Abstract
Spinach (Spinacia oleracea L.) is a dioecious plant with male heterogametic sex determination and homomorphic sex chromosomes (XY). The dioecism is utilized for producing commercial hybrid seeds, and hence understanding the molecular-genetic basis of the species’ sex determining locus is an important issue for spinach breeding. In this study, seven dominant DNA markers were shown to completely co-segregate with the male-determining gene in segregating spinach populations comprising > 1500 plants. In addition, these seven dominant DNA markers were completely associated with the male-determining gene in over 100 spinach germplasm accessions and cultivars. These observations suggest that, in spinach, a Y-chromosomal region around the male-determining locus does not (or almost not) recombine with a counterpart region on the X chromosome. Using five of the seven DNA markers, five bacterial artificial chromosome (BAC) clone contigs with a total length of approximately 690 kbp were constructed. Full sequencing of six representative BAC clones (total insert length 504 kbp) from the five contigs and a transcriptome analysis by RNA-seq revealed that the Y-chromosomal region around the male-determining locus contains large amounts of repetitive elements, suggesting that the region might be poor in gene content. Most of the repeats found in this region are novel Ty1-copia-like and its derivative elements that accumulate predominantly in heterochromatic regions. Our findings may provide valuable insight into spinach genome structure and clues for future research into the evolution of the sex determining locus.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cosexuality (hermaphroditism or monoecy) is the most prevalent sexual systems in angiosperms. Only a small percentage of angiosperms are dioecious. However, dioecious plants are distributed among over 40 angiosperm families, suggesting that dioecy has evolved recently and independently from cosexual ancestors in diverse taxa (Charlesworth 2016; Renner and Ricklefs 1995; Westergaard 1958).
Sex determination in dioecious plants is often controlled by genetic factors, i.e., sex-determining genes and sex chromosomes. In Silene latifolia, there are heteromorphic sex chromosomes, and the Y-chromosomes have extensive non-recombining regions (Charlesworth 2016; Vyskot and Hobza 2015). Absent or reduced recombination in the sex-determining locus is also inferred using genetic approaches in papaya, a species with slight sex-chromosome heteromorphism (Liu et al. 2004). Among dioecious plants with no apparent sex-chromosome heteromorphism, grapevine and poplar have small sex-linked non-recombining regions (~ 155 and ~ 100 kbp, respectively) that account for less than 1% of their sex chromosomes (Charlesworth 2016; Geraldes et al. 2015; Picq et al. 2014). The evolutionary transition from cosexuality to dioecy predicts at least two mutations that affect male and female functions. Evolutionary theory predicts that the mutations occurred at closely linked loci, and that suppression of recombination between the loci may be favored to maintain a stable fully sexually dimorphic population without cosexual or neuter individuals (Charlesworth and Charlesworth 1978).
Spinach (Spinacia oleracea) is a leafy vegetable belonging to the goosefoot family (Chenopodiaceae s.s.). It is dioecious and its populations generally have equal numbers of female and male plants. However, some varieties and genotypes produce monoecious plants with various proportions of pistillate and staminate flowers or, in some cases, hermaphroditic flowers (Janick and Stevenson 1955; Katayama and Shida 1956; Onodera et al. 2008). The dioecism and monoecism in spinach are utilized for producing commercial hybrid seeds, and hence the elucidation of the mechanisms controlling the plant’s sex expression is important for spinach breeding (Janick 1998; Onodera et al. 2011).
In spinach, males are the heterogametic (XY) sex and females are homogametic (XX) (Janick and Stevenson 1954). Gender in dioecious spinach lines is controlled by an allelic pair, designated X and Y, which are located on the largest chromosome (Iizuka and Janick 1962). This chromosome pair shows no obvious sex-associated difference in size (Ellis and Janick 1960). The sister genus of Spinacia, Blitum, consists of cosexual (hermaphroditic and gynomonoecious) species (Fuentes-Bazan et al. 2012a, b; Kadereit et al. 2010; Naeger and Golenberg 2016). As mentioned above, spinach can have intermediate sexual conditions, e.g., monoecy and gynomonoecy (Janick and Stevenson 1955; Katayama and Shida 1956; Onodera et al. 2008), one of which may be the ancestral sexual condition of Spinacia. Therefore, dioecism in spinach may have evolved via at least two mutations producing males and females. Inconstancy of sex expression has also been observed in the Long Standing Bloomsdale cultivar, in which male plants produced occasional seeds (ovules) (Janick and Stevenson 1954). However, no neuter individuals have been reported so far in crosses between female and male or monoecious plants (Janick and Stevenson 1955; Onodera et al. 2011; Yamamoto et al. 2014). If the male-determining locus (Y) consists of two (or more) factors, meiotic recombination may be suppressed across the region. Our previous study suggested that recombination rates are low around the male-determining locus (Y), although the chromosome pair recombines across most of their length (Takahata et al. 2016). Highly reliable sex diagnostic markers have now been developed and implemented in spinach breeding programs (Akamatsu et al. 1998; Tohoku Seed Co., personal communication). However, it is still not clear whether the spinach male-determining locus (region) is fully recombinationally suppressed.
In this research, spinach sex-linked amplified fragment length polymorphism (AFLP) markers developed by Onodera et al. (2011) were converted into sequence characterized amplified region (SCAR) markers, most of which amplified male-specific DNA fragments. Here, we describe linkage and association analyses using male-specific DNA fragments that suggest complete (or severe) recombination suppression of the region around the male-determining locus. Furthermore, we fully sequenced over 500 kbp in total from six BAC genomic clones representing the region around the male-determining locus, estimated the minimum size of the fully sex-linked region, and investigated novel Ty1-copia-like and its derivative elements predominantly found in these BAC clones.
Materials and methods
Plant materials
Two populations (03-009-sib-cross A and B, consisting of 191 males and 213 females, respectively) were generated by crosses between members of the dioecious breeding line 03-009 (Tohoku Seed Co. Ltd., Utsunomiya, Tochigi, Japan), for linkage analysis between DNA markers and the male-determining factor(s). Two backcross progeny populations (BC1F1 and BC2F1 generations) from a cross between an all-female (true-breeding highly female monoecious) breeding line 03-259 (Tohoku Seed Co. Ltd.) and the same dioecious breeding line (03-009) were also analyzed (BC1F1, n = 121, Onodera et al. 2011; BC2F1, n = 473, present study). We also used further backcross progeny populations (03-336 × 03-009 BC1F1 and BC2F1) (Yamamoto et al. 2014) generated from a cross between another monoecious breeding line 03-336 (Tohoku Seed Co. Ltd.) and dioecious breeding line 03-009 to test linkage: the BC1F1 and BC2F1 populations correspond to the test-cross populations A and B in Yamamoto et al. (2014), respectively. The spinach populations, 03-259 × 03-009 BC2F1 and 03-009-sib-cross A and B, generated in the present study were grown in a plastic greenhouse during March–June in 2013, 2014 and 2015, respectively.
A total of 109 Spinacia (S. oleracea, S. turkestanica, and S. tetrandra), Blitum (B. bonus-henricus, B. californicum, B. capitatum, B. virgatum, and B. nuttallianum), and Chenopodium (C. album and C. quinoa) germplasm accessions were obtained from the National Institute of Agrobiological Sciences (NIAS) Genebank (Tsukuba, Japan), the United States Department of Agriculture (USDA) National Plant Germplasm System, and Center for Genetic Resources, The Netherlands (CGN) (Table S1). The germplasm accessions and four spinach cultivars (cv. Atlas, Sakata Seed Co., Ltd., Yokohama, Japan; cv. Mazeran and Okame, Takii and Co., Ltd., Kyoto, Japan; cv. SPI588, Syngenta Vegetables, Boise, ID, USA) were grown in a plastic greenhouse during March–July 2015, and used for DNA marker and/or Southern blot analysis. Beta vulgaris line TK81-MS (Onodera et al. 1999) used for the Southern blot analysis was kindly provided as leaf tissue from Prof. Tomohiko Kubo (Hokkaido University). Total cellular DNA was prepared from leaf tissue of individual plants using the method of Sassa (2007).
For Spinacia plants bearing over 100 flowers, we estimated the percentage of female flowers per plant. Plants with percentage > 95–100% were classified as female. Plants with no female flowers, and the > 0–95% femaleness classes were considered male and monoecious, respectively.
DNA marker development and analysis
SCAR, cleaved amplified polymorphic sequences (CAPS) and derived cleaved amplified polymorphic sequence (dCAPS) markers were designed from DNA sequences of sex-linked AFLP loci identified in a previous study (Table S2; Onodera et al. 2011). DNA sequencing of the AFLP loci, and marker design, were carried out as described in a previous study (Yamamoto et al. 2014). SCAR markers were also developed from insert ends of spinach genomic BAC clones (Table S3).
This study used newly developed markers, and three commercially used spinach male-specific SCAR markers, T11A (5′-CCCTAATTAACTCCTCTTTACCCAA-3′ and 5′-TACAAGCCCCATTATCATAACAGTC-3′, accession number E15132, Akamatsu et al. 1998), V20A (5′-TACCGTTGAATCAGTGTTGTAAGTG-3′ and 5′-GGTCGACAACACAGCCAATTAT-3′, accession number E15133, Akamatsu et al. 1998) and SP_0018 developed by Tohoku Seed Co., Ltd. (whose primer sequence information was not disclosed). Amplification reactions for the markers were performed according to Yamamoto et al. (2014).
BAC-end sequencing and whole-BAC sequencing
An arrayed genomic BAC library of 03-009 male with 5.6× coverage of the spinach genome (unpublished) was screened with the DNA markers. The insert ends of the BAC clones were sequenced using T7 Promoter Primer (5′-TAATACGACTCACTATAGGG-3′) and Reverse Sequencing Primer (RP) (5′-CTCGTATGTTGTGTGGAATTGTGAGC-3′) of pCC1BAC vector (EpiCentre, Madison, WI, USA). For whole-BAC sequencing, shotgun subcloning and Sanger sequencing of the BAC clones were carried out at approximately 10× coverage.
BAC contig construction
Overlaps between BAC clones identified as carrying the SCAR markers were determined by PCR with primer pairs targeting BAC-end sequences, in addition to BAC-end and whole-BAC sequence data. To estimate the size of BAC inserts and BAC contigs, BAC clone DNA (1 µg) digested with NotI (Takara, Kusatsu, Japan) was separated by pulsed field gel electrophoresis (PFGE) in 1.0% agarose gel (0.5× Tris–Borate–EDTA) on CHEF-DRII system (Bio-Rad, Hercules, CA, USA) for 14 h at 6 V/cm and 14 °C, with pulses ranging from 1 to 10 s.
Southern blot analysis
Total cellular DNA (5 µg) digested with EcoRI was separated by electrophoresis in 0.8% agarose gel, transferred to nylon membranes (Hybond N+, GE Healthcare, Piscataway, NJ, USA), and hybridized with digoxigenin (DIG) labeled-DNA probes prepared by use of PCR DIG Synthesis Kit (Roche Diagnostics, Basel, Switzerland).
Identifying interspersed repeats and estimating copy numbers of repeated sequences
De novo identification of full-length long terminal repeat (LTR) retrotransposons was carried out using the software LTR_FINDER (Xu and Wang 2007). Searches for interspersed repeat motifs and low complexity DNA sequences were performed using RepeatMasker (http://www.repeatmasker.org) with a database consisting of RepBase (http://www.girinst.org) and elements (named SpRE, see below) found in the present study.
To estimate the copy numbers of repeated sequences included in the BAC clones (AP017636–AP017641), we aligned the BAC clones to the draft spinach genome sequence (spinach_genome_v1.fa, ftp://www.spinachbase.org/pub/spinach; Xu et al. 2017) using the NUCmer program (Kurtz et al. 2004). The number of genome segments with similarity > 80 or 90% to BAC clones was counted at each nucleotide position on the BAC clones.
Fluorescence in situ hybridization (FISH)
Chromosome observations in mitotic root tips and fluorescence in situ hybridization (FISH) mapping of 45S rDNA loci were carried out as described in Fujito et al. (2015). LTR retrotransposons were mapped using a LTR fragment (457 bp) as a probe, which was generated by PCR with the following primer sets: 5′-CGTCACAGGCAAGAAATGAA-3′ and 5′-CGTACGATACGCTTCCGATT-3′. Individual spinach chromosomes were identified with reference to the karyotypic features reported by (Ito et al. 2000).
Phylogenetic analysis
Amino acid sequences of reverse transcriptase from plant retroelements were aligned using MUSCLE software implemented in MEGA 7.0 (Kumar et al. 2016). Phylogenetic trees were constructed with the neighbor-joining (NJ) method using MEGA 7.0. The reliability of the NJ trees was assessed by 1000 bootstrap replicates.
Insertion time estimate of LTR retroelements
The divergence (K) between 5′- and 3′-LTR of each retroelement was calculated in MEGA7.0, using the Jukes and Cantor (1969) method. Insertion times, T, were estimated as T = K/2r, where K is the divergence, and r is a rate of 7 × 10−9 base substitution per site per generation for Arabidopsis thaliana (Ossowski et al. 2010), according to the method of Xu et al. (2017).
Gene prediction and annotation
Prediction of genes on BAC sequences masked by RepeatMasker was carried out by AUGUSTUS (Stanke et al. 2008) using RNA-seq data (DRA004582; Takahata et al. 2016) obtained from spinach breeding line 03-336. The predicted genes were annotated using BLAST (Altschul et al. 1997) with the NCBI non-redundant protein sequence (NR) database (ftp://ftp.ncbi.nlm.nih.gov/blast/db/FASTA/nr.gz) and hmmscan in HMMER 3.0 (http://hmmer.org) against the Pfam 31.0 database with the cutoff defined by the gathering thresholds. To verify expression of the predicted genes, total cellular RNA was isolated from male inflorescences (03-009) using an RNeasy Plant Mini Kit in combination with an RNase-Free DNase Set (QIAGEN, Venlo, The Netherlands), and analyzed by mRNA sequencing, as described by Takahata et al. (2016). RNA-seq reads (DRA002981, DRA006107) from 03-009-males were mapped to the BAC clone sequences (AP017636–AP017641) using the TopHat program (Trapnell et al. 2009). The Integrative Genomics Viewer (IGV) software (Robinson et al. 2011) was used to visualize the aligned reads.
Results
Linkage analysis between the male-determining locus (denoted here by Y) and seven DNA markers using a large scale population
Four SCAR markers converted from the four AFLP markers previously found to be tightly linked to the male-determining locus, and three commercially available male-specific SCAR markers (T11A, V20A and SP_0018; see “Materials and methods”), were found to amplify male-specific fragments from genomic DNA from the dioecious stock 03-009, the monoecious 03-336 stock, and the highly female monoecious line 03-259 stock (Table S2, Figure S1). Two markers (CAPS SP_0006a and dCAPS SP_0006b) were developed from an AFLP marker, E26M13, to detect single nucleotide polymorphisms at different sites that were located 93 base pairs apart. The 03-009-male had the CT genotype for SP_0006a, while the 03-009-female, the monoecious line 03-336, and the highly female monoecious line 03-259 were CC. For SP_0006b, the monoecious line 03-336 had the AA genotype, and the 03-009-male, 03-009-female, and highly female monoecious line 03-259 were GG (Table S2, Figure S2).
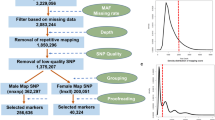
Nine DNA markers were examined for linkage with the male-determining locus using the six segregating populations (n = 1554 in total) derived from sib-crosses and back-crosses of the spinach breeding lines 03-009, 03-336, and 03-259 (see “Materials and methods”). As summarized in Table 1, seven of the nine markers (excluding SP_0006a and SP_0006b) showed complete sex-linkage, while five recombinants were found between SP_0006a/b and the male-determining locus (Table 1).
Although the SCAR markers SP_0003 and SP_0015, and SP_0006a, showed complete linkage with the male-determining locus in the 03-259 × 03-009 BC2F1 family (n = 121), Onodera et al. (2011) reported that the AFLP markers from which the SCAR and CAPS markers were derived were not completely linked with the locus in the same population. The inconsistency may be due to lower reproducibility of the AFLP as compared with the SCAR markers.
Association analysis between the male phenotype and the seven DNA markers
To examine the association between the male phenotype and the seven dominant male-specific markers showing complete sex linkage, 105 males, 103 females, and six monoecious plants were selected from unrelated material including four spinach cultivars and 101 Spinacia germplasm accessions. Among the 214 plants genotyped for the markers (Table S4), the region could be classified into three haplotypes (Table 2). The haplotype lacking the dominant alleles (the male-specific DNA fragments) of the seven markers was inferred to be X-linked, and named Hap_X, since all females and monoecious plants were homozygotes for the absence of the markers. Hap_Y1 carried all seven markers, while Hap_Y2 was identical, except that it lacked the SP_0014 marker. Hap_Y1 and Hap_Y2 were found in 52 and 53 males, respectively, but no females and monoecious plants with these two haplotypes were found. The results suggest that presence of the markers is strongly associated with the male phenotype (Fisher’s exact test, p < 2.2e−16; Table 2), and that a fully Y-linked region exists around the male-determining locus.
Construction of BAC contigs associated with the male-determining region on the spinach Y chromosome
As shown in Fig. 1, 18 BAC clones, ranging from 49 to 110 kbp in size, were isolated from our male spinach genome BAC library by screening with five of the seven markers (excluding SP_0003 and SP_0015); 2–5 positive clones were found per marker, and no clone was positive for more than one of the markers; hence they were assembled into five contigs.
BAC clone contigs covering chromosomal regions associated with the spinach male-determining locus (Y). Gray boxes represent spinach genomic BAC clones isolated using markers fully linked to male-determining locus and using BAC-end-derived markers. Solid black boxes indicate BAC-end sequences, based on which the markers were designed. Dashed lines link the BAC clones and their associated markers
A BLAST search (Camacho et al. 2009) against the draft spinach genome sequence (Xu et al. 2017) revealed that 25 of the 36 BAC-end sequences were highly repetitive (> 100 BLAST hits with > 90% identity) in the spinach genome. Primer pairs targeting three BAC-ends (78-1K_RP, 64-3P_T7 and 95-21D_T7; Fig. 1 and Table S3), with no BLAST hits showing > 90% identity, amplified male-specific DNA fragments, as did a primer pair designed from a presumably low-copy (or single-copy) region found in BAC-end 78-1K_T7 (Figure S3). The primer sequences of the BAC-end derived marker 64-3p-T7 are located on the contig associated with the SP_0018 marker (Fig. 1; Table 2 and Table S3). These primer pairs derived from BAC-ends allowed us to isolate four further BAC clones from the library, which extended the size of the BAC contigs associated with V20A and SP_0018. However, the repetitive nature of the BAC-end sequences prevented further extension of the five contigs and we were unable to fill the gaps between them. Altogether, a total of 22 BAC clones associated with the male-determining region were assembled into the five contigs, ranging from 106 to 180 kbp (692 kbp in total length; Fig. 1).
Sequence characterization of chromosomal regions associated with the male-determining region
To characterize the genetic content of the chromosomal region associated with the male-determining region, six BAC clones (79–110 kbp) representing the five BAC contigs were fully sequenced (Fig. 1). Two of the six clones [26–14K (81,519 bp) and 41–10L (78,321 bp)] were derived from the same contig and overlapped by 43,505 bp with one nucleotide mismatch (which was not verified by direct sequencing of genomic PCR products, and could be an artifact e.g., a replication error in E. coli). We, therefore, provisionally concatenated the 26–14K and 41–10L sequences. The total length of the completely sequenced BAC clones was 503,533 bp.
Identification of LTR retroposon-like sequences
As summarized in Table 3, the LTR_finder software (Xu and Wang 2007) identified ten LTR retroposon-like sequences from five of the six BAC clones sequenced. The sequences are approximately 10 kbp in length, and carry 1.4 kbp LTR sequences at both ends. They were classified into two groups designated Spinach-REtroposon-1 (SpRE1) and -2 (SpRE2), based on sequence differences, although their LTR regions are highly similar (Fig. 2).
Two novel LTR retrotransposons found in the chromosomal region associated with the male-determining region. a Schematic structures of novel LTR retrotransposons SpRE1 and SpRE2. LTR long terminal repeat, PR protease, INT integrase, RT reverse transcriptase, RH RNase H. Three arrow pairs facing each other (P1–P6) represent primers used for RT-PCR analysis shown in Fig. 4. b Alignment of nucleotide sequences for the primer binding site (PBS) and polypurine tract (PPT) of LTR retrotransposons in angiosperms. c Amino acid alignment of conserved motifs and a domain in copia-like retrotransposons from angiosperms. RIRE1 (D85597), SORE-1 (AB370254), Ta1-3 (X13291), and Tto-1 (D83003) are copia-like elements derived from Oryza australiensis, Glycine max, A. thaliana, and Nicotiana tabacum respectively
As shown in Fig. 2, two conserved motifs, primer binding site (PBS) and polypurine tract (PPT), are found downstream of the 5′LTR and upstream of the 3′LTR in both retroposons. A polypeptide encoded by SpRE1 was found to have significant homology with Ty1-copia-type gag and pol polyproteins, and displayed the highest homology (E value = 0.0) with a polyprotein (BAA22288) encoded by a retrotransposon from wild rice, RIRE1 (D85597.1). This gene also retains RNA binding, D[S/T]G, YXDD motifs and a domain (GKGY) conserved among retroposons’ polyproteins (Fig. 2). Furthermore, phylogenetic analyses of the reverse transcriptase domains of LTR retrotransposons placed SpRE1 in a Ty1-copia cluster (Figure S4). We found no evidence that SpRE2 encodes proteins associated with retrotransposons, and concluded that SpRE1 is a member of Ty1-copia family, and that SpRE2 could be a non-autonomous retroelement associated with SpRE1, assigned to the class of LArge Retrotransposon Derivatives (LARDs) (Kalendar et al. 2004). The estimated insertion times of the SpRE copies ranged from 0.1 to 1.3 (average 0.6) million years ago (Table 3).
Furthermore, as summarized in Table 4, RepeatMasker analysis revealed that 50.4% of the six BAC clone sequences can be accounted for by retroposon-like sequences, most of which (46.2% of the BAC clone sequences) were assigned to SpRE elements. The retroposon-like sequences other than SpRE consist of LTR/Copia, LTR/Gypsy, LTR/Pao, and LINE/L1. DNA-mediated transposon like, simple repeat, and low complexity sequences accounted for only a small fraction (1.4, 0.8, and 0.3%, respectively) of the BAC clone sequences.
Southern blots of EcoRI-digested genomic DNA from Spinacia species (S. oleracea, S. turkestanica, and S. tetrandra) gave strong smeared hybridization signals when probed with a SpRE-LTR fragment, but no signals were detected from other Chenopodiaceae s.s. species from the genera Blitum, Chenopodium, and Beta (Fig. 3).
Southern blot analysis of SpRE elements. Genomic DNA digested with EcoRI was probed with SpRE-LTR fragment. The same blot was rehybridized with a chloroplast gene [large subunit of RUBISCO (rbcL)] probe as a loading control. Lane 1, male S. oleracea (line 03-009); lane 2, female S. oleracea (line 03-009); lane 3, monoecious S. oleracea (03-336); lane 4, male S. turkestanica (CGN09594); lane 5, female S. turkestanica (CGN09594); lane 6, male S. tetrandra (PI 647859); lane 7, female S. tetrandra (PI 647859); lane 8, B. bonus-henricus (PI 662294); lane 9, B. californicum (Ames 28033); lane 10, B. capitatum (PI 658745); lane 11, B. virgatum (PI 658753); lane 12, B. nuttallianum (PI 662303); lane 13, Chenopodium album (Ames 28345); lane 14, Chenopodium quinoa (Ames 13215); lane 15, B. vulgaris (TK-81MS)
To determine the chromosomal distribution of SpRE elements, metaphase chromosome spreads from male and female 03-009 plants were hybridized in situ with a SpRE-specific LTR probe and a 45S rDNA probe. Using FISH, 45S rDNA loci were mapped on the short arms of chromosomes 5 and 6, but not on the short arms of chromosome 2, on which 45S rDNA locus has previously been reported to be located using different spinach materials (e.g., cv. Mazeran and Minsterland) (Fujito et al. 2015; Ito et al. 2000). The observed discrepancy may be due to intraspecific copy number variation of ribosomal RNA genes, as well as intraspecific numerical variation of rDNA loci that have been observed in a wide range of plant species (Kataoka et al. 2012; Rogers and Bendich 1987). SpRE-LTR signals showed a dispersed distribution along all chromosomes, with a greater density in the heterochromatic regions that were strongly stained by DAPI (Fig. 4). It is worth noting that there was no or reduced signal intensity of SpRE-LTR at 45S rDNA loci and most centromeric regions, as has previously been observed in sugar beet and barley (Brandes et al. 1997; Heslop-Harrison et al. 1997). Furthermore, we found no obvious differences in the hybridization pattern of SpRE-LTR in male plants between the largest chromosome (chromosome 1) pair, which carries the sex-determining locus (Fig. 4).
Fluorescence in situ hybridization (FISH) mapping of SpRE retrotransposons on mitotic metaphase chromosomes in male and female spinach plants from dioecious line 03-009. Chromosomes were counterstained with 49,6-diamidino-2-phenylindole (DAPI). Heterochromatic regions were stained brightly with DAPI. FITC (green) signals represent SpRE-LTR, and triangles show the locations of 45S rDNA loci (Cy3, red). The largest chromosome pair (chromosome 1) corresponds to the sex chromosome pair (Iizuka and Janick 1962). Bars = 5 µm
Estimation of repeated sequence copy number included in the BAC clones associated with the male-determining locus
As shown in Fig. 5 and Figures S5–S8, NUCMER-alignment analysis (see “Materials and methods”) showed that a large part of the BAC clone sequences was repetitive: 88% of the BAC clone sequences showed > 80% sequence similarity to more than ten segments in the draft genome, and only ~ 4% of the BAC clone sequences showed homology to one or no segment of the draft genome. This analysis also showed that both RepeatMasker-annotated sequences (e.g., SpRE elements and Ty3-gypsy like elements) and unannotated sequences were highly repetitive (Fig. 5 and Figures S5–S8), suggesting that the latter might contain novel repeat element(s). Strong smeared hybridization signals were detected when Southern blots of EcoRI-digested spinach genomic DNA were probed with a high-copy region (nucleotide position 1023–2134) of BAC clone 5-19F (showing homology to ~ 3700 genome sequence segments), which was not annotated by RepeatMasker (Figure S9).
Number of spinach genome segments with similarity to BAC clone 5-19F. Black and gray lines represent the number of genome segments that show > 80 and > 90% similarity, respectively. To plot the zero values on a log scale, 1 was added to all values. Repetitive and low complexity elements identified by RepeatMasker are indicated under the histogram showing the alignment result. Arrows indicate the positions of the high and low single-copy sequences used as probes for Southern blot analysis shown in Figure S9. Nucleotide positions of the sequences are shown in the parentheses
Using a low-copy region of 5-19F (position 96108–96699; an estimated 2–6 copies per genome) as a probe, several bands were detected in the Southern blots, and one was male-specific (Figure S9). Furthermore, the probe that was prepared from a non-repetitive sequence of 5-19F (position, 48721–49226) gave a hybridization signal with a single band of the expected size (4.1 kbp) on the Southern blot of EcoRI-digested male genomic DNA, but failed to detect the hybridization signal in female and monoecious genomic DNA. This observation suggests that the BAC clone associated with the male-determining region contains a single copy sequence (or sequences) unique to males, in addition to the repetitive sequences described above.
Low gene density of the chromosomal region associated with the male-determining locus
To identify the genes located close to the male-determining locus, we predicted genes using the entire sequences of all six BAC clones. This analysis yielded a total of 45 genes (Table S6). BLASTX searching revealed that 14 had no significant homology to any sequences in the NCBI non-redundant protein (nr) database. Although one of the remaining 31 predicted genes showed significant similarity (E value = 2e−20) to protein translocase subunit SecA2 (KNA09622.1), the others had similarity to retroelement-related, hypothetical, and uncharacterized proteins. An HMMscan using the Pfam database showed that the polypeptides encoded by 36 of the 45 predicted genes did not match any domains, and only nine were annotated with functional domains. However, seven of these matched transposon- and retroposon-related domains, and the other two polypeptides were annotated with SecA DEAD-like domain and RNA polymerase Rpb3/Rpb11 dimerization domain.
To confirm the expression of the predicted genes, 27,752,167 and 82,268,893 RNA-seq reads from the whole vegetative aerial parts and inflorescences of male spinach plants were mapped to each of the BAC clone sequences. Overall, only 0.016–0.025% of the RNA-seq reads were aligned to the Y-linked BAC clone sequences. However, most of the alignments mapped to RepeatMasker-annotated regions, and the coding regions of approximately half of the predicted genes aligned to no or very few reads. Some of the RNA-seq reads mapped to the remaining half of the genes, but covered only parts of the coding regions, or did not sufficiently support the predicted gene models (i.e., exon–intron boundaries) (data not shown). Considering the highly repetitive nature of the BAC clones, a substantial proportion of the RNA-seq reads aligned to the predicted genes probably represents transcripts from loci irrelevant to the clones. Therefore, we conclude that the sequenced BAC clones carry no or only a few functional genes.
Discussion
Meiotic recombination is clearly fully or almost fully suppressed around the spinach male-determining locus, although the sex chromosomes are homomorphic. These results are consistent with our previous observations obtained from a comparative analysis between spinach sex chromosomes and sugar beet autosomes (Takahata et al. 2016), and suggest that the region in spinach is larger than those in grapevine and poplar (see “Introduction”). However, we failed to find candidates for sex-determining factors in the BAC clone sequences identified, probably because they account for only a part of the sex-linked non-recombining region. In papaya, with cytologically almost homomorphic sex chromosomes, the fully sex-linked region extends over about eight megabases (Wang et al. 2012). Further experiments are needed to determine the size in spinach. The haplotype variations in the spinach non-recombining region suggest very rare recombination, although mutation is also a possible explanation (e.g., a deletion causing loss of a marker from the Y-linked region). It differs from the sex chromosome haplotype variants in papaya and grapevine, where one Y-linked haplotype is associated with hermaphroditism, and may be an ancestral type (Picq et al. 2014; VanBuren et al. 2015). Haplotype analysis based on more variant sites would allow a better understanding of the situation in spinach.
The draft spinach genome sequence has been suggested to have a transposable element (TE) content of > 70% (Xu et al. 2017). The draft genome assembly suggests that Ty1-copia and Ty3-gypsy retroelements are predominant among spinach TEs (Xu et al. 2017), as in many plant genomes (Kumar and Bennetzen 1999; Wicker and Keller 2007), but only about half of the assembly is currently assigned to the six chromosomes, and repeat-rich regions are liable to be poorly represented. However, our estimate for the male-associated BAC clone sequences (~ 500 kbp in total length) yielded a value of only 51.8%. Given that 25,495 protein-coding genes were identified from the spinach draft genome sequence of ~ 870 Mb (total length of scaffolds), the gene density of the spinach genome can be estimated to be ~ 34 kbp/gene. As our study found no clear evidence of genes in the BAC clones, it is likely that the spinach non-recombining region may be high in repetitive elements and its gene density even lower than the genome-wide value, as in papaya (Wang et al. 2012), consistent with theoretical models predicting that suppressed recombination permits an accumulation of repetitive sequences (Bachtrog 2013; Charlesworth et al. 1994).
FISH experiments revealed the distribution of the SpRE1 (Ty1-copia) and SpRE2 (LARD) elements identified here that included all chromosomes, with higher concentrations in heterochromatin, which is similar to results for plant chromosomes in general, where Ty1-copia elements are distributed throughout most of the length of chromosomes, with regions of higher and lower relative concentration (Brandes et al. 1997; Heslop-Harrison et al. 1997). An analysis using a technique to separately detect SpRE1 and SpRE2 elements is needed to obtain an accurate distribution pattern of Ty1-copia retroelements in spinach. Specific repetitive elements were found to accumulate preferentially in the sex chromosomes of hemp, Silene latifolia, and papaya (Cermak et al. 2008; Hobza et al. 2015, 2006; Na et al. 2014; Sakamoto et al. 2000), and comparisons between the spinach Y- and X-linked regions would be valuable.
The evolutionary age of the spinach male-determining region is currently unknown. In sugar beet, a hermaphroditic member of the Chenopodiaceae s.s., the syntenic regions to previously identified spinach sex-linked regions recombine at relatively low rates, relative to other autosomal regions, and are high in repetitive elements and low in gene density (Dohm et al. 2014; Takahata et al. 2016). Given that spinach and sugar beet are, respectively, dioecious and hermaphroditic members of the Chenopodiaceae s.s., it is possible that the spinach male-determining region might have evolved in a repetitive region with an ancestrally low gene density. Nevertheless, considering the rather long divergence time (~ 38.4 MYA) of these species (Xu et al. 2017), this hypothesis needs to be verified using a more suitable outgroup, such as species from the sister genus of Spinacia, Blitum (see “Introduction”).
The estimated insertion times of SpRE copies (0.1–1.3 MYA) are much later than the divergence time (~ 15.4 MYA) between Spinacia and its sister genus, Blitum, inferred from rbcL data (Fuentes-Bazan et al. 2012a, b; Kadereit et al. 2010); this is consistent with the observation that SpRE elements are not detectable in Chenopodiaceae s.s. members other than Spinacia. The genus Spinacia includes two wild relatives (S. turkestanica and S. tetrandra) of cultivated spinach (S. oleracea), and S. turkestanica is thought to be the closest relative of S. oleracea (Hammer 2001). We recently showed that S. tetrandra has heteromorphic sex chromosomes, is phylogenetically distinct from the other Spinacia members, and differs distinctly in genome size and karyotype from other species (Fujito et al. 2015). The present data suggest that the ancestral copy of SpRE elements emerged before S. tetrandra and S. turkestanica (or S. oleracea), though its amplification may have occurred independently in each of these lineages.
References
Akamatsu T, Suzuki T, Uchimiya H (1998) Determination of male or female of spinach by using DNA marker. Japanese patent JP 1998052284
Altschul SF, Madden TL, Schäer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Bachtrog D (2013) Evolution of Sex Chromosomes. In: Losos J (ed) The Princeton guide to evolution. Princeton University Press, Princeton, pp 387–396
Brandes A, HeslopHarrison J, Kamm A, Kubis S, Doudrick R, Schmidt T (1997) Comparative analysis of the chromosomal and genomic organization of Ty1-copia-like retrotransposons in pteridophytes, gymnosperms and angiosperms. Plant Mol Biol 33:11–21
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden T (2009) BLAST plus: architecture and applications. Bmc Bioinformatics 10:421
Cermak T, Kubat Z, Hobza R, Koblizkova A, Widmer A, Macas J, Vyskot B, Kejnovsky E (2008) Survey of repetitive sequences in Silene latifolia with respect to their distribution on sex chromosomes. Chromosome Res 16:961–976
Charlesworth D (2016) Plant sex chromosomes. Annu Rev Plant Biol 67:397–420
Charlesworth B, Charlesworth D (1978) Model for evolution of dioecy and gynodioecy. Am Nat 112:975–997
Charlesworth B, Sniegowski P, Stephan W (1994) The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371:215–220
Dohm JC, Minoche AE, Holtgräwe D, Capella-Gutiérrez S, Zakrzewski F, Tafer H, Rupp O, Sörensen TR, Stracke R, Reinhardt R, Goesmann A, Kraft T, Schulz B, Stadler PF, Schmidt T, Gabaldón T, Lehrach H, Weisshaar B, Himmelbauer H (2014) The genome of the recently domesticated crop plant sugar beet (Beta vulgaris). Nature 505:546–549
Ellis JR, Janick J (1960) The chromosomes of Spinacia oleracea. Am J Bot 47:210–214
Fuentes-Bazan S, Mansion G, Borsch T (2012a) Towards a species level tree of the globally diverse genus Chenopodium (Chenopodiaceae). Mol Phylogenet Evol 62:359–374
Fuentes-Bazan S, Uotila P, Borsch T (2012b) A novel phylogeny-based generic classification for Chenopodium sensu lato, and a tribal rearrangement of Chenopodioideae (Chenopodiaceae). Willdenowia 42:5–24
Fujito S, Takahata S, Suzuki R, Hoshino Y, Ohmido N, Onodera Y (2015) Evidence for a common origin of homomorphic and heteromorphic sex chromosomes in distinct Spinacia species. G3 (Bethesda) 5:1663–1673
Geraldes A, Hefer CA, Capron A, Kolosova N, Martinez-Nuñez F, Soolanayakanahally RY, Stanton B, Guy RD, Mansfield SD, Douglas CJ, Cronk QC (2015) Recent Y chromosome divergence despite ancient origin of dioecy in poplars (Populus). Mol Ecol 24:3243–3256
Hammer K (2001) Chenopodiaceae, Spinacia oleracea L. In: Hanelt P, Institut für Pflanzengenetik und Kulturpflanzenforschung Gatersleben Germany (eds) Mansfeld’s encyclopedia of agricultural and horticultural crops (except ornamentals), 1st English edn. Springer, Berlin, pp 249–251
Heslop-Harrison J, Brandes A, Taketa S, Schmidt T, Vershinin A, Alkhimova E, Kamm A, Doudrick R, Schwarzacher T, Katsiotis A, Kubis S, Kumar A, Pearce S, Flavell A, Harrison G (1997) The chromosomal distributions of Ty1-copia group retrotransposable elements in higher plants and their implications for genome evolution. Genetica 100:197–204
Hobza R, Lengerova M, Svoboda J, Kubekova H, Kejnovsky E, Vyskot B (2006) An accumulation of tandem DNA repeats on the Y chromosome in Silene latifolia during early stages of sex chromosome evolution. Chromosoma 115:376–382
Hobza R, Kubat Z, Cegan R, Jesionek W, Vyskot B, Kejnovsky E (2015) Impact of repetitive DNA on sex chromosome evolution in plants. Chromosome Res 23:561–570
Iizuka M, Janick J (1962) Cytogenetic analysis of sex determination in Spinacia oleracea. Genetics 47:1225–1241
Ito M, Ohmido N, Akiyama Y, Fukui K, Koba T (2000) Characterization of spinach chromosomes by condensation patterns and physical mapping of 5S and 45S rDNAs by FISH. J Am Soc Hortic Sci 125:59–62
Janick J (1998) Hybrids in horticultural crops. In: Lamkey KR, Staub JE (eds) Concepts and breeding of heterosis in crop plants. Crop Science Society of America, Madison, pp 45–56
Janick J, Stevenson E (1954) A genetic study of the heterogametic nature of the staminate plant in spinach. Proc Am Soc Hortic Sci 63:444–446
Janick J, Stevenson E (1955) Genetics of the monoecious character in spinach. Genetics 40:429–437
Jukes T, Cantor C (1969) Evolution of protein molecules. In: Munro H (ed) Mammalian protein metabolism. Academic Press, New York, pp 21–132
Kadereit G, Mavrodiev E, Zacharias E, Sukhorukov A (2010) Molecular phylogeny of Atripliceae (Chenopodioideae, Chenopodiaceae): implications for systematics, biogeography, flower and fruit evolution, and the origin of C4 photosynthesis. Am J Bot 97:1664–1687
Kalendar R, Vicient CM, Peleg O, Anamthawat-Jonsson K, Bolshoy A, Schulman AH (2004) Large retrotransposon derivatives: abundant, conserved but nonautonomous retroelements of barley and related genomes. Genetics 166:1437–1450
Kataoka R, Hara M, Kato S, Isobe S, Sato S, Tabata S, Ohmido N (2012) Integration of linkage and chromosome maps of red clover (Trifolium pratense L.). Cytogenet Genome Res 137:60–69
Katayama Y, Shida S (1956) Development of intersexual flowers and their location on the stalk in spinach. Jpn J Breed 6:19–22
Kumar A, Bennetzen J (1999) Plant retrotransposons. Annu Rev Genet 33:479–532
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Kurtz S, Phillippy A, Delcher A, Smoot M, Shumway M, Antonescu C, Salzberg S (2004) Versatile and open software for comparing large genomes. Genome Biol 5:R12
Liu Z, Moore PH, Ma H, Ackerman CM, Ragiba M, Yu Q, Pearl HM, Kim MS, Charlton JW, Stiles JI, Zee FT, Paterson AH, Ming R (2004) A primitive Y chromosome in papaya marks incipient sex chromosome evolution. Nature 427:348–352
Na JK, Wang J, Ming R (2014) Accumulation of interspersed and sex-specific repeats in the non-recombining region of papaya sex chromosomes. BMC Genom 15:335
Naeger J, Golenberg E (2016) Mode and tempo of sequence and floral evolution within the Anserineae. Plant Syst Evol 302:385–398
Onodera Y, Yamamoto PM, Kubo T, Tetsuo M (1999) Heterogeneity of the atp6 presequences in normal and different sources of male-sterile cytoplasms of sugar beet. J Plant Physiol 155:656–660
Onodera Y, Yonaha I, Niikura S, Yamazaki S, Mikami T (2008) Monoecy and gynomonoecy in Spinacia oleracea L.: morphological and genetic analyses. Sci Hortic 118:266–269
Onodera Y, Yonaha I, Masumo H, Tanaka A, Niikura S, Yamazaki S, Mikami T (2011) Mapping of the genes for dioecism and monoecism in Spinacia oleracea L.: evidence that both genes are closely linked. Plant Cell Rep 30:965–971
Ossowski S, Schneeberger K, Lucas-Lledó JI, Warthmann N, Clark RM, Shaw RG, Weigel D, Lynch M (2010) The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 327:92–94
Picq S, Santoni S, Lacombe T, Latreille M, Weber A, Ardisson M, Ivorra S, Maghradze D, Arroyo-Garcia R, Chatelet P, This P, Terral JF, Bacilieri R (2014) A small XY chromosomal region explains sex determination in wild dioecious V. vinifera and the reversal to hermaphroditism in domesticated grapevines. BMC Plant Biol 14:229
Renner S, Ricklefs R (1995) Dioecy and its correlates in the flowering plants. Am J Bot 82:596–606
Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP (2011) Integrative genomics viewer. Nat Biotechnol 29:24–26
Rogers S, Bendich A (1987) Ribosomal-RNA genes in plants—variability in copy number and in the intergenic spacer. Plant Mol Biol 9:509–520
Sakamoto K, Ohmido N, Fukui K, Kamada H, Satoh S (2000) Site-specific accumulation of a LINE-like retrotransposon in a sex chromosome of the dioecious plant Cannabis sativa. Plant Mol Biol 44:723–732
Sassa H (2007) A technique to isolate DNA from woody and herbaceous plants by using a silica-based plasmid extraction column. Anal Biochem 363:166–167
Stanke M, Diekhans M, Baertsch R, Haussler D (2008) Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24:637–644
Takahata S, Yago T, Iwabuchi K, Hirakawa H, Suzuki Y, Onodera Y (2016) Comparison of spinach sex chromosomes with sugar beet autosomes reveals extensive synteny and low recombination at the male-determining locus. J Hered 107:679–685
Trapnell C, Pachter L, Salzberg S (2009) TopHat: discovering splice junctions with RNA-SEq. Bioinformatics 25:1105–1111
VanBuren R, Zeng F, Chen C, Zhang J, Wai CM, Han J, Aryal R, Gschwend AR, Wang J, Na JK, Huang L, Zhang L, Miao W, Gou J, Arro J, Guyot R, Moore RC, Wang ML, Zee F, Charlesworth D, Moore PH, Yu Q, Ming R (2015) Origin and domestication of papaya Yh chromosome. Genome Res 25:524–533
Vyskot B, Hobza R (2015) The genomics of plant sex chromosomes. Plant Sci 236:126–135
Wang J, Na JK, Yu Q, Gschwend AR, Han J, Zeng F, Aryal R, VanBuren R, Murray JE, Zhang W, Navajas-Pérez R, Feltus FA, Lemke C, Tong EJ, Chen C, Wai CM, Singh R, Wang ML, Min XJ, Alam M, Charlesworth D, Moore PH, Jiang J, Paterson AH, Ming R (2012) Sequencing papaya X and Yh chromosomes reveals molecular basis of incipient sex chromosome evolution. Proc Natl Acad Sci USA 109:13710–13715
Westergaard M (1958) The mechanism of sex determination in dioecious flowering plants. Adv Genet Incorp Mol Genet Med 9:217–281
Wicker T, Keller B (2007) Genome-wide comparative analysis of copia retrotransposons in Triticeae, rice, and Arabidopsis reveals conserved ancient evolutionary lineages and distinct dynamics of individual copia families. Genome Res 17:1072–1081
Xu Z, Wang H (2007) LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res 35:W265–W268
Xu C, Jiao C, Sun H, Cai X, Wang X, Ge C, Zheng Y, Liu W, Sun X, Xu Y, Deng J, Zhang Z, Huang S, Dai S, Mou B, Wang Q, Fei Z (2017) Draft genome of spinach and transcriptome diversity of 120 Spinacia accessions. Nat Commun 8:15275
Yamamoto K, Oda Y, Haseda A, Fujito S, Mikami T, Onodera Y (2014) Molecular evidence that the genes for dioecism and monoecism in Spinacia oleracea L. are located at different loci in a chromosomal region. Heredity 112:317–324
Acknowledgements
The authors thank Tohoku Seed Co. Ltd. (Utsunomiya, Tochigi, Japan) very much for providing the spinach breeding lines used in this study. We appreciate the technical assistance provided by Mrs. H. Yokomoto. The work presented here was supported by the Interuniversity Bio-Backup Project (IBBP) as an application code (Hokkaido 0001). We used the DNA Sequencing Facility of Research Faculty of Agriculture, Hokkaido University.
Funding
This work was supported by the Japan Society for the Promotion of Science (Grants-in-Aid for Scientific Research (KAKENHI) Grant Numbers 26292001 and 16H06279), Ministry of Education, Culture, Sports, Science and Technology (KAKENHI Grant Number 221S0002), and the Takeda Science Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Data availability
The BAC clone sequences (AP017636–AP017641) and the RNA-seq reads (DRA006107) generated during and/or analyzed during the current study are available in the DNA Data Bank of Japan (DDBJ). The primer sequences of the SP_0018 marker developed by Tohoku Seed Co., Ltd. is not publicly available because it remains a trade secret.
Additional information
Communicated by S. Hohmann.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kudoh, T., Takahashi, M., Osabe, T. et al. Molecular insights into the non-recombining nature of the spinach male-determining region. Mol Genet Genomics 293, 557–568 (2018). https://doi.org/10.1007/s00438-017-1405-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-017-1405-2