Abstract
Tobacco smoking is a severe health hazard worldwide, as nearly one-third of the global adult population smokes tobacco products. This high prevalence highlights the importance of studying the genetic determinants of nicotine dependence (ND). To identify such genetic factors, more than 20 genome-wide linkage studies have been conducted across different populations using a variety of ND assessment approaches, including smoking quantity (SQ), Heaviness of Smoking Index (HSI), Fagerström Test for Nicotine Dependence (FTND), ever-smoking, habitual smoking, or maximum number of cigarettes smoked in a 24-h period. This review provides a critical update on the progress during the years since our last review, published in 2004, in identifying susceptibility loci for ND. Although a significant number of reported genomic regions did not reach the level of “suggestive” or “significant” linkage and failed to be replicated in other independent studies, thirteen regions, located on chromosomes 3–7, 9–11, 17, 20, and 22, have been found to be suggestive or significant in at least two independent samples. Among them, the regions on chromosomes 9 (91.9–136.5 cM), 10, 11, and 17 have received the strongest support. A summary of eight regions that have been nominated for “significant” linkage to ND is provided.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Despite increasing public awareness of the health risks of using tobacco products, approximately 1.2 billion people worldwide smoke tobacco daily. It is estimated that every year, 4.2 million people die from tobacco-related diseases: this number is predicted to approach 10 million by the year 2020 (WHO 2002). In the USA, approximately 70.3 million Americans aged 12 or older use tobacco products (SAMHSA 2001), and 20.8% of adults aged 18 and up smoke (CDC 2007). Thus, tobacco is one of the most widely abused substances, and it kills more than 438,000 US citizens each year (CDC 2005). Economically, smoking is responsible for about 7% of total US health care costs, an estimated $157.7 billion each year, of which $75.8 billion is direct medical costs.
Over the last decade, many large-sample twin studies in the US and other countries have demonstrated that genetic factors contribute to the risk of becoming a regular smoker (Carmelli et al. 1992; Heath et al. 1993; Heath and Martin 1993; Kendler et al. 1999; Madden et al. 1999; Maes et al. 1999; Swan et al. 1996, 1990a, b; True et al. 1999; Vink et al. 2005). Initial evidence for a genetic influence on nicotine dependence (ND) came from cross-sectional twin studies that showed a mean heritability of 0.53 (range 0.28–0.84) for cigarette smoking (Hughes 1986). Recently, we conducted a meta-analysis of genetic parameter estimates for ND based on 17 twin studies and determined that the weighted mean heritability for ND is 0.59 in male and 0.46 in female smokers, with an average of 0.56 for all smokers (Li et al. 2003a). Complex segregation analyses of smoking behavior in 493 three-generation families support a dominant major gene effect with residual familial correlation (Cheng et al. 2000). Together, these findings strongly suggest that ND, like many other physical and behavioral human disorders, is a complex trait (or disorder) that involves multiple genes and environmental risk factors, as well as interactions between genes or between genes and the environment. Further, the proportion of genetic versus environmental influences on different smoking stages differs by sex (Madden et al. 1999; Morley et al. 2006; Vink et al. 2006), with genetic factors appearing to have a larger role in initiation than in persistence in women, whereas the opposite is observed in men (Li et al. 2003a).
To identify susceptibility loci for ND, significant recent efforts have been made using an approach that tests for linkage of the disorder to polymorphic markers across the entire genome. We are aware of more than 20 published genome-wide linkage studies for smoking behavior (Bergen et al. 1999, 2003; Bierut et al. 2004; Duggirala et al. 1999; Ehlers and Wilhelmsen 2006, 2007; Gelernter et al. 2004, 2007; Goode et al. 2003; Li et al. 2003b, 2007a, 2006; Loukola et al. 2007; Morley et al. 2006; Pomerleau et al. 2007; Saccone et al. 2003, 2007; Swan et al. 2006; Vink et al. 2004, 2006; Wang et al. 2005). However, only a limited number of putative genomic linkages have been replicated in independent studies. A significant limiting factor in replicating these linkages is genetic heterogeneity, especially when the sample size is relatively small or participants of various ethnic origins are included. In addition, the size of the genetic effect, the density of markers, the definition and assessment of the phenotypes, and the statistical approaches might contribute to difficulty in replicating the findings of genome-wide linkage scans. Despite these concerns and limitations, significant progress has been made, particularly in the last few years. The primary purpose of this review is to provide an update on the progress made in identifying susceptibility loci for ND since our last review (Li et al. 2004).
Populations used in the reported linkage studies
Table 1 provides a detailed description of the populations employed in previous linkage studies for all ND-related measures. Collectively, these investigations have relied on 13 populations: the Collaborative Studies on the Genetics of Alcoholism (COGA), the Framingham Heart Study (FHS), the Mid-South Tobacco Family (MSTF) study, the Nicotine Addiction Genetics (NAG) project, the Finnish Twin Families (FTF), the Mission Indians in Southwest California, the Genetic Epidemiology Network of Arteriopathy (GENOA) study, the Smoking in Families Study (SMOFAM), the Netherlands Twin Register (NTR) study, the Genetics of Cocaine or Opioid Dependence (GCOD) study, the Christchurch sample of New Zealand, the Australian Twin Registry (ATR), and the Family Study of Panic Disorder (FSPD). Except for the MSTF (Li et al. 2006, 2007b), GCOD (Gelernter et al. 2007), and GEONA (Pomerleau et al. 2007) studies, which include substantial numbers of African-Americans (AA), and a Native American Mission Indian cohort (Ehlers and Wilhelmsen 2006, 2007), the participants of the other studies are of primarily European or European-American (EA) origin. The methods of assessing ND differ greatly from study to study, and include habitual smoking, regular and persistent tobacco use, smoking quantity (SQ), maximum number of cigarettes smoked in a 24-h period (MaxCigs24), the Heaviness of Smoking Index (HSI), the Fagerström Tolerance Questionnaire (FTQ), the Fagerström Test for ND (FTND), and DSM-IV or DSM-IV-like criteria.
Inclusion criteria for the current review
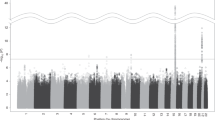
As described above, more than 20-linkage scans for various ND-related behaviors have been reported during the past several years, most since 2005. In reviewing the linkage regions nominated on the basis of these studies, one finds that almost all autosomal chromosomes (except for chromosome 15) have been implicated as harboring susceptibility loci for ND-related phenotypes. Considering that: (1) numerous genomic regions have been linked to various smoking phenotypes; (2) relatively detailed lists of these nominated regions (except for those loci reported during the past year or so) can be found in recent reviews (Ho and Tyndale 2007; Li et al. 2004; Vink et al. 2004); and (3) many of these results have not been replicated in independent studies, this review focuses primarily on those regions that show “suggestive linkage” in at least two independent samples or “significant” linkage in one study according to the rigorous criteria proposed by Lander and Kruglyak (1995), which define an LOD of >3.6 or a P value of <2.2 × 10−5 as a “significant linkage” and an LOD of >2.2 but <3.6 or a P value of 7.4 × 10−4 as a “suggestive linkage.” For those reports in which genome-wide empirical P values were provided using the permutation approach, “significant linkage” was declared if the genome-wide P value was ≤0.05 and “highly significant linkage” if the P value was ≤0.001. Under such criteria, many regions that show “suggestive” linkage to ND-related measures in a single study will not be discussed in the following sections. However, this is not to suggest that the linkage peaks reported in a single study are all false positives and should be ignored in future study. To be comprehensive and helpful for other researchers in the genetic study on tobacco and other substance abuses, Fig. 1 provides a graphical summary of most, if not all, of the regions that have been nominated for “suggestive” or “significant” linkage to ND in the literature. For detailed information on these nominated regions in a single study, please refer to Table 2. Also, to ensure the comparability of these loci across studies, the map position of each marker or marker pair that defines the linkage region in the original study was checked against the most recent version of the human linkage map through the website (http://www.ncbi.nlm.nih.gov/mapview/static/humansearch.html#marsh).
A graphical illustration of the chromosomal locations of peaks or intervals with “significant” or “suggestive linkage” to all ND-related measures in individual or repeated analyses of data from the Collaborative Studies on the Genetics of Alcoholism (COGA), the Framingham Heart Study (FHS), the Mid-South Tobacco Family (MSTF) study, the Nicotine Addiction Genetics (NAG) project, the Finnish Twin Families (FTF), the Mission Indians in Southwest California, the Genetic Epidemiology Network of Arteriopathy (GENOA) study, the Smoking in Families Study (SMOFAM), the Netherlands Twin Register (NTR) study, the Genetics of Cocaine or Opioid Dependence (GCOD) study, the Christchurch sample of New Zealand, the Australian Twin Registry (ATR), and the Family Study of Panic Disorder (FSPD)
“Significant” or “suggestive” susceptibility loci for ND found in at least two independent studies
According to the criteria described above, 13 linkage regions on 11 chromosomes have been identified. These linkage regions are summarized in Table 3 and Fig. 2. On inspection, several features become evident. First, except for chromosomes 5 and 9, for each of which two regions have been identified (defined as Regions 1 and 2), only one region was identified for other chromosomes. Second, the regions on chromosomes 9 (from 91.9 to 136.5 cM based on the Marshfield map), 10, 11, and 17 have received greater independent replication than the other regions. For example, the linkage region from 91.9 to 136.5 cM on chromosome 9 has been detected in four independent samples, namely, the FHS (Li et al. 2003b), COGA (Bergen et al. 1999), the EA sample of GCOD (Gelernter et al. 2007), and the AA sample of MSTF (Li et al. 2006). Within this linkage region, three genes, namely, γ-aminobutyric acid type B (GABAB) receptor subunit 2 (BABAB2), neurotrophic tyrosine kinase receptor 2 (NTRK2), and Src homology 2 domain-containing transforming protein C3 (SHC3), have been identified using family based association analysis and demonstrated to be significantly associated with ND in the MSTF sample (Beuten et al. 2005, 2007b; Li et al. 2007b). Also, the genomic region from 62 to 158 cM on chromosome 10 has been linked to ND in five independent populations: the Christchurch sample of New Zealand (Straub et al. 1999; Sullivan et al. 2004), FTF (Loukola et al. 2007), the EA sample of GCOD (Gelernter et al. 2007), the AA sample of MSTF (Li et al. 2006), and the EA sample of MSTF (Li et al. 2007a). Further, the region on chromosome 11 was detected by my research group in the FHS sample (Li et al. 2003b; Wang et al. 2005) and in both the AA (Li et al. 2006) and EA (Li et al. 2007a) samples of the MSTF cohort, as well as by Gelernter et al. (2004) in the FSPD sample, by Loukola et al. (2007) in the FTF sample, and by Morley et al. (2006) in the ATR sample. Because β-arrestin 1 is located in this region and is an important regulator of signal transduction mediated by opioid receptors through promotion of receptor desensitization and internalization (Bradaia et al. 2005; Cen et al. 2001; Gainetdinov et al. 2004), we were motivated to determine whether the β-arrestins 1 and 2 (located in a linked region to ND on chromosome 17; see below for details) are associated with ND. Our results indicated that these two genes are significantly associated with ND in European smokers (Sun et al. 2007). Furthermore, we found the strength of these associations to be higher after removal of the smoking quantity component from HSI and FTND scores in both the AA and EA samples, suggesting that these two genes play a critical role in biological processes involved in the regulation of smoking urgency (Sun et al. 2007).
Summary of chromosomal locations of nominated regions for all ND-related measures with “significant” or “suggestive” linkage score by at least two independent studies. Only chromosomes with positive linkages are shown. The linkage results were obtained from the following studies: AA/MSTF (Li et al. 2006); EA/MSTF (Li et al. 2007a); FHS (Li et al. 2003b; Wang et al. 2005); AA/GCOD and EA/GCOD (Gelernter et al. 2007), COGA (Bergen et al. 1999; Bierut et al. 2004; Duggirala et al. 1999), SMOFAM (Swan et al. 2006), FTF (Loukola et al. 2007), Mission Indians (Ehlers and Wilhelmsen 2007), FSPD (Gelernter et al. 2004), Christchurch (Straub et al. 1999; Sullivan et al. 2004), ATR (Morley et al. 2006), and Finnish/NAG and Australia/NAG (Saccone et al. 2007). Abbreviations: AA/MSTF African-American sample of the Mid-South Tobacco Family study, EA/MASTF European-American sample of the Mid-South Tobacco Family study, FHS Framingham Heart Study, GOCA Collaborative Studies on the Genetics of Alcoholism, Australia/NAG the Australia family sample of the Nicotine Addiction Genetics project, Finnish/NAG the Finnish family sample of the Nicotine Addiction Genetics project, FTF Finnish Twin Families, AA/GCOD African-American sample of Genetics of Cocaine or Opioid Dependence study, EA/GHCOD European-American sample of Genetics of Cocaine or Opioid Dependence study, Mission Indians Mission Indians in Southwest California, SMOFAM Smoking in Families Study, NTR Netherlands Twin Register study, Christchurch Christchurch sample of New Zealand, ATR Australian Twin Registry, and FSPD Family Study of Panic Disorder. Linkage peak marked with * on chromosomes 5, 10, 11, and 20 indicates a “significant linkage,” as reported in original study
The region from 31.9 to 65 cM on chromosome 17 has been linked to ND in four studies based on three independent samples, FHS (Li et al. 2003b; Wang et al. 2005), COGA (Duggirala et al. 1999), and the EA sample of MSTF (Li et al. 2007a). Since the identification of linkage of the region on chromosome 17 to ND in our genome-wide linkage scan for SQ in the FHS sample, we have conducted candidate gene-based association analysis of this region as we did for the linked region on chromosomes 9 and 11. Our family-based association analysis revealed that GABA-A receptor-associated protein (GABARAP) (Lou et al. 2007), Discs, large homolog 4 (DLG4) or post-synaptic density protein-95 (Lou et al. 2007), protein phosphatase regulatory subunit B1 (PPP1R1B) or dopamine- and cAMP-regulated phosphoprotein, 32-KD; DARPP32 (Beuten et al. 2007a), and β-arrestin 2 (Sun et al. 2007) are significantly associated with ND in at least one of the two MSTF samples.
Third, of the 13 nominated loci listed in Table 3 and Fig. 2, four showed evidence of “significant” linkage to ND. They are located on chromosomes 5 with a genome-wide P value of 0.037 for FTND in the AA sample of GCOD (Gelernter et al. 2007), 10 with a maximum LOD score of 4.17 for SQ in the AA sample of MSTF (Li et al. 2006), 11 with a pointwise P value of 0.000001 for SQ in FHS (Li et al. 2003b), and 20 with a maximum LOD score of 4.22 for MaxCigs24 in the Finnish family sample of NAG (Saccone et al. 2007). Finally, although 13 susceptibility loci on 11 chromosomes are nominated here, we should not assume the regions identified in different populations are same set of genes or genetic variants. Rather, although these regions are more likely to harbor susceptibility loci for ND, the nature of the genetic variants may differ across samples.
Nominated “significant” susceptibility loci for ND in one study
As shown in Table 4, eight genomic regions, on chromosomes 1, 5, 10, 11, 12, 16, 20, and 22, have been nominated as “significant” loci for ND-related phenotypes. Of these loci, regions on chromosomes 1 and 5 were detected with the empirically genome-wide significance level determined by permutation analysis of at least 1,000 simulated genome-wide scans (Gelernter et al. 2007; Wang et al. 2005). The other six regions were detected with conventional one-round linkage analysis according to the theoretical threshold (Li et al. 2003b, 2007a, 2006; Saccone et al. 2007; Swan et al. 2006). Unlike the regions on chromosomes 1, 12, and 16, the regions on chromosomes 5, 10, 11, 20, and 22 have been replicated by other independent studies, although the LOD score or P value from other studies did not reach the threshold for “significant” linkage.
Interestingly, although the significant region from 151.9 to 175.6 cM (based on the Marshfield map) on chromosome 1 (Wang et al. 2005) has received only limited support from two independent human studies (Bergen et al. 1999; Goode et al. 2003), it receives strong support from a linkage study for oral nicotine consumption in C57BL/6J × C3H/HeJ F2 intercross mice (Li et al. 2007c). Among the four detected significant quantitative trait loci (QTL), the locus with the largest LOD score, 15.7, was located at around 96 cM on chromosome 1 (Li et al. 2007c). This region of the mouse genome is syntenic with human chromosome 1 at around 169 cM. As for the “significant” linkage for ND on chromosomes 12 and 22, it has been detected only in the combined AA and EA samples of the MSTF cohort (Li et al. 2007a) and in the combined Australian and Finnish samples of NAG (Saccone et al. 2007). Given that plausible candidate genes with known biological functions in the etiology of dependence on nicotine and other substances of abuse are located within these regions, including ionotropic N-methyl d-aspartate glutamate receptor (NMDA) subunit 2B, and neurotrophin 3, GABA-A receptor-associated protein-like protein 1 on chromosome 12 and β-adrenergic receptor kinase 2 on chromosome 22, more linkage and position-based association studies are greatly needed to validate these linkage results.
Conclusions and perspectives
Despite inherent difficulties in conducting genetic studies on complex traits, significant progress has been made in recent years in the search for susceptibility loci for ND. By applying the same rigorous criteria for determination of “significant” or “suggestive” linkage to all reported linkage peaks for ND-related phenotypes, and requiring evidence from at least two independent studies, 13 regions on 11 chromosomes have been identified. Of these, the regions on chromosomes 9 (between 91.9 and 136.5 cM), 10, 11, and 17 have been detected by the greatest number of independent studies. In addition, a list of eight “significant” linkages on chromosomes 1, 5, 10, 11, 12, 16, 20, and 22 is provided. Considering that these regions have received the most support, it is suggested they be afforded the highest priority in searching for vulnerability genes for ND in future studies.
Several other issues deserve mention. Although different measures have been used to assess ND across studies, the linkage results appear similar, suggesting this phenomenon is robust. The genetic underpinnings of ND may not be critical to the ultimately observed phenotypic variation in ND characteristics. Although various linkage peaks were identified using different ND measures, smoking quantity (i.e., daily smoking rate or its variants) has yielded the most reproducible and strongest findings. Evidence that different genomic regions are definitively associated with specific ND qualities awaits further research. Second, sample size is likely to be a significant consideration in some of the failure to replicate across studies. Early investigations of genetic linkage or association studies of ND commonly included only a few to several hundred subjects, a sample size now considered unlikely to have the power required to yield conclusive results. Finally, although it has been common to conduct genetic studies in samples consisting of participants of various origins, recent findings suggest this may not represent an optimal approach, potentially producing results that are confusing or misleading. This point is well illustrated in a recent study in which several linkage peaks were detected in one ethnic sample, and pooling subjects across ethnic groups did not improve the statistical power to detect linkage (Li et al. 2007b). For example, by comparing the linkage results identified from the AA and EA samples and their combination, we found four overlapping regions on chromosomes 9 (two regions), 11, and 18 in the two samples. Furthermore, we identified five regions, on chromosomes 2, 4, 10, 12, and 17, that showed linkage to ND only in the EA sample and two regions, on chromosomes 10 and 13, that showed linkage only in the AA sample. This indicates that genetic differences underlying ND exist in these two ethnic populations. Given that populations of primarily European origin have been used in most reported studies on ND, more genetic studies with other ethnic groups such as AAs and Asians are greatly needed in the future.
References
Bergen AW, Korczak JF, Weissbecker KA, Goldstein AM (1999) A genome-wide search for loci contributing to smoking and alcoholism. Genet Epidemiol 17(Suppl 1):S55–S60
Bergen AW, Yang XR, Bai Y, Beerman MB, Goldstein AM, Goldin LR (2003) Genomic regions linked to alcohol consumption in the Framingham Heart Study. BMC Genet 4(Suppl 1):S101
Beuten J, Ma JZ, Payne TJ, Dupont RT, Crews KM, Somes G, Williams NJ, Elston RC, Li MD (2005) Single- and multilocus allelic variants within the GABAB receptor subunit 2 (GABAB2) gene are significantly associated with nicotine dependence. Am J Hum Genet 76:859–864
Beuten J, Ma JZ, Lou XY, Payne TJ, Li MD (2007a) Association analysis of the protein phosphatase 1 regulatory subunit 1B (PPP1R1B) gene with nicotine dependence in European- and African-American smokers. Am J Med Genet B Neuropsychiatr Genet 144:285–290
Beuten J, Ma JZ, Payne TJ, Dupont RT, Lou XY, Crews KM, Elston RC, Li MD (2007b) Association of Specific Haplotypes of Neurotrophic Tyrosine Kinase Receptor 2 Gene (NTRK2) with Vulnerability to Nicotine Dependence in African-Americans and European-Americans. Biol Psychiatry 61:48–55
Bierut LJ, Rice JP, Goate A, Hinrichs AL, Saccone NL, Foroud T, Edenberg HJ, Cloninger CR, Begleiter H, Conneally PM, Crowe RR, Hesselbrock V, Li TK, Nurnberger JI Jr, Porjesz B, Schuckit MA, Reich T (2004) A genomic scan for habitual smoking in families of alcoholics: common and specific genetic factors in substance dependence. Am J Med Genet 124A:19–27
Bradaia A, Berton F, Ferrari S, Luscher C (2005) Beta-Arrestin2, interacting with phosphodiesterase 4, regulates synaptic release probability and presynaptic inhibition by opioids. Proc Natl Acad Sci USA 102:3034–3039
Carmelli D, Swan GE, Robinette D, Fabsitz R (1992) Genetic influence on smoking—a study of male twins. N Engl J Med 327:829–833
CDC (2005) Annual smoking-attributable mortality, years of potential life lost, and productivity losses—United States, 1997–2001. Morb Mortal Wkly Rep 54:625–628
CDC (2007) Cigarette smoking among adults—United States, 2006. Morb Mortal Wkly Rep 56:1157–1161
Cen B, Yu Q, Guo J, Wu Y, Ling K, Cheng Z, Ma L, Pei G (2001) Direct binding of beta-arrestins to two distinct intracellular domains of the delta opioid receptor. J Neurochem 76:1887–1894
Cheng LS, Swan GE, Carmelli D (2000) A genetic analysis of smoking behavior in family members of older adult males. Addiction 95:427–435
Duggirala R, Almasy L, Blangero J (1999) Smoking behavior is under the influence of a major quantitative trait locus on human chromosome 5q. Genet Epidemiol 17(Suppl 1):S139–S144
Ehlers CL, Wilhelmsen KC (2006) Genomic screen for loci associated with tobacco usage in Mission Indians. BMC Med Genet 7:9
Ehlers CL, Wilhelmsen KC (2007) Genomic screen for substance dependence and body mass index in southwest California Indians. Genes Brain Behav 6:184–191
Gainetdinov RR, Premont RT, Bohn LM, Lefkowitz RJ, Caron MG (2004) Desensitization of G protein-coupled receptors and neuronal functions. Annu Rev Neurosci 27:107–144
Gelernter J, Liu X, Hesselbrock V, Page GP, Goddard A, Zhang H (2004) Results of a genomewide linkage scan: support for chromosomes 9 and 11 loci increasing risk for cigarette smoking. Am J Med Genet 128B:94–101
Gelernter J, Panhuysen C, Weiss R, Brady K, Poling J, Krauthammer M, Farrer L, Kranzler HR (2007) Genomewide linkage scan for nicotine dependence: identification of a chromosome 5 risk locus. Biol Psychiatry 61:119–126
Goode EL, Badzioch MD, Kim H, Gagnon F, Rozek LS, Edwards KL, Jarvik GP (2003) Multiple genome-wide analyses of smoking behavior in the Framingham Heart Study. BMC Genet 4(Suppl 1):S102
Heath AC, Martin NG (1993) Genetic models for the natural history of smoking: evidence for a genetic influence on smoking persistence. Addict Behav 18:19–34
Heath AC, Cates R, Martin NG, Meyer J, Hewitt JK, Neale MC, Eaves LJ (1993) Genetic contribution to risk of smoking initiation: comparisons across birth cohorts and across cultures. J Subst Abuse 5:221–246
Ho MK, Tyndale RF (2007) Overview of the pharmacogenomics of cigarette smoking. Pharmacogenomics J 7:81–98
Hughes JR (1986) Genetics of smoking: a brief review. Behav Therapy 17:335–345
Kendler KS, Neale MC, Sullivan P, Corey LA, Gardner CO, Prescott CA (1999) A population-based twin study in women of smoking initiation and nicotine dependence. Psychol Med 29:299–308
Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247
Li MD, Ma JZ, Beuten J (2004) Progress in searching for susceptibility loci and genes for smoking-related behaviour. Clin Genet 66:382–392
Li MD, Cheng R, Ma JZ, Swan GE (2003a) A meta-analysis of estimated genetic and environmental effects on smoking behavior in male and female adult twins. Addiction 98:23–31
Li MD, Ma JZ, Cheng R, Dupont RT, Williams NJ, Crews KM, Payne TJ, Elston RC (2003b) A genome-wide scan to identify loci for smoking rate in the Framingham Heart Study population. BMC Genet 4(Suppl 1):S103
Li MD, Payne TJ, Ma JZ, Lou XY, Zhang D, Dupont RT, Crews KM, Somes G, Williams NJ, Elston RC (2006) A genomewide search finds major susceptibility loci for nicotine dependence on chromosome 10 in African Americans. Am J Hum Genet 79:745–751
Li MD, Ma JZ, Payne TJ, Lou XY, Zhang D, Dupont RT, Elston RC (2007a) Genome-wide linkage scan for nicotine dependence in European Americans and its converging results with African Americans in the Mid-South Tobacco Family sample. Mol Psychiatry 2007 June 19 [Epub ahead of print]
Li MD, Sun D, Lou XY, Beuten J, Payne TJ, Ma JZ (2007b) Linkage and association studies in African- and Caucasian-American populations demonstrate that SHC3 is a novel susceptibility locus for nicotine dependence. Mol Psychiatry 12:462–473
Li XC, Karadsheh MS, Jenkins PM, Brooks JC, Drapeau JA, Shah MS, Lautner MA, Stitzel JA (2007c) Chromosomal loci that influence oral nicotine consumption in C57BL/6J × C3H/HeJ F2 intercross mice. Genes Brain Behav 6:401–410
Lou XY, Ma JZ, Sun D, Payne TJ, Li MD (2007) Fine mapping of a linkage region on chromosome 17p13 reveals that GABARAP and DLG4 are associated with vulnerability to nicotine dependence in European-Americans. Hum Mol Genet 16:142–153
Loukola A, Broms U, Maunu H, Widen E, Heikkila K, Siivola M, Salo A, Pergadia ML, Nyman E, Sammalisto S, Perola M, Agrawal A, Heath AC, Martin NG, Madden PA, Peltonen L, Kaprio J (2007) Linkage of nicotine dependence and smoking behavior on 10q, 7q and 11p in twins with homogeneous genetic background. Pharmacogenomics J 2007 June 5 [Epub ahead of print]
Madden PA, Heath AC, Pedersen NL, Kaprio J, Koskenvuo MJ, Martin NG (1999) The genetics of smoking persistence in men and women: a multicultural study. Behav Genet 29:423–431
Maes HH, Woodard CE, Murrelle L, Meyer JM, Silberg JL, Hewitt JK, Rutter M, Simonoff E, Pickles A, Carbonneau R, Neale MC, Eaves LJ (1999) Tobacco, alcohol and drug use in eight- to sixteen-year-old twins: the Virginia Twin Study of Adolescent Behavioral Development. J Stud Alcohol 60:293–305
Morley KI, Medland SE, Ferreira MA, Lynskey MT, Montgomery GW, Heath AC, Madden PA, Martin NG (2006) A possible smoking susceptibility locus on chromosome 11p12: evidence from sex-limitation linkage analyses in a sample of Australian Twin Families. Behav Genet 36:87–99
Pomerleau OF, Pomerleau CS, Chu J, Kardia SL (2007) Genome-wide linkage analysis for smoking-related regions, with replication in two ethnically diverse populations. Nicotine Tob Res 9:955–958
Saccone NL, Neuman RJ, Saccone SF, Rice JP (2003) Genetic analysis of maximum cigarette-use phenotypes. BMC Genet 4(Suppl 1):S105
Saccone SF, Pergadia ML, Loukola A, Broms U, Montgomery GW, Wang JC, Agrawal A, Dick DM, Heath AC, Todorov AA, Maunu H, Heikkila K, Morley KI, Rice JP, Todd RD, Kaprio J, Peltonen L, Martin NG, Goate AM, Madden PA (2007) Genetic linkage to chromosome 22q12 for a heavy-smoking quantitative trait in two independent samples. Am J Hum Genet 80:856–866
SAMHSA (2001) Summary of findings from the 2000 national household survey on drug abuse, Rockville
Straub RE, Sullivan PF, Ma Y, Myakishev MV, Harris-Kerr C, Wormley B, Kadambi B, Sadek H, Silverman MA, Webb BT, Neale MC, Bulik CM, Joyce PR, Kendler KS (1999) Susceptibility genes for nicotine dependence: a genome scan and followup in an independent sample suggest that regions on chromosomes 2, 4, 10, 16, 17 and 18 merit further study. Mol Psychiatry 4:129–144
Sullivan PF, Neale BM, van den Oord E, Miles MF, Neale MC, Bulik CM, Joyce PR, Straub RE, Kendler KS (2004) Candidate genes for nicotine dependence via linkage, epistasis, and bioinformatics. Am J Med Genet B Neuropsychiatr Genet 126:23–36
Sun D, Ma JZ, Payne TJ, Li MD (2007) Beta-Arrestins 1 and 2 are associated with nicotine dependence in European American smokers. Mol Psychiatry 2007 June 19 [Epub ahead of print]
Swan GE, Carmelli D, Reed T, Harshfield GA, Fabsitz RR, Eslinger PJ (1990a) Heritability of cognitive performance in aging twins. The National Heart, Lung, and Blood Institute Twin Study. Arch Neurol 47:259–262
Swan GE, Carmelli D, Rosenman RH, Fabsitz RR, Christian JC (1990b) Smoking and alcohol consumption in adult male twins: genetic heritability and shared environmental influences. J Subst Abuse 2:39–50
Swan GE, Carmelli D, Cardon LR (1996) The consumption of tobacco, alcohol, and coffee in Caucasian male twins: a multivariate genetic analysis. J Subst Abuse 8:19–31
Swan GE, Hops H, Wilhelmsen KC, Lessov-Schlaggar CN, Cheng LS, Hudmon KS, Amos CI, Feiler HS, Ring HZ, Andrews JA, Tildesley E, Benowitz N (2006) A genome-wide screen for nicotine dependence susceptibility loci. Am J Med Genet B Neuropsychiatr Genet 141:354–360
True WR, Xian H, Scherrer JF, Madden PA, Bucholz KK, Heath AC, Eisen SA, Lyons MJ, Goldberg J, Tsuang M (1999) Common genetic vulnerability for nicotine and alcohol dependence in men. Arch Gen Psychiatry 56:655–661
Vink JM, Beem AL, Posthuma D, Neale MC, Willemsen G, Kendler KS, Slagboom PE, Boomsma DI (2004) Linkage analysis of smoking initiation and quantity in Dutch sibling pairs. Pharmacogenomics J 4:274–282
Vink JM, Willemsen G, Boomsma DI (2005) Heritability of smoking initiation and nicotine dependence. Behav Genet 35:397–406
Vink JM, Posthuma D, Neale MC, Eline Slagboom P, Boomsma DI (2006) Genome-wide linkage scan to identify loci for age at first cigarette in Dutch sibling pairs. Behav Genet 36:100–111
Wang D, Ma JZ, Li MD (2005) Mapping and verification of susceptibility loci for smoking quantity using permutation linkage analysis. Pharmacogenomics J 5:166–172
WHO (2002) The World Health Report 2002. World Health Organization
Acknowledgments
The author thanks Drs. David Bronson, Thomas J Payne, and Jamie Mangold for their critical editing and Drs. Daolong Wang, Jennie Z. Ma, Xiang-Yang Lou, and Weihua Huang for their help in the preparation of the tables and figures used for the report. The preparation of this article was in part supported by NIH grants DA-12844 and DA-13783.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Li, M.D. Identifying susceptibility loci for nicotine dependence: 2008 update based on recent genome-wide linkage analyses. Hum Genet 123, 119–131 (2008). https://doi.org/10.1007/s00439-008-0473-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-008-0473-0