Abstract
The tobacco cultivar Nicotiana tabacum is a natural amphidiploid that is thought to be derived from ancestors of Nicotiana sylvestris and Nicotiana tomentosiformis. To compare these chloroplast genomes, DNA was prepared from isolated chloroplasts from green leaves of N. sylvestris and N. tomentosiformis, and subjected to whole-genome shotgun sequencing. The N. sylvestris chloroplast genome comprises of 155,941 bp and shows identical gene organization with that of N. tabacum, except one ORF. Detailed comparison revealed only seven different sites between N. tabacum and N. sylvestris; three in introns, two in spacer regions and two in coding regions. The chloroplast DNA of N. tomentosiformis is 155,745 bp long and possesses also identical gene organization with that of N. tabacum, except four ORFs and one pseudogene. However, 1,194 sites differ between these two species. Compared with N. tabacum, the nucleotide substitution in the inverted repeat was much lower than that in the single-copy region. The present work confirms that the chloroplast genome from N. tabacum was derived from an ancestor of N. sylvestris, and suggests that the rate of nucleotide substitution of the chloroplast genomes from N. tabacum and N. sylvestris is very low.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chloroplasts are the plant-specific organelles that contain their own genetic system. A wide variety of chloroplast genomes has been completely sequenced and all these sequences can be assembled into single circular forms (Palmer 1991; Sugiura 1992; Wakasugi et al. 2001). The sequenced genomes vary considerably in size from 35 to 204 kbp, and the number of genes differs from 63 to 252 (http://www.ncbi.nlm.nih.gov). Based on the completely sequenced chloroplast genomes, a set of common protein-coding regions was concatenated and used for phylogenetic analysis (Martin et al. 1998), and genome-wide dot-plot analysis of a wide range of plant and algal species was performed (Maul et al. 2002). On the other hand, it is also important to compare closely related species, using not only coding sequences but also spacer regions, in view of microevolution. This line of research is still scanty though an attempt has started for the genus Oenothera (Hupfer et al. 2000) and sequencing of plastid DNA has been performed for Atropa belladonna, a closely related species to tobacco (Schmitz-Linneweber et al. 2002), in the light of nuclear-plastid incompatibilities. Detailed comparison of complete chloroplast DNA sequences has been made among related cereal species, rice, maize and wheat (Tsudzuki et al. 2004). The spacer (the intergenic region) includes elements necessary for gene expression, for example, promoters, termination signals and ribosomal-binding sites. In addition, it is possible that some of the so-called “spacers” include genes for non-coding RNAs. The sprA gene encoding a 218 nt small RNA was identified in such a region (Vera and Sugiura 1994). It is extremely difficult to predict RNA-coding genes except highly conserved RNA species, e.g. tRNAs and rRNAs. One of the ways to predict genes for non-coding RNAs would be to compare intergenic regions among closely related species. This method was successfully applied for the prediction of small RNA-coding genes in Escherichia coli (Hershberg et al. 2003). Nicotiana tabacum (tobacco) is a natural amphidiploid derived from two progenitors. Ancestors of Nicotiana sylvestris and Nicotiana tomentosiformis were the likely progenitors (e.g. Smith 1974; Fulnecek et al. 2002). The chloroplast genome is believed to have originated from a species close to N. sylvestris (reviewed by Smith 1974; Gray et al. 1974; Kung et al. 1982; Olmstead and Palmer 1991). N. tabacum is a relatively recent amphidiploid species as it was estimated to arise only 6 Myr ago (Okamuro and Goldberg 1985) or even 0.2 Myr ago by the more recent calculation (Clarkson et al. 2005). Therefore, it affords the opportunity to investigate chloroplast DNA changes after an amphidiploidization event as the descendants of its progenitors are available. The analysis of restriction patterns of chloroplast DNAs was extensively performed for a variety of Nicotiana species and a phylogenetic tree of chloroplast DNA evolution was proposed (Kung et al. 1982), whereas complete sequences of chloroplast DNAs from Nicotiana species other than N. tabacum (Shinozaki et al. 1986) were not determined. Here, we report the complete nucleotide sequences of chloroplast DNAs from both N. sylvestris and N. tomentosiformis and close comparison throughout the entire sequences. Our present study provides unambiguous evidence that the ancestor of N. sylvestris is the maternal progenitor of N. tabacum.
Materials and methods
Nicotiana sylvestris and N. tomentosiformis green leaves were harvested from 4- to 5-week-old plants in a growth chamber at 28°C under 16 h light/8 h dark conditions. N. sylvestris plants were transferred in the dark for 72 h at 28°C to consume starch before chloroplast isolation. Intact chloroplasts were prepared essentially as described (Tanaka et al. 1987), using 20-50-80% discontinuous Percoll gradients instead of linear gradients. Chloroplast DNA was isolated with Plant DNAzol Reagent (Invitrogen, USA). DNA concentrations were measured using a Molecular Imager FX (Bio-Rad, USA) with PicoGreen dsDNA quantification kit (Molecular Probes, USA). Purity of DNA preparations was monitored by 0.8% agarose gel electrophoreses after EcoR I digestion, and only DNA preparations with high quality were used (Fig. S1). Shotgun sequencing was performed by Shimadzu Co., Ltd. (Kyoto, Japan). Briefly, chloroplast DNA was fluid-mechanically fragmented by a HydroShear (Gene Machine, USA) and 1.5~3.5 kbp DNA fragments were recovered after agarose gel electrophoresis. To construct random DNA libraries, the DNA fragments recovered were blunt-ended and cloned into the HincII site of pUC118. DNA templates were isolated from individual clones using MagExtractor plasmid kit (TOYOBO, Japan), and single-pass sequencing was performed for both ends of inserts (1~3 kbp) with M13 and M13 reverse primers. Sequence data were first subjected to BLAST search and then were assembled and analyzed using Sequencher v. 4.1 (Gene Codes Corporation, USA). Direct sequencing of PCR-amplified fragments was used to fill large gaps and the primer walking on shotgun clones to close small gaps. Primers were prepared by Hokkaido System Science (Sapporo, Japan). Sequencing was performed with the BigDye terminator v. 3.0 cycle sequencing ready reaction kit and a 3100 Genetic Analyzer (Applied Biosystems, USA). CEQ DTCS-Quick Start kit and a CEQ 8000 sequencer (Beckman Coulter, USA) were also used. The RNA sequencing method was adopted for two regions with short inverted repeats using the CUGA sequencing kit (Nippon Gene, Japan). Sequence data have been deposited with the DDBJ/GenBank/EMBL DNA databases under accession numbers Z00044 (N. tabacum), AB237912 (N. sylvestris) and AB240139 (N. tomentosiformis).
Results and discussion
The N. sylvestris chloroplast genome
A total of 2,257 random sequences (ca. 600 bp long each) were provided by shotgun sequencing. To access the purity of the DNA preparation, the random sequences were subjected to similarity searches with the sequence of N. tabacum chloroplast DNA (accession number Z00044), N. tabacum mitochondrial DNA (Sugiyama et al. 2005, accession numbers AP006340-2) and Escherichia coli K-12 DNA (accession number U00096) as a marker of contaminating bacteria. Five hundred and forty sequences were found to be similar to chloroplast DNA (24%), six to mitochondrial DNA (0.4%) and three to E. coli DNA (0.1%), and hence the remaining 1,708 sequences were most likely to be from nuclear DNA. Therefore, the DNA preparation was practically free from mitochondrial DNA and bacterial DNA. The 540 chloroplast-related sequences, which correspond to ca. two genome-equivalents, were assembled into a draft circle. Eight large gaps (ca. 3 kbp) remained to be determined and these regions were amplified by PCR for further sequencing. Short gaps and ambiguous positions were sequenced by the primer walking on the corresponding pUC118 clones. Four hundred and seventy six sequences were determined in our laboratory. The complete sequence was assembled by the combination of random shotgun sequencing and site-oriented sequencing. During the course of comparison of the N. tabacum chloroplast DNA sequence, we found some errors in the 1998 version (details will be reported elsewhere).
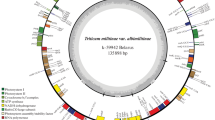
N. sylvestris chloroplast DNA is only 2 bp shorter than N. tabacum chloroplast DNA (Table 1). The gene content (except one ORF) and order are identical to those of N. tabacum (Fig. 1, color version in Fig. S2). We found only seven different sites (9 bp differences) between N. sylvestris and N. tabacum (Table 2). The evolutionary rate was calculated to be only 0.75 nucleotide changes per genome and per Myr when comparing the chloroplast genomes of the two species which share an evolutionary distance of 12 Myr ago. Although this low evolutionary rate may be specific for Nicotiana, the data indicate that evolutionary rate calculations to predict the age and history of genera and species may be subject to methodological errors as neither evolutionary rates are constant across lineages nor may be constant along the life history of a given species. The low frequency of nucleotide changes may also be explained by the recent calculation about an origin of tobacco just 0.2 MYr ago (Clarkson et al. 2005). It should be noted that these different sites are localized in the large and small single-copy regions (LSC and SSC regions, respectively) but not in the inverted repeat (IR). With respect to N. sylvestris, addition is found in two spacers and deletion is observed in one intron. Interestingly, the deletion and addition occurred at A–stretches or T–stretches. Nucleotide changes occurred in two introns. In rpoC2, an isoleucine codon ATC in N. sylvestris was substituted by ATT, resulting in no amino acid change, whereas a glutamic acid codon CAA of ndhF in N. sylvestris was changed to a proline codon CCA in N. tabacum, that is, only one amino acid change between these chloroplast genomes.
Gene map of the chloroplast genome from Nicotiana sylvestris. Genes shown on the inside of the circle are transcribed clockwise and those on the outside are transcribed counterclockwise. Genes for tRNAs are represented by one-letter code of amino acids with anticodons. Asterisks denote split genes. ORFs are shown by orf plus codon number (70 codons or more). Overlapping genes are displayed by low–high boxes for downstream or inside genes. Gene names are explained in Table 2 of Wakasugi et al. (1998). Filled triangles inside the circle indicate sites different between N. sylvestris and Nicotiana tabacum. This map is the same as that of N. tabacum except orf63 overlapped with trnS and psbI (orf90 in N. tabacum)
The N. tomentosiformis chloroplast genome
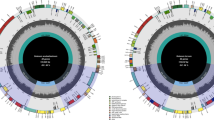
A total of 2,448 random sequences were provided, among which 2,001 (82%) showed similarity to N. tabacum chloroplast DNA. Therefore, the chloroplast DNA preparation was practically pure enough for shotgun sequencing. The reason for low purity of the chloroplast DNA sample from N. sylvestris is not known but its nuclei may be fragile. We assembled the 2,001 sequences (at a sevenfold to eightfold redundancy) into a draft circle, which left one gap (ca. 100 bp) and several dozens of small spaces and ambiguous positions. We sequenced 78 such regions to cover all these sites. The chloroplast DNA of N. tomentosiformis is 198 bp shorter than that of N. tabacum (Table 1). The IR-LSC junctions were compared among 13 Nicotiana species and found to have expanded and contracted during the evolution of this genus (Goulding et al. 1996). Gene conversions were proposed to account for these small and apparently random IR expansions. As shown in Fig. 2, the IR of N. tomentosiformis expanded 64 bp towards the LSC and hence JLA lies within rps19, resulting in the occurrence of its truncated copy (referred to as ψrps19) on the IRA as observed in several Nicotiana species and other dicot plants (Goulding et al. 1996). The IR also expanded 13 bp towards the SSC. ORF338 in IRB is a shortened version of ORF350 in N. tabacum due to a frame-shift (these ORF portions within IRB are identical in sequence to the 5′ part of ycf1). Multiple insertions and deletions occurred also within the internal IR, resulting in further expansion of 10 bp.
We found 1,194 different sites (2,272 bp differences) between N. tabacum and N. tomentosiformis; however, the gene content (except several ORFs) and order are again identical to those of N. tabacum (color gene map in Fig. S3). Distribution of differences (bp) between N. tomentosiformis and N. tabacum is summarized in Table 3. The nucleotide substitution within the IR (0.55%) is much lower than that in the single-copy regions (1.78~1.92%), which is consistent with previous reports (e.g. Wolfe et al. 1987). The rate of nucleotide substitution was the lowest in RNA-coding regions (0.12%), while it was the highest in intergenic regions (3.48%), approx. 30-fold higher than the former, indicating the high accumulation of mutations in spacers.
Genes and ORFs in the three Nicotiana chloroplast genomes
Table 4 lists genes and conserved ORFs (ycfs) with differences in sequence among the three Nicotiana species. Among the 79 protein-coding genes (including 6 ycfs) so far identified, 37 N. tomentosiformis genes are not identical in amino acid sequences predicted from DNA sequences to those of N. tabacum. The ycf10 gene is an exception and low in amino acid identity (86.2%), while the others are highly similar to each other (96.4~99.6% identities). The ycf10 product is involved in efficient inorganic carbon uptake into the chloroplast of Chlamydomonas but is not required for cell viability (Rolland et al. 1997), suggesting that the sequence divergence is tolerable and hence the low similarity is observed even in the closely related species. Among the 35 genes encoding stable RNA species, five genes show sequence differences.
Previously we reported 12 of the ORFs with 70 codons or more unique to N. tabacum (Wakasugi et al. 1998). Here, we add five additional ORFs that were not listed due to overlapping with known genes and locating within introns, because possible transcripts were detected from some of these ORFs (unpublished results). Table 5 lists these ORFs together with those of Atropa (Schmitz-Linneweber et al. 2002). Among the newly annotated ORFs, four are conserved between N. sylvestris and N. tabacum, whereas ORF90 that overlaps with psbI and trnS was shortened to ORF63 in N. sylvestris due to nucleotide deletion (see Table 2, site 1). ORFs mapped on the IR are well conserved as IRs are stable during evolution (see Table 3). In the LSC, ORF99, ORF70C and ORF71A are well conserved among the three Nicotianas and Atropa, and hence these may be protein-coding genes or regions overlapping with these ORFs may encode non-coding RNAs. Furthermore, an extensive sequence comparison of the spacer regions from the three Nicotiana and related species would be useful to predict additional non-coding RNAs. In N. tomentosiformis, a new ORF of 73 codons appeared by a nucleotide substitution, which overlaps in part with rps4 on the opposite strand (not listed in Table 5). This ORF is unique to N. tomentosiformis, suggesting again that it is unlikely to be a protein-coding gene.
Conclusion
This is the first example that the chloroplast genomes of an allopolyploid and its present-day progenitors were completely sequenced. These sequences offer basic information regarding microevolution among closely related species. Overall identities with N. tabacum are 99.99% for N. sylvestris and 98.54% for N. tomentosiformis. Based on detailed comparison between the three chloroplast DNA sequences, it is obvious that the N. tabacum chloroplast genome originated from an ancestor of the present-day N. sylvestris.
References
Clarkson JJ, Lim KY, Kovarik A, Chase MW, Knapp S, Leitch AR (2005) Long-term genome diploidization in allopolyploid Nicotiana section Repandae (Solanaceae). New Phytol 168:241–252
Fulnecek J, Lim KY, Leitch AR, Kovarik A, Matyasek R (2002) Evolution and structure of 5S rDNA loci in allotetraploid Nicotiana tabacum and its putative parental species. Heredity 88:19–25
Goulding SE, Olmstead RG, Morden CW, Wolfe KH (1996) Ebb and flow of the chloroplast inverted repeat. Mol Gen Genet 252:195–206
Gray JC, Kung SD, Wildman SG, Sheen SJ (1974) Origin of Nicotiana tabacum L. detected by polypeptide composition of Fraction 1 proteins. Nature 252:226–227
Hershberg R, Altuvia S, Margalit H (2003) A survey of small RNA-encoding genes in Escherichia coli. Nucleic Acids Res 31(7):1813–1820
Hupfer H, Swiatek M, Hornung S, Herrmann RG, Maier RM, Chiu W-L, Sears B (2000) Complete nucleotide sequence of the Oenothera elata plastid chromosome, representing plastome I of the five distinguishable Euoenothera plastomes. Mol Gen Genet 263:581–585
Kung SD, Zhu YS, Shen GF (1982) Nicotiana chloroplast genome III. Chloroplast DNA evolution. Theor Appl Genet 61:73–79
Martin W, Stoebe B, Goremykin V, Hansmann S, Hasegawa M, Kowallik KV (1998) Gene transfer to the nucleus and the evolution of chloroplasts. Nature 393:162–165
Maul JE, Lilly JW, Cui L, dePamphilis CW, Miller W, Harris EH, Stern DB (2002) The Chlamydomonas reinhardtii plastid chromosome: islands of genes in a sea of repeats. Plant Cell 14:2659–2679
Okamuro JK, Goldberg RB (1985) Tobacco single-copy DNA is highly homologous to sequences present in the genomes of its diploid progenitors. Mol Gen Genet 198:290–298
Olmstead R, Palmer JD (1991) Chloroplast DNA and systematics in the Solanaceae. In: Hawkes JG, Lester RN, Nee M, Estrada N (eds) Solanaceae III taxonomy, chemistry and evolution. Kew, Royal Botanic Gardens, Linnean Society of London, pp 301–320
Palmer JD (1991) Plastid chromosomes: structure and evolution, In: Bogorad L, Vasil IK (eds) The molecular biology of plastids, Academic, San Diego, pp 5–53
Rolland N, Dorne A-J, Amoroso G, Sültemeyer DF, Joyard J, Rochaix J-D (1997) Disruption of the plastid ycf10 open reading frame affects uptake of inorganic carbon in the chloroplast of Chlamydomonas. EMBO J 16:6713–6726
Sasaki T, Yukawa Y, Miyamoto T, Obokata J, Sugiura M (2003) Identification of RNA editing sites in chloroplast transcripts from the maternal and paternal progenitors of tobacco (Nicotiana tabacum): comparative analysis shows the involvement of distinct trans-factors for ndhB editing. Mol Biol Evol 20:1028–1035
Schmitz-Linneweber C, Tillich M, Herrmann RG, Maier RM (2001) Heterologous, splicing-dependent RNA editing in chloroplasts: allotetraploidy provides trans-factors. EMBO J 20:4874–4883
Schmitz-Linneweber C, Regel R, Du TG, Hupfer H, Herrmann RG, Maier RM (2002) The plastid chromosome of Atropa belladonna and its comparison with that of Nicotiana tabacum: the role of RNA editing in generating divergence in the process of plant speciation. Mol Biol Evol 19:1602–1612
Shinozaki K, Ohme M, Tanaka M, et al (1986) The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J 5:2043–2049
Smith HH (1974) Nicotiana. In: King RC (eds) Handbook of Genetics 2. Plenum Press, New York, pp 281–314
Sugiura M (1992) The chloroplast genome. Plant Mol Biol 18:149–168
Sugiyama Y, Watase Y, Nagase M, Makita N, Yagura S, Hirai A, Sugiura M (2005) The complete nucleotide sequence and multipartite organization of the tobacco mitochondrial genome: comparative analysis of mitochondrial genomes in higher plants. Mol Genet Genomics 272:603–615
Tanaka M, Obokata J, Chunwongse J, Shinozaki K, Sugiura M (1987) Rapid splicing and stepwise processing of a transcript from the psbB operon in tobacco chloroplasts. Mol Gen Genet 209:427–431
Tsudzuki J, Tsudzuki T, Wakasugi T, Kinoshita K, Kondo T, Ito Y, Sugiura M (2004) Comparative analysis of the whole chloroplast genomes from rice, maize and wheat. Endocytobiosis Cell Res 15:339–344
Vera A, Sugiura M (1994) A novel RNA gene in the tobacco plastid genome: its possible role in the maturation of 16S rRNA. EMBO J 13:2211–2217
Wakasugi T, Sugita M, Tsudzuki T, Sugiura M (1998) Updated gene map of tobacco chloroplast DNA. Plant Mol Biol Rep 16:231–241
Wakasugi T, Tsudzuki T, Sugiura M (2001) The genomics of land plant chloroplasts: gene content and alteration of genomic information by RNA editing. Photosynth Res 70:107–118
Wolfe KH, Li W-H, Sharp PM (1987) Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci USA 84:9054–9058
Acknowledgments
We thank Dr. Yasushi Yukawa and Dr. Tatsuya Wakasugi for suggestions and discussion. This work was performed as one of the technology development projects of the “Green Biotechnology Program” supported by NEDO (New Energy and Industrial Technology Development Organization).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by R. Herrmann
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Yukawa, M., Tsudzuki, T. & Sugiura, M. The chloroplast genome of Nicotiana sylvestris and Nicotiana tomentosiformis: complete sequencing confirms that the Nicotiana sylvestris progenitor is the maternal genome donor of Nicotiana tabacum . Mol Genet Genomics 275, 367–373 (2006). https://doi.org/10.1007/s00438-005-0092-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-005-0092-6