Abstract
U-box proteins are widely distributed among eukaryotic organisms and show a higher prevalence in plants than in other organisms. Plant U-box (PUB) proteins play crucial regulatory roles in various developmental and physiological processes. Previously, 64 and 77 PUB genes have been identified in Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa), respectively. In this study, 101 putative PUB genes were identified in the Chinese cabbage (Brassica rapa ssp. pekinensis line Chiifu-401-42) genome and compared with other 15 representative plants. By specific protein domains and a phylogenetic analysis, the B. rapa PUB (BrPUB) gene family was subdivided into 10 groups. Localization of BrPUB genes showed an uneven distribution on the ten chromosomes of B. rapa. The orthologous and co-orthologous PUB gene pairs were identified between B. rapa and A. thaliana. RNA-seq transcriptome data of different tissues revealed tissue-specific and differential expression profiles of the BrPUBs, and quantitative real-time PCR analysis showed inverse gene expression patterns of the BrPUB-ARMs in response to cold and heat stresses. Altogether, the identification, classification, phylogenetic analysis, chromosome distribution, conserved motifs, and expression patterns of BrPUBs were predicted and analysed. Importantly, this study of BrPUBs provides a rich resource that will aid in the determination of PUB functions in plant development.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The ubiquitin proteolytic pathway plays an important role in the degradation of target proteins. No part of the cell is beyond the reach of the ubiquitin–proteasome regulatory system (Glickman and Ciechanover 2002). Three enzymes are involved in the ubiquitination process: ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin–protein ligase (E3). E1 transfers ubiquitin to E2, and E2 binds the ubiquitin. E3 removes the ubiquitin molecule from E2 and attaches it to the target substrate, forming a covalent bond between ubiquitin and the target. Finally, target proteins can be tagged by multiple ubiquitin molecules and then degraded by the 26S proteasome (Ciechanover and Schwartz 1998). The ubiquitin proteolytic pathway is involved in various cellular processes, such as cell cycle control, antigen processing, transcription, and receptor desensitization (Kirschner 1999; Hershko et al. 2000). Of the three enzymes involved in the ubiquitination process, E3 enzymes plays a central role in determining the specificity of the ubiquitination system and have been classified into two families (the HECT family and the RING finger family; Ciechanover 1998). Recently, the U-box protein family has been added as a third family of E3 enzymes. Moreover, the U-box protein–ubiquitin fusion degradation protein 2 (UFD2)—the first U-box protein identified in yeast—is thought to have E4 enzyme ability. E4 can mediate the assembly of polyubiquitin chains on proteins after the action of E3 and are commonly considered a specialized type of E3 enzyme (Koegl et al. 1999; Cyr et al. 2002). Previous studies of the U-box genes have included a systematic analysis of their E3 ligase activity and various functions in plant growth and development. However, there are still relatively few analyses of the response of these genes to stress conditions. Brassica rapa ssp. pekinensis (Chinese cabbage) is one of the most important B. rapa crops. This subspecies, which originated in China, has undergone thousands of years of cultivation and artificial selection. Moreover, Chinese cabbage is an increasingly important leaf vegetable worldwide due to its high yield and quality, and the growth, and development of this plant are significant for its yield. Recently, the Chinese cabbage (Chiifu-401-42) genome was sequenced and assembled, and this sequence provided the opportunity for analysis of U-box genes across the entire genome (Wang et al. 2011).
U-box proteins are characterized by the presence of a U-box domain of approximately 70 amino acids (Aravind and Koonin 2000; Ohi et al. 2003). The U-box domain is structurally similar to the RING domain but lacks the zinc-chelating residues of the RING finger and instead stabilizes its structure by forming salt-bridges (Ohi et al. 2003). In previous studies, the largest U-box protein family was identified in plants (Patterson 2002), and the U-box proteins found in plants were designated as Plant U-boxes (PUBs). In comparison with the two U-box genes identified in the yeast genome and the 21 U-box genes annotated in the human genome (Koegl et al. 1999; Hatakeyama et al. 2001; Ohi et al. 2003), 64 and 77 PUB genes have been predicted in the Arabidopsis genomes (Azevedo et al. 2001; Wiborg et al. 2008) and rice genomes (Zeng et al. 2008), respectively. The PUB proteins found in plants can be classified into various groups based on the presence of common domains (Azevedo et al. 2001; Andersen et al. 2004; Mudgil et al. 2004; Zeng et al. 2008; Yee and Goring 2009). ARM domain-containing PUB proteins and Kinase domain-containing PUB proteins are the two most prevalent groups found in plants. In addition to these common PUB groups, there are specific groups found only in certain species. For example, two PUB-MIF4G proteins and one PUB-TPR-Kinase protein were found to be specific to Arabidopsis and rice, respectively (Azevedo et al. 2001; Wiborg et al. 2008; Zeng et al. 2008).
Increasing lines of evidence suggest that the PUB proteins play crucial regulatory roles in various plant physiological and developmental processes, such as self-incompatibility (Stone et al. 1999, 2003; Indriolo et al. 2012, 2014), defence (Durrant et al. 2000; Kirsch et al. 2001; González-Lamothe et al. 2006; Yang et al. 2006; Yee and Goring 2009) and abiotic stress response (Yee and Goring 2009; Seo et al. 2012). Interestingly, the majority of PUB proteins with an elucidated biological function are from the PUB-ARM group (Yee and Goring 2009). However, the role of PUB-ARM genes in Chinese cabbage under abiotic stress is unknown. To assess how PUB-ARM genes in Chinese cabbage respond to different conditions, six abiotic stresses (GA, ABA, salinity, PEG, cold and heat stresses) were utilized. Plants are frequently subjected to salinity, osmotic, cold and heat stresses in their natural environments, and GA and ABA can regulate many aspects of plant growth and development (Monte et al. 2003; Seo et al. 2012).
B. rapa and A. thaliana both belong to the family Brassicaceae (Cruciferae). They have a close evolutionary relationship, providing a useful resource for comparative genomics analysis. During evolution, the PUB family underwent a large gene expansion, and the process of gene duplication and evolution of PUB genes is complex. More work should be done to elucidate the evolution and potential functions of PUB genes. To better understand the role of PUB proteins in Chinese cabbage, we took advantage of the available genome sequences to perform a genome-wide analysis of PUB genes in B. rapa. In total, we identified 101 PUB genes in the B. rapa genome and classified them using an improved method that differs from previous classifications. We further analysed the phylogenetic relationships, conserved motifs, chromosome distribution and gene duplication of these BrPUB genes. Moreover, we utilized publicly available gene expression data to analyse the expression patterns of these genes in different tissues. In addition, 41 PUB-ARM genes were selected for an examination of their comprehensive expressional profile under 6 different abiotic stresses. Our results provide useful information for in-depth studies of the PUB family in Brassica.
Materials and methods
Sequence retrieval in database
The genome sequences of B. rapa were downloaded from the Brassica database (BRAD, http://brassicadb.org/brad/) (Wang et al. 2011). To identify all of the members of the PUB family in B. rapa, we analysed the domains of all the B. rapa proteins using a Hidden Markov Model (HMM) profile (Finn et al. 2011) of the U-box domain (PF04564) retrieved from the Pfam 27.0 database (http://Pfam.sanger.ac.uk/) (Bateman et al. 2004) with an expected value (e value) cut-off of 1.0. Then, we verified these sequences using the Pfam database (http://Pfam.sanger.ac.uk/), the SMART program (http://smart.embl-heidelberg.de/) (Letunic et al. 2012) and the NCBI database (http://www.ncbi.nlm.nih.gov/). The genome sequences of Arabidopsis thaliana were retrieved from the TAIR database (http://www.arabidopsis.org/). A. thaliana PUB (AtPUB) proteins were obtained using the same algorithms.
The rice PUB proteins were retrieved from previous analyses by Zeng et al. (Zeng et al. 2008). The Pfam 27.0 database (http://pfam.sanger.ac.uk/) was used to screen the genome assemblies of Malus domestica, Cucumis sativus, Medicago truncatula, Populus trichocarpa, Carica papaya, Theobroma cacao, Vitis vinifera, Solanum lycopersicum, Sorghum bicolor, Selaginella moellendorffii, Physcomitrella patens, Chlamydomonas reinhardtii and Cyanidioschyzon merolae. The genome data were downloaded from the Phytozome database (http://www.phytozome.net/), and the evolutionary relationships of these species were determined according to the PGDD database (http://chibba.agtec.uga.edu/duplication/) (Lee et al. 2013).
Phylogenetic analysis
Phylogenetic trees were produced individually using the full-length sequences of PUB proteins. The identified PUB proteins were aligned by the Muscle program (Edgar 2004). Phylogenetic analyses were conducted using MEGA6 (http://www.megasoftware.net/) with the neighbour-joining (NJ) method (Tamura et al. 2013). The bootstrap value was set at 1000, and the numbers of the clades mean bootstrap support values were expressed as percentages.
Physico-chemical characterization and conserved motifs analysis of PUB proteins
The physical and chemical characteristics of BrPUB proteins were predicted by Protparam (http://web.expasy.org/protparam) (Gasteiger et al. 2005). The conserved motifs of PUB proteins were detected by Multiple EM for motif elicitation (MEME, http://meme.sdsc.edu/meme/) (Bailey et al. 2009). The DNA sequences and coding domain sequences (CDS) of BrPUB genes were analysed by the tool GSDS (http://gsds.cbi.pku.edu.cn/) (Hu et al. 2014).
Physical locations of BrPUB genes and BrPUB gene duplications in the Chinese cabbage genome
To determine the physical locations of PUB genes in the Chinese cabbage genome, the starting and ending positions of all BrPUB genes on each chromosome were obtained from the BRAD database. Based on this information, a distribution map of the positions of BrPUB genes on 10 chromosomes was drawn using a Perl script. To identify gene duplications, all of the CDS sequences of the BrPUB genes were BLAST searched against each other (identity >85 %, e value <1e−10), and then, gene alignment coverage was obtained by pair-wise alignment using the date previously calculated by BLAST. Gene alignment coverage = (alignment length − mismatches)/length of the larger gene. The pairs that had a gene alignment coverage that was more than 0.75 were considered to be duplications. The Ks values were calculated by the method of Nei and Gojobori as implemented by the KaKs calculator (Zhang et al. 2006), and the divergence time was computed according to the synonymous substitution rate of 1.5 × 10−8 substitutions per site per year (Koch et al. 2000). Purple lines were used to link the duplicated genes between different chromosomes.
Identification of orthologous and paralogous genes
Homologous PUB genes between B. rapa and A. thaliana were identified by the OrthoMCL program (http://www.orthomcl.org/cgi-bin/OrthoMclWeb.cgi) (Li et al. 2003). BLASTP (e value ≤1e−10), and orthoMCL Pairs were used to find orthologs, in-paralogs and co-orthologs between the two species. Circos software (Krzywinski et al. 2009) was used to link these genes to their chromosomal positions. Cytoscape software was applied to build a network of their relationships (Shannon et al. 2003).
RNA-Seq data analysis
Previously generated and analysed Illumina RNA-seq data were used for this study (Tong et al. 2013). Four tissues (root, stem, leaf and flower) of B. rapa accession Chiifu-401-42 were analysed. The abundance of transcriptional data is expressed as fragments per kilobase of exon per million fragments mapped (FPKM). The expression cluster of BrPUB genes from the aforementioned tissues was analysed by Cluster 3.0 (http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm). The heat maps were constructed by Tree View (http://jtreeview.sourceforge.net/).
Plant materials, growth conditions and abiotic stress treatments
Chinese cabbage (Chiifu-401–42) grown in plastic pots in a 3:1 soil–vermiculite mixture in a controlled-environment growth chamber was used as starting material. The artificial growth conditions were set at 24/16 °C, with a photoperiod of 16/8 h for day/night and a relative humidity of 65–70 %. Five-leafed plants were used for different abiotic stress treatments. The plants were irrigated with 100 µM gibberellic acid (GA), 100 µM abscisic acid (ABA), 15 % (w/v) polyethylene glycol (PEG) 6000 and 250 mM NaCl. The above plants were watered thoroughly and deeply. In addition, some five-leaf stage plants were transferred from the normal growth condition to 4 and 38 °C for cold and heat treatments, respectively. All treatments were performed over a continuous time course (0, 1, 6, 12 h). Five-leaf stage plants exposed to normal artificial growth conditions were used as controls. Young leaves from control and stress-treated plants were collected as samples in three biological replicates for RNA preparation, and the samples were quickly frozen in liquid nitrogen and stored at −70 °C until use.
RNA isolation, reverse transcription and quantitative real-time PCR (qRT-PCR) analysis
Total RNA was extracted from plant leaves with an RNA kit (TaKaRa, Dalian, China). RNA was reverse transcribed into cDNA using a Prime Script RT reagent kit (TaKaRa, Dalian, China). Then, the cDNA was diluted 1:20 with ddH2O to be used as a template for qRT-PCR. Quantitative real-time PCR was performed using the SYBR Premix Ex Taq kit® (TaKaRa, Dalian, China) on an ABI Prism 7500 detection system (Applied Biosystems, Foster City, CA, USA) with the following cycling profile: 95 °C for 30 s, 95 °C for 5 s (40 cycles), 60 °C for 30 s, and melting curve analysis at 65 °C for 10 s with 61 cycles. The reaction mixture volume for qRT-PCR is 20 µL. Experimental repeat runs for three biological and three technical replicates were included in the analysis. The actin gene Bra028615 was used as an internal control to normalize the expression level of the target genes. Specific primers used for qRT-PCR were designed by Beacon Designer 7 and are shown in Table S10. An analysis of the relative gene expression using the comparative Ct value method was performed (Heid et al. 1996). The results were calculated by the 2−ΔΔCt according to previous reports (Livak and Schmittgen 2001).
Results
Identification, classification and phylogenetic analysis of BrPUB proteins in Chinese cabbage and comparative analyses
To identify PUB proteins in Chinese cabbage, we searched the entire B. rapa genome sequence for genes containing a U-box domain using an HMM profile of the U-box domain PF04564 with an e value cut-off of 1e−1. A total of 101 putative PUB genes in B. rapa were identified and were designated as BrPUB 1–101 according to the generic system (Table 1). Using the same algorithms, 62 PUB genes in the Arabidopsis database were also identified. Compared with the recent report by Wiborg et al. (2008), 2 AtPUB proteins were absent from this study because At3g49065 was removed by TAIR and At5g05230 lacked a clear U-box domain.
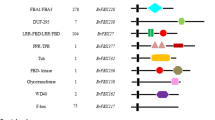
PUB proteins can be further classified on the basis of other domains present. The amino acid sequences of the U-box proteins in B. rapa and A. thaliana were used to search against the SMART program and the NCBI protein database to identify other domains. In addition to the U-box, a variety of other protein domains are present in these proteins (Table 2). Based on the presence of these other domains, PUB proteins in B. rapa and A. thaliana were divided into 10 and 8 groups, respectively. Notably, in addition to the U-box and the other domains used for classification, proteins within a group sometimes contained other domains (Zeng et al. 2008).
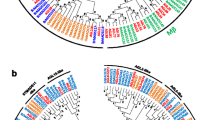
Due to the structure of PUB protein domains and the low support values for informative characters, we used phylogenetic analysis to support our group designations (Fig. S1, Fig. 1). First, a phylogenetic tree was drawn by means of MEGA 6 software (using the NJ method and bootstrapping 1000 times) to identify the phylogenetic relationships among all AtPUB proteins (Fig. S1). The topology of the phylogenetic tree allowed us to divide the AtPUBs into eight subfamilies (Groups III–X). The classifications based on protein domains and phylogenetic analyses were completely identical, indicating that the two methods were in strong agreement. In our subfamily classification of AtPUB proteins, we also referred to the classification model constructed by Azevedo et al. and Heise et al. (http://www.arabidopsis.org/browse/genefamily/pub.jsp) (Azevedo et al. 2001; Heise et al. 2002). In a previous study, AtPUBs were divided into seven subfamilies (Class I–VII), so we labelled the previously defined clades in the trees shown in Fig. S1 to facilitate a comparison. Some of the groups (e.g. Group III, VI, IX and X) were supported by previous studies, while others (e.g. Group IV, V, VII and VIII) changed. The differences were as follows: the former Class V was further divided into two parts, one defined as a new group (Group IX) and the other combined with Class IV to form a new group (Group VII); the former Class VI was further divided into two groups (Groups IV and V).
Phylogenetic tree of PUB proteins of Chinese cabbage and Arabidopsis. Multiple sequence alignment of the full-length sequences of the PUB proteins was done using MUSCLE, and the phylogenetic tree was constructed using MEGA 6 by the NJ method with 1000 bootstrap replicates. The tree was divided into ten phylogenetic subgroups, designated as Group I–X.10 clades with different colours are marked. Members of Chinese cabbage and Arabidopsis are denoted by circles and triangles, respectively (color figure online)
Next, 101 BrPUBs and 62 AtPUBs were chosen for a combined phylogenetic analysis (Fig. 1). Based on the phylogenetic relationship, 101 BrPUBs and 62 AtPUBs were clustered into 10 groups, which approach was identical with the above classification results. Every group contained BrPUBs, but no AtPUBs were found in Groups I and II. Group V, with 41 BrPUBs and 28 AtPUBs, was found to be the largest group and consisted of proteins containing the ARM domain. The ARM repeat block is an approximately 40-amino acid motif that was first identified in Drosophila (Riggleman et al. 1989). It was found to take part in protein–protein interactions and in the regulation of cell death and defence (Huber and Weis 2001; Zeng et al. 2004). Group VII, with 27 BrPUBs and 13 AtPUBs, was the second largest group, consisting of proteins containing one or more kinase domain(s) in the N-terminal region. Group VI, with 24 BrPUBs and 14 AtPUBs, was the third largest group and only contained the U-box domain. In addition to these three classes, there are other smaller subfamilies characterized by specific domains such as WD40 repeats, TPR motifs, and MIF4G motifs. Both WD40 and TPR domains have been shown to function in protein–protein interactions (Das et al. 1998; Blatch and Lässle 1999; Espejo et al. 2002), and MIF4G has been shown to function in RNA metabolism (Ponting 2000; Kraft et al. 2013).
For comparative genomic analyses, we searched for PUB protein-coding sequences in the genomes of 16 representative species (Fig. 2). Our findings showed that the number of PUB genes in Algae, Bryophyta or Lycophytes is less than that in Angiospermae which occurred during the whole-genome duplication events (Fig. 2). Surprisingly, we found that the number of PUB genes in C. reinhardtii was much more than that in C. merolae, and the number of PUB genes in higher plants was relatively stable, which indicated that the PUB proteins may have expanded before divergence of higher plants and lower plant species. The density of PUB proteins in the whole Chinese cabbage genome (0.356) was less than that in A. thaliana (0.459), while it was greater than that of other 14 species, suggesting that they may have important roles in Brassicaceae plants. Although there were more PUB proteins in M. domestica than in Chinese cabbage, its PUB protein densities (0.136) were lower than that in Chinese cabbage because of its large genome sizes.
Characterization of PUB proteins and conserved motif identification in Chinese cabbage
The physical and chemical characteristics of all BrPUB proteins were analysed (Table S1). The number of amino acid residues encoded by the BrPUB proteins ranged from 238 to 1661. The molecular weights (MWs) of BrPUB proteins varied from 26.8 to 223.9 kD. The theoretical isoelectric points (pI) of the 101 family members varied from 5.01 to 9.24. The protein characteristics of three biggest groups (Group V, VI and VII) were compared using the MWs and pI of their BrPUB members (Fig. 3). Comparative analysis of different groups indicated that most of the proteins from Group V (PUB-ARM, 73.2 %) and Group VII (PUB-Kinase, 70.4 %) were acidic, with a wide fluctuation in MWs. A majority of Group VI (U-box only, 87.5 %) proteins showed a wider fluctuation in pI values and lower MWs than the proteins in Groups V and VII.
The instability index is used to evaluate the stability of protein structure. The average instability index of the 87 BrPUB proteins was less than 40.0, indicating that most BrPUBs were unstable proteins. Only 14 proteins were determined to be stable, with instability indices varying from 29.75 to 39.99. Most BrPUBs contained numerous aliphatic amino acids, and the aliphatic index reached an average of 93.78. The hydropathicity value of + of the proteins was less than zero, suggesting that most BrPUBs are hydrophilic.
A phylogenetic tree of all BrPUBs was drawn with MEGA 6 (using the NJ method and bootstrapping 1,000 times) to identify phylogenetic relationships among all of the BrPUB proteins (Fig. 4a). The conserved motifs were searched by the MEME program (Fig. 4b), and the LOGOs of the protein motifs were also obtained by MEME (Fig. S2). The exon and intron distribution map showing the gene structure of BrPUBs was analysed using the GSDS2.0 program (Fig. 4c). In general, 101 BrPUBs were clustered into ten large groups identical with our classification results. Most members of a given group shared a similar motif composition and similar exon and intron distributions. This strong concordance with phylogenetic relationship may indicate functional similarities within a subgroup.
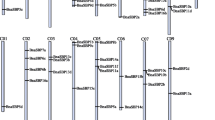
Phylogenetic relationships, conserved motif compositions and gene structures of BrPUB proteins. The NJ tree of BrPUB proteins, their motif locations and gene structure. a The multiple sequence alignment of 101 full-length BrPUB proteins was done using MUSCLE, and the phylogenetic tree was constructed using MEGA 6 by the NJ method with 1000 bootstrap replicates. The tree was divided into ten phylogenetic subgroups designated as Group I–X marked with different colour backgrounds. b The schematic representation of the conserved motifs in the BrPUB proteins detected by MEME analysis. Each motif is represented by a colour box numbered at the bottom. c The gene structures of BrPUBs were analysed by the tool GSDS. Introns and exons are represented by black lines and coloured boxes, respectively (color figure online)
Furthermore, with the exception of the U-box domain, different groups showed different motif profiles, but only some of the motifs could be annotated by the tool SMART. All of the BrPUBs contained motifs 1 and 3, which were parts of U-box domain. The majority of BrPUBs contained the highly conserved motifs 4, 8, 12, 13 and 14, which may be essential for the functions of BrPUBs. In addition, an inspection of motif distribution indicated that some motifs were only present in specific classes of the BrPUB family. For example, motifs 22 and 24 were characteristic of Group V and corresponded to the ARM domain. Motif 5 corresponding to the Kinase domain, and motif 29, was specific to Group VII. Moreover, motif 28 existed mainly in Groups I, V and VI, while motif 20 was specific to Group VI. An analysis of the primary gene structure showed that members in one group usually shared a similar genetic structure. Interestingly, with the exception of Groups V and VI, all of the other groups had at least six exons.
Chromosomal distribution, copy number variation and differential retention of PUB genes in Chinese cabbage
Genome chromosomal distribution analysis revealed that Chinese cabbage PUBs were distributed across all 10 chromosomes and all 3 subgenomes (Fig. 5; Table S3). In total, 101 BrPUB genes were mapped separately onto chromosomes A01–A10. On average, one PUB gene was present every 0.9 Mb. These BrPUB genes were distributed unevenly on the 10 chromosomes (A1–A10) of Chinese cabbage with the largest number—14 PUB genes each—detected on chromosomes 3 and 9 (Fig. 4). Every chromosome had at least six PUB genes. Furthermore, the 101 BrPUB genes were mapped onto the chromosomes in relation to the three subgenomes (LF, MF1, and MF2): 44 in LF, 33 in MF1, and 24 in MF2 (Table S3).
Distribution of the BrPUB genes on 10 Chinese cabbage chromosomes. The 101 BrPUB genes unevenly located on each conserved collinear blocks of the chromosomes. The chromosome numbers are marked above each chromosome. The PUB genes present on duplicated chromosomal segments are connected by purple lines between the two relevant chromosomes. The conserved collinear blocks on each chromosome are labeled A to X and are colour coded according to inferred ancestral chromosomes following an established convention (color figure online)
A comparison of homologous PUB genes in Arabidopsis and the three B. rapa subgenomes (LF, MF1 and MF2) revealed that most BrPUB genes in the collinear blocks were conserved throughout the divergent evolution of Arabidopsis and B. rapa (Table S3). The gene dosage hypothesis predicts that genes whose products interact with other gene products in a dose-sensitive manner, creating a web of dependency, are more likely to be retained (Thomas et al. 2006; Birchler and Veitia 2007). Interestingly, genes of Groups VI and VII were retained following triplication and fractionation in B. rapa at a higher rate than Group V genes (Fig. S3). At 46.2 %, Group VI and Group VII genes were retained at two or three copies, which is greater than the retention of Group V genes (38.7 %), while more of the Group V genes (17.9 %) were completely lost.
Orthologous groups and the duplication of PUB genes
Angiosperms have a propensity for chromosomal duplication and occasionally triplication (Lee et al. 2013). Extensive genome replication, especially gene-level triplication after divergence from Arabidopsis, could increase the flexibility of B. rapa (Wang et al. 2011). In fact, the number of BrPUB genes was notably lower than the triplicate number observed for AtPUBs, indicating that gene loss occurred during polyploid speciation. In this study, we analysed ortholog PUB gene groups between Chinese cabbage and Arabidopsis using the OrthoMCL program. A total of 166 orthologous gene pairs and 17 co-orthologous gene pairs were identified in the PUB proteins of these two species (Table S4). In addition, 117 and 24 paralogous PUB gene pairs were identified in Chinese cabbage and Arabidopsis, respectively (Table S4). The relationships among the orthologous, co-orthologous and paralogous PUB genes between B. rapa and Arabidopsis were assessed by Circos (Fig. 6). The numbers of homologous BrPUB genes on Arabidopsis chromosomes 5 and 1 were higher than those on other chromosomes among the orthologous gene pairs of B. rapa and Arabidopsis. Only four Arabidopsis U-box genes had no orthologous BrPUB gene; these genes have been duplicated in Arabidopsis after the split. Thirteen AtPUB genes were found to have only one ortholog in B. rapa; these genes were present before the divergence, but two of the three copies were lost after B. rapa genome triplication (Fig. S4a). Forty-five AtPUB genes had at least two orthologs in B. rapa, and these genes were preferentially retained after triplication (Fig. S4b, c).
Ortholog groups of PUB genes in B. rapa and Arabidopsis Genome. Ten Chinese cabbage chromosomes and five Arabidopsis chromosomes are marked with different colors with their names on the periphery. The lines in the figure represent four pairs. The lines of orthologous gene pairs are coloured in red; the lines of co-orthologous gene pairs are coloured in black; the lines of paralogous gene pairs in Chinese cabbage are coloured in blue and the lines of paralogous gene pairs in Arabidopsis are coloured in yellow. The figure was created by the software Circos (color figure online)
Gene duplication events are important for the study of the evolutionary mechanisms utilized by plant genomes; both tandem and segmental gene duplications have had a significant influence on the expansion and evolution of gene families in plant genomes (De Bodt et al. 2005). Typically, one (sometimes two) gene duplicates in tandem as the original event, which is defined as tandem duplication and often produces gene clusters or hotspots. Segmental duplication occurs when genes are duplicated as a segment, resulting from massive genome-scale events, such as polyploidy or duplications of large chromosome-level regions. Segmental duplications often result in many homologs on different chromosomes (Cannon et al. 2004; Freeling 2009). The large size of the BrPUB gene family suggests that this family may have undergone frequent duplication events during evolution. The duplicated genes were identified based on amino acid identity and gene alignment coverage (Tables S5, S6). As shown in Fig. 5, a total of 36 duplicated genes were identified in B. rapa (amino acid identity >85 %, gene alignment coverage >0.75), including 34 segmental duplication events between chromosomes as well as two tandem duplication events on the same chromosome (BrPUB48 and BrPUB49). The divergence time of duplicated BrPUB gene pairs ranged from 8.43 to 19.83 million years ago (MYA) and averaged 13.13 MYA, which indicates that the divergence of duplicated PUB genes in B. rapa occurred largely before the triplication events (5–9 MYA) (Table S6).
Expressional pattern analysis of BrPUBs in different tissues
We analysed Illumina RNA-seq transcriptomic data in four different tissues (roots, stems, leaves and flowers). The FPKM values were taken to represent the transcriptional level. More than 29,000 Chinese cabbage genes were extracted from the four tissues. The transcriptional levels of 93.0 % of the BrPUBs gene were obtained from at least one tissue, with an overall coverage of 78.2 % among the four tissues. However, 7 BrPUBs (BrPUB32, 34, 39, 48, 49, 62 and 82) were not detected (Fig. 7; Table S7). These seven BrPUBs may not be expressed or may have spatially or temporally restricted expressional patterns (Table S7). Some BrPUBs exhibited tissue-specific expression. For instance, 3 PUB-ARM genes (BrPUB41, 69, and 87) were expressed specifically in flowers, while BrPUB94 was expressed specifically in roots. BrPUB24, 31 and 42 were not expressed in leaves, and BrPUB16 had a low expression level in roots. Interestingly, the FPKM values of four BrPUBs (BrPUB11, 42, 53 and 91) in root were greater than 100, demonstrating that these genes may be important in B. rapa root development. In addition, BrPUB6, mostly expressed in the root, was homologous with AtPUB19, a negative regulator of drought response mediated by abscisic acid (Seo et al. 2012).
Expression patterns of BrPUB-ARMs under different abiotic conditions
All 41 PUB-ARM genes were selected for an examination of their comprehensive expressional profiles under cold, heat, GA, ABA, PEG, and salt treatments by qRT-PCR (Table S8). The results are shown using the program TreeView (Figs. 8, 9).
Expression analysis of B. rapa PUB-ARM genes under heat and cold abiotic stresses. a Heat map representation and hierarchical clustering of B. rapa PUB-ARM genes under heat and cold stresses. b The relative expression ratios of B. rapa PUB-ARM genes under heat stresses. c The relative expression ratios of B. rapa PUB-ARM genes under cold stresses. The relative expression levels of BrPUB in leaves under these stresses were quantified against the control transcript levels. The bar at the bottom of each heat map represents relative expression values
Expression analyses of B. rapa PUB-ARM genes under four abiotic stresses. a Heat map representation and hierarchical clustering of BrPUB genes during GA, ABA, NaCl and PEG stresses. b The relative expression ratios of B. rapa PUB-ARM genes under GA stresses. c The relative expression ratios of B. rapa PUB-ARM genes under ABA stresses. d The relative expression ratios of B. rapa PUB-ARM genes under NaCl stresses. e The relative expression ratios of B. rapa PUB-ARM genes under PEG stresses. Every stress contains three durations: 1, 4 and 12 h. The relative expression levels of BrPUB in leaves under these stresses were quantified against the control transcript levels. The bar at the bottom of each heat map represents relative expression values
The results showed that 41BrPUB-ARM genes responded to the treatments differently. Interestingly, some BrPUB-ARM genes displayed inverse gene expression patterns in response to cold and heat treatments. For instance, BrPUB41, 54, 69, and 71 were up-regulated under heat treatment, but were down-regulated under cold treatment. In contrast, BraPUB18, 19, 21, 33, 62, 67, 79, 86, and 100 were up-regulated under cold treatment, but were down-regulated under heat treatment (Fig. 8). Moreover, the expressions of BrPUB19, 21, 53, 56, 66, 83 and 90 were down-regulated or barely altered under all the treatments, except under cold treatment, while BraPUB26 did not respond to treatments other than cold and heat (Fig. S5). Under GA stress, the expressions of BrPUB72 and 95 were up-regulated at 4, 6 and 12 h (Fig. 8a). Moreover, the expression of BrPUB72 was 20 times grater than that of the control at 6 and 12 h (Fig. 9b). Under ABA stress, the expressions of two BrPUB genes (BrPUB71 and 100) were 20 times greater than that of the control at 6 and 12 h (Fig. 9c). BrPUB6, 54, 67, and 86 were up-regulated at 1 h and then decreased rapidly in later stages (Fig. 9a). Under NaCl stress, the expressions of six BrPUB genes (BrPUB2, 7, 19, 41, 65, and 87) were up-regulated at 1 h and then decreased in later stages (Fig. 9a). Under PEG stress, most BrPUBs were down-regulated or showed no change, while only BrPUB72 was obviously up-regulated at 6 and 12 h (Fig. 9a, e). With the BrPUB-ARM genes showing increased expression in response to a range of abiotic stresses (Figs. 8, 9, Fig. S5), it seems likely that these proteins play a role in mediating ubiquitin-directed protein degradation in adaptation-related responses to various environmental stresses.
Eleven duplicated BrPUB-ARM genes were investigated under different stresses in B. rapa, and a line chart was used to show the trends among them (Fig. 10; Table S9). Similar expression patterns were found in two gene pairs, including BrPUB (78: 89; 33: 62) and BrPUB (66: 90; 11: 26: 53). Overall, the most duplicated gene pairs showed similar expression, except for the BrPUB67 and 80 gene pair.
Discussion
Chinese cabbage is an important vegetable that is cultivated worldwide. The PUB family proteins, which have E3 ligase activity, play crucial regulatory roles in various plant developmental and physiological processes. With the rapid development of high-speed sequencing technologies and the implementation of many whole-genome-sequencing projects, PUB family diversity is likely to expand, and species-specific functions of PUB genes, including conserved members of this family, are likely to emerge (Yee and Goring 2009). In this study, we analyse the PUB gene family in Chinese cabbage and in 15 other species, including 13 higher plants and 2 lower plants. A total of 938 PUB genes are identified and analysed in our research. After careful comparison, we found that the PUB genes in C. reinhardtii have already expanded compared with C. merolae and its PUB protein density (0.270) was even greater than that of most of the high plants. Thus, the important of PUB proteins in various physiological processes, particularly basic function, can be shown through the evolution of the PUB gene family in lower plant. The density of PUB proteins in Brassicales was greater than that of other species used in our analyses, which indicated that the PUB proteins may have very important roles in Brassicales species.
A total of 101 genes were identified as members of the Chinese cabbage PUB family in our research. These genes were classified into ten groups based on both structural and phylogenetic analyses. It is noteworthy that one PUB protein (AtPUB5) in Arabidopsis and three PUB proteins (BrPUB1, 75 and 76) in B. rapa were classified as U-box-only proteins based on a domain analysis, but they appeared more like Group V (PUB-ARM) proteins based on the phylogenetic analysis. This could be due to deletion of the ARM domain during evolutionary development. This classification is further supported by protein and gene structural analyses; the protein and gene structures of these four proteins more closely resemble PUB-ARM proteins (Fig. 5); the result of AtPUB5 was not shown. Compared with Arabidopsis and rice, two new types of PUB proteins were found in B. rapa, including proteins in Groups I (U-box + TIR + NB-ARC + LRR3 + RPT) and II (U-box + GDA1_CD39), which arose during the triplication that occurred after the divergence from Arabidopsis. Moreover, the numbers of members in each group in B. rapa are different compared with Arabidopsis and rice. Thus, the PUB genes in these species are present in different groups with different gene numbers.
In some taxa—especially in higher plant lineages—all chromosomes double on the genomic level, which initiates the process of fractionation among the new homologs and is known as whole-genome duplication (WGD) (Freeling 2009). The Brassicaceae genome experienced two WGD events—Arabidopsis beta and Arabidopsis alpha—during the phylogeny of the species (Bowers et al. 2003). Later, the Chinese cabbage genome experienced one WGT event against Arabidopsis (Wang et al. 2011). WGD results in gene duplication and is typically followed by substantial gene loss (Lee et al. 2013). The gene balance hypothesis predicts that genes that have products that participate in macromolecular complexes or in transcriptional or signalling networks are more likely to be retained (Birchler and Veitia 2007). The PUB-ARM proteins have been shown to function in large complexes during flower development (Liu et al. 2012), salt stress (Ni et al. 2010; Salt et al. 2011) and drought stress (Drechsel et al. 2011), and the majority of PUB proteins with an elucidated biological functions belong to this group. In this study, more Group VI (U-box only) and VII (Kinase + U-box) PUB genes were found to be retained in duplicate or triplicate than Group V (U-box + ARM) genes, indicating that Group VI and VII PUB genes may have important roles during plant growth and development. We speculate that the PUB genes in Group V were preferentially lost compared with other groups, possibly due to the functional divergence of these genes between A. thaliana and B. rapa. The genome of B. rapa is almost a complete triplication of the genome of A. thaliana, and both segmental and partial tandem gene duplications play important roles in the expansion and evolution of gene families in plant genomes. In this study, each PUB gene in Arabidopsis had one–ten orthologous genes derived from Chinese cabbage, indicating that genomic duplications and partial tandem duplications were accompanied by the triplication. Further analysis identified 36 duplicated genes in the B. rapa genome, of which only two had undergone tandem duplication, while 34 BrPUB genes had undergone segmental duplication.
The largest PUB group in B. rapa is the ARM domain-containing group of proteins, and all members in this group were found to have E3 ligase activity (Mudgil et al. 2004; Samuel et al. 2006). The PUB-ARM genes may be affected by various abiotic stresses. In previous studies, the PUB-ARM genes in Capsicum annuum were shown to be affected by salinity stress (Cho et al. 2006), and other plants were affected by the application of hormones, such as GA (Monte et al. 2003) and ABA (Samuel et al. 2008). However, how PUB-ARM genes in Chinese cabbage respond to abiotic stresses had not been studied previously. In this study, all 41 B. rapa PUB-ARM genes were identified, and their expression patterns under different stress treatments were analysed. Many BrPUB-ARM genes responded to temperature stress. In particular, some had inverse expression profiles under cold and heat treatments. BrPUB41, 54, 69, and 71 are strongly up-regulated under heat treatment, but are down-regulated under cold treatment. Conversely, BrPUB18, 19, 21, 33, 62, 67, 79, 86 and 100 are strongly up-regulated under cold treatment, but are down-regulated under heat treatment. Two PUB-ARM genes, AtPUB18 and 19 in Arabidopsis, were found to be expressed transiently under ABA treatment (Yee and Goring 2009). In this study, several BrPUB-ARM genes were up-regulated under ABA treatment (Fig. 2b, c), including BrPUB6 and 54, which are most closely related to AtPUB19 and 18, respectively. Interestingly, other BrPUB genes were strongly up-regulated under ABA treatment, including BrPUB71 and 100 (Fig. 2c). BrPUB72 was expressed at relatively high levels under GA and PEG treatments (6 and 12 h). These results further support the hypothesis that PUB genes play important roles in the face of various environmental stresses.
In summary, a total of 101 PUB genes were identified in the Chinese cabbage genome. We used an improved method which was based on protein domain and phylogenetic analyses to classify the PUB proteins from Chinese cabbage and A. thaliana—a method that is of value in the study of other species. This classification is likely to assist in clarifying a molecular genetic basis for determining the functions of PUB proteins in the ubiquitin-mediated proteolysis pathway and to provide functional gene resources for transgenic research. Our results suggest that Group VI and VII PUB genes, which have received little attention, are preferentially retained compared with Group V genes based on the gene-dosage hypothesis. In addition, the expansion of the PUB genes in B. rapa was attributed to segmental duplications rather than tandem duplications. Tissue expression pattern analysis of the PUB genes, together with the expression patterns of BrPUB genes under different abiotic stresses, provided a basic resource for the examination of the molecular regulation of Chinese cabbage development and stress resistance. Our study is the first systematic and comprehensive analysis of PUB genes in Chinese cabbage, but the functional mechanisms of BrPUBs need to be further explored. Nevertheless, this study provides a preliminary exploration of BrPUBs and lays the foundation for the study of PUB genes in other species.
References
Andersen P, Kragelund BB, Olsen AN, Larsen FH, Chua NH, Poulsen FM, Skriver K (2004) Structure and biochemical function of a prototypical Arabidopsis U-box domain. J Biol Chem 279:40053–40061
Aravind L, Koonin EV (2000) The U box is a modified RING finger—a common domain in ubiquitination. Curr Biol 10:R132–R134
Azevedo C, Santos-Rosa MJ, Shirasu K (2001) The U-box protein family in plants. Trends Plant Sci 6:354–358
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res:gkp335
Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL (2004) The Pfam protein families database. Nucleic Acids Res 32:D138–D141
Birchler JA, Veitia RA (2007) The gene balance hypothesis: from classical genetics to modern genomics. Plant Cell 19:395–402
Blatch GL, Lässle M (1999) The tetratricopeptide repeat: a structural motif mediating protein–protein interactions. BioEssays 21:932–939
Bowers JE, Chapman BA, Rong J, Paterson AH (2003) Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422:433–438
Cannon SB, Mitra A, Baumgarten A, Young ND, May G (2004) The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol 4:10
Cho SK, Chung HS, Ryu MY, Park MJ, Lee MM, Bahk YY, Kim J, Pai HS, Kim WT (2006) Heterologous expression and molecular and cellular characterization of CaPUB1 encoding a hot pepper U-Box E3 ubiquitin ligase homolog. Plant Physiol 142:1664–1682
Ciechanover A (1998) The ubiquitin–proteasome pathway: on protein death and cell life. EMBO J 17:7151–7160
Ciechanover A, Schwartz AL (1998) The ubiquitin-proteasome pathway: the complexity and myriad functions of proteins death. Proc Natl Acad Sci USA 95:2727–2730
Cyr DM, Höhfeld J, Patterson C (2002) Protein quality control: U-box-containing E3 ubiquitin ligases join the fold. Trends Biochem Sci 27:368–375
Das AK, Cohen PT, Barford D (1998) The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions. EMBO J 17:1192–1199
De Bodt S, Maere S, Van de Peer Y (2005) Genome duplication and the origin of angiosperms. Trends Ecol Evol 20:591–597
Drechsel G, Bergler J, Wippel K, Sauer N, Vogelmann K, Hoth S (2011) C-terminal armadillo repeats are essential and sufficient for association of the plant U-box armadillo E3 ubiquitin ligase SAUL1 with the plasma membrane. J Exp Bot 62:775–785
Durrant WE, Rowland O, Piedras P, Hammond-Kosack KE, Jones JD (2000) cDNA-AFLP reveals a striking overlap in race-specific resistance and wound response gene expression profiles. Plant Cell 12:963–977
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Espejo A, Côté J, Bednarek A, Richard S, Bedford M (2002) A protein-domain microarray identifies novel protein-protein interactions. Biochem J 367:697–702
Finn RD, Clements J, Eddy SR (2011) HMMER web server: interactive sequence similarity searching. Nucleic Acids Res:gkr367
Freeling M (2009) Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Annu Rev Plant Biol 60:433–453
Gasteiger E, Hoogland C, Gattiker A, Wilkins MR, Appel RD, Bairoch A (2005) Protein identification and analysis tools on the ExPASy server. In: The proteomics protocols handbook. Springer, Totowa, NJ, USA, pp 571–607
Glickman MH, Ciechanover A (2002) The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 82:373–428
González-Lamothe R, Tsitsigiannis DI, Ludwig AA, Panicot M, Shirasu K, Jones JD (2006) The U-box protein CMPG1 is required for efficient activation of defense mechanisms triggered by multiple resistance genes in tobacco and tomato. Plant Cell 18:1067–1083
Hatakeyama S, Yada M, Matsumoto M, Ishida N, Nakayama KI (2001) U box proteins as a new family of ubiquitin-protein ligases. J Biol Chem 276:33111–33120
Heid CA, Stevens J, Livak KJ, Williams PM (1996) Real time quantitative PCR. Genome Res 6:986–994
Heise A, Lippok B, Kirsch C, Hahlbrock K (2002) Two immediate-early pathogen-responsive members of the AtCMPG gene family in Arabidopsis thaliana and the W-box-containing elicitor-response element of AtCMPG1. Proc Natl Acad Sci USA 99:9049–9054
Hershko A, Ciechanover A, Varshavsky A (2000) The ubiquitin system. Nat Med 6:1073–1081
Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G (2014) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics:btu817
Huber AH, Weis WI (2001) The structure of the beta-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by beta-catenin. Cell 105:391–402
Indriolo E, Tharmapalan P, Wright SI, Goring DR (2012) The ARC1 E3 ligase gene is frequently deleted in self-compatible Brassicaceae species and has a conserved role in Arabidopsis lyrata self-pollen rejection. Plant Cell 24:4607–4620
Indriolo E, Safavian D, Goring DR (2014) The ARC1 E3 ligase promotes two different self-pollen avoidance traits in Arabidopsis. Plant Cell 26:1525–1543
Kirsch C, Logemann E, Lippok B, Schmelzer E, Hahlbrock K (2001) A highly specific pathogen-responsive promoter element from the immediate-early activated CMPG1 gene in Petroselinum crispum. Plant J 26:217–227
Kirschner M (1999) Intracellular proteolysis. Trends Cell Biol 9:M42–M45
Koch MA, Haubold B, Mitchell-Olds T (2000) Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol Biol Evol 17:1483–1498
Koegl M, Hoppe T, Schlenker S, Ulrich HD, Mayer TU, Jentsch S (1999) A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 96:635–644
Kraft JJ, Treder K, Peterson MS, Miller WA (2013) Cation-dependent folding of 3′ cap-independent translation elements facilitates interaction of a 17-nucleotide conserved sequence with eIF4G. Nucleic Acids Res 41:3398–3413
Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645
Lee TH, Tang H, Wang X, Paterson AH (2013) PGDD: a database of gene and genome duplication in plants. Nucleic Acids Res 41:D1152–D1158
Letunic I, Doerks T, Bork P (2012) SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res 40:D302–D305
Li L, Stoeckert CJ, Roos DS (2003) OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13:2178–2189
Liu J, Li W, Ning Y, Shirsekar G, Cai Y, Wang X, Dai L, Wang Z, Liu W, Wang GL (2012) The U-box E3 ligase SPL11/PUB13 is a convergence point of defense and flowering signaling in plants. Plant Physiol 160:28–37
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408
Monte E, Amador V, Russo E, Martínez-García J, Prat S (2003) PHOR1: a U-Box GA signaling component with a role in proteasome degradation? J Plant Growth Regul 22:152–162
Mudgil Y, Shiu S-H, Stone SL, Salt JN, Goring DR (2004) A large complement of the predicted Arabidopsis ARM repeat proteins are members of the U-box E3 ubiquitin ligase family. Plant Physiol 134:59–66
Ni X, Tian Z, Liu J, Song B, Li J, Shi X, Xie C (2010) StPUB17, a novel potato UND/PUB/ARM repeat type gene, is associated with late blight resistance and NaCl stress. Plant Sci 178:158–169
Ohi MD, Vander Kooi CW, Rosenberg JA, Chazin WJ, Gould KL (2003) Structural insights into the U-box, a domain associated with multi-ubiquitination. Nat Struct Mol Biol 10:250–255
Patterson C (2002) A new gun in town: the U box is a ubiquitin ligase domain. Sci Signal 2002:pe4-pe4
Ponting CP (2000) Novel eIF4G domain homologues linking mRNA translation with nonsense-mediated mRNA decay. Trends Biochem Sci 25:423–426
Riggleman B, Wieschaus E, Schedl P (1989) Molecular analysis of the armadillo locus: uniformly distributed transcripts and a protein with novel internal repeats are associated with a Drosophila segment polarity gene. Genes Dev 3:96–113
Salt JN, Yoshioka K, Moeder W, Goring DR (2011) Altered germination and subcellular localization patterns for PUB44/SAUL1 in response to stress and phytohormone treatments. PLoS One 6:e21321
Samuel MA, Salt JN, Shiu SH, Goring DR (2006) Multifunctional arm repeat domains in plants. Int Rev Cytol 253:1–26
Samuel MA, Mudgil Y, Salt JN, Delmas F, Ramachandran S, Chilelli A, Goring DR (2008) Interactions between the S-domain receptor kinases and AtPUB-ARM E3 ubiquitin ligases suggest a conserved signaling pathway in Arabidopsis. Plant Physiol 147:2084–2095
Seo DH, Ryu MY, Jammes F, Hwang JH, Turek M, Kang BG, Kwak JM, Kim WT (2012) Roles of four Arabidopsis U-box E3 ubiquitin ligases in negative regulation of abscisic acid-mediated drought stress responses. Plant Physiol 160:556–568
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504
Stone SL, Arnoldo M, Goring DR (1999) A breakdown of Brassica self-incompatibility in ARC1 antisense transgenic plants. Science 286:1729–1731
Stone SL, Anderson EM, Mullen RT, Goring DR (2003) ARC1 is an E3 ubiquitin ligase and promotes the ubiquitination of proteins during the rejection of self-incompatible Brassica pollen. Plant Cell 15:885–898
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Thomas BC, Pedersen B, Freeling M (2006) Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homolog leaving clusters enriched in dose-sensitive genes. Genome Res 16:934–946
Tong C, Wang X, Yu J, Wu J, Li W, Huang J, Dong C, Hua W, Liu S (2013) Comprehensive analysis of RNA-seq data reveals the complexity of the transcriptome in Brassica rapa. BMC Genom 14:689
Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, Bai Y, Mun JH, Bancroft I, Cheng F (2011) The genome of the mesopolyploid crop species Brassica rapa. Nat Genet 43:1035–1039
Wiborg J, O’Shea C, Skriver K (2008) Biochemical function of typical and variant Arabidopsis thaliana U-box E3 ubiquitin-protein ligases. Biochem J 413:447–457
Yang CW, González-Lamothe R, Ewan RA, Rowland O, Yoshioka H, Shenton M, Ye H, O’Donnell E, Jones JD, Sadanandom A (2006) The E3 ubiquitin ligase activity of Arabidopsis PLANT U-BOX17 and its functional tobacco homolog ACRE276 are required for cell death and defense. Plant Cell 18:1084–1098
Yee D, Goring DR (2009) The diversity of plant U-box E3 ubiquitin ligases: from upstream activators to downstream target substrates. J Exp Bot 60:1109–1121
Zeng LR, Qu S, Bordeos A, Yang C, Baraoidan M, Yan H, Xie Q, Nahm BH, Leung H, Wang GL (2004) Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell 16:2795–2808
Zeng LR, Park CH, Venu R, Gough J, Wang GL (2008) Classification, expression pattern, and E3 ligase activity assay of rice U-box-containing proteins. Mol Plant 1:800–815
Zhang Z, Li J, Zhao XQ, Wang J, Wong GK, Yu J (2006) KaKs_calculator: calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinform 4:259–263
Acknowledgments
This work is supported by the Specialized Research Fund for the Doctoral Program of Higher Education (SRFDP, BO201300666), the National Basic Research Program of China (973 Program, 2012CB113903), the National Natural Science Foundation of China (31471886) and the Jiangsu Province Natural Science Foundation (BK20130673). This study was funded by the Specialized Research Fund for the Doctoral Program of Higher Education (SRFDP, BO201300666), the National Basic Research Program of China (973 Program, 2012CB113903), the National Natural Science Foundation of China (31471886) and the Jiangsu Province Natural Science Foundation (BK20130673).
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical standard
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Hohmann.
C. Wang and W. Duan contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
438_2015_1075_MOESM1_ESM.png
Supplementary material 1 (PNG 202 kb) Supplementary Fig. 1. Phylogenetic relationships and subgroup designations in U-box proteins from Arabidopsis
438_2015_1075_MOESM2_ESM.png
Supplementary material 2 (PNG 1156 kb) Supplementary Fig. 2. Sequence logos of PUB domains in B. rapa. The overall height of the stack indicates the level of sequence conservation. The height of residues within the stack indicates the relative frequency of each residue at that position
438_2015_1075_MOESM3_ESM.png
Supplementary material 3 (PNG 44 kb) Supplementary Fig. 3. Retention by number of homologous copies in the syntenic region between Arabidopsis and B. rapa
438_2015_1075_MOESM4_ESM.png
Supplementary material 4 (PNG 528 kb) Supplementary Fig. 4. The networks of PUB genes in B. rapa and Arabidopsis. This interrelation network has been constructed using B. rapa and Arabidopsis orthologous gene pairs. (a) One orthologous gene pair between B. rapa and Arabidopsis. (b) Two orthologous gene pairs between B. rapa and Arabidopsis. (c) Three or more than three orthologous gene pairs between B. rapa and Arabidopsis
438_2015_1075_MOESM5_ESM.png
Supplementary material 5 (PNG 197 kb) Supplementary Fig. 5. Expression analyses of B. rapa PUB-ARM genes under six abiotic stresses. Heat map representation and hierarchical clustering of BrPUB-ARM genes during GA, ABA, NaCl, PEG, heat and cold stresses. Every stress contains three durations: 1 h, 4 h and 12 h. The relative expression levels of BrPUB-ARMs in leaves under these stresses were quantified against the control transcript levels. The bar at the bottom of each heat map represents relative expression values
Rights and permissions
About this article
Cite this article
Wang, C., Duan, W., Riquicho, A.R. et al. Genome-wide survey and expression analysis of the PUB family in Chinese cabbage (Brassica rapa ssp. pekinesis). Mol Genet Genomics 290, 2241–2260 (2015). https://doi.org/10.1007/s00438-015-1075-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-015-1075-x