LY, Horng JL, Kunkel JG, Hwang PP (2006) Proton pump-rich cell secretes acid in skin of zebrafish larvae. Am J Physiol Cell Physiol 290:C371–C378
Lin CC, Lin LY, Hsu HH, Thermes V, Prunet P, Horng JL, Hwang PP (2012) Acid secretion by mitochondrion-rich cells of medaka (Oryzias latipes) acclimated to acidic freshwater. Am J Physiol Regul Integr Comp Physiol 302:R283–R291
Liu ST, Tsung L, Horng JL, Lin LY (2013) Proton-facilitated ammonia excretion by ionocytes of medaka (Oryzias latipes) acclimated to seawater. Am J Physiol Regul Integr Comp Physiol 305:R242–R251
Marshall WS, Bryson SE (1998) Transport mechanisms of seawater teleost chloride cells: an inclusive model of a multifunctional cell. Comp Biochem Physiol A 119:97–106
Pan TC, Liao BK, Huang CJ, Lin LY, Hwang PP (2005) Epithelial Ca2+ channel expression and Ca2+ uptake in developing zebrafish. Am J Physiol Regul Integr Comp Physiol 289:R1202–R1211
Sardet C, Pisam M, Maetz J (1979) The surface epithelium of teleostean fish gills. Cellular and junctional adaptations of the chloride cell in relation to salt adaptation. J Cell Biol 80:96–117
Shahsavarani A, McNeill B, Galvez F, Wood CM, Goss GG, Hwang PP, Perry SF (2006) Characterization of a branchial epithelial calcium channel (ECaC) in freshwater rainbow trout (Oncorhynchus mykiss). J Exp Biol 209:1928–1943
Shen WP, Horng JL, Lin LY (2011) Functional plasticity of mitochondrion-rich cells in the skin of euryhaline medaka larvae (Oryzias latipes) subjected to salinity changes. Am J Physiol Regul Integr Comp Physiol 300:R858–R868
Shih TH, Horng JL, Liu ST, Hwang PP, Lin LY (2012) Rhcg1 and NHE3b are involved in ammonium-dependent sodium uptake by zebrafish larvae acclimated to low-sodium water. Am J Physiol Regul Integr Comp Physiol 302:R84–R93
Suzuki Y, Itakura M, Kashiwagi M, Nakamura N, Matuki T, Sauta H, Naito N, Takano K, Fujita T, Hirose S (1999) Identification by differential display of a hypertonicity-inducible inward rectifier potassium channel highly expressed in chloride cells. J Biol Chem 274:11376–11382
Takeda H, Shimada A (2010) The art of medaka genetics and genomics: what makes them so unique? Annu Rev Genet 44:217–241
Tang CH, Hwang LY, Shen ID, Chiu YH, Lee TH (2011) Immunolocalization of chloride transporters to gill epithelia of euryhaline teleosts with opposite salinity-induced Na+/K+-ATPase responses. Fish Physiol Biochem 37:709–724
Tse WKF, Au DWT, Wong CKC (2006) Characterization of ion channel and transporter mRNA expression in isolated gill chloride and pavement cells of seawater acclimating eels. Biochem Biophys Res Commun 346:1181–1190
van der Heijden AJ, Verbost PM, Bijvelds MJ, Atsma W, Wendelaar Bonga SE, Flik G (1999) Effects of sea water and stanniectomy on branchial Ca2+ handling and drinking rate in eel (Anguilla anguilla L.). J Exp Biol 202:2505–2511
Wagner CA, Mohebbi N, Uhlig U, Giebisch GH, Breton S, Brown D, Geibel JP (2011) The anion exchanger pendrin (SLC26A4) and renal acid–base homeostasis. Cell Physiol Biochem 28:497–504
Wang YF, Tseng YC, Yan JJ, Hiroi J, Hwang PP (2009) Role of SLC12A10.2, a Na+-Cl− cotransporter-like protein, in a Cl uptake mechanism in zebrafish (Danio rerio). Am J Physiol Regul Integr Comp Physiol 296:R1650–R1660
Wilson JM, Laurent P, Tufts B, Benos DJ, Donowitz M, Vogl AW, Randall DJ (2000) NaCl uptake by the branchial epithelium in freshwater teleost fish: an immunological approach to ion-transport protein localization. J Exp Biol 203:2279–2296
Wittbrodt J, Shima A, Schartl M (2002) Medaka–a model organism from the far East. Nat Rev Genet 3:53–64
Wu SC, Horng JL, Hwang PP, Wen ZH, Lin CS, Lin LY (2010) Ammonium-dependent sodium uptake in mitochondrion-rich cells of medaka (Oryzias latipes) larvae. Am J Physiol Cell Physiol 298:C237–C250
Yan JJ, Chou MY, Kaneko T, Hwang PP (2007) Gene expression of Na+/H+ exchanger in zebrafish H+-ATPase-rich cells during acclimation to low-Na+ and acidic environments. Am J Physiol Cell Physiol 293:C1814–C1823
Acknowledgements
This study was financially supported by grants to P.P.H. from Academia Sinica and the National Science Council, Taiwan, R.O.C. We extend our thanks to Ms. Y.C. Tung for her assistance during the experiments.
Disclosures
The authors declare no conflicts of interest, financial or otherwise.
Author Contributions
Author contributions: H.-H.H., L.-Y.L., Y.-C.T. and J.-L.H. performed experiments; H.-H.H., L.-Y.L., Y.-C.T. and J.-L.H. analyzed data; H.-H.H., L.-Y.L., Y.-C.T., J.-L.H. and P.-P.H. interpreted experimental results; H.-H.H., Y.-C.T., J.-L.H. and P.-P.H. prepared figures; H.-H.H and J.-L.H. drafted the manuscript; L.-Y.L., J.-L.H. and P.-P.H. edited and revised the manuscript; J.-L.H. and P.-P.H. conceived and designed the study; P.-P.H. approved the final version of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Pung-Pung Hwang and Jiun-Lin Horng contributed equally to this study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 170 kb)
Rights and permissions
About this article
Cite this article
Hsu, HH., Lin, LY., Tseng, YC. et al. A new model for fish ion regulation: identification of ionocytes in freshwater- and seawater-acclimated medaka (Oryzias latipes). Cell Tissue Res 357, 225–243 (2014). https://doi.org/10.1007/s00441-014-1883-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00441-014-1883-z