Abstract
Planarians represent an insufficiently explored group of aquatic invertebrates that might serve as hosts of histophagous ciliates belonging to the hymenostome genus Tetrahymena. During our extensive research on freshwater planarians, parasitic tetrahymenas were detected in two of the eight planarian species investigated, namely, in Dugesia gonocephala and Girardia tigrina. Using the 16S and 18S rRNA genes as well as the barcoding cytochrome oxidase subunit I, one ciliate species was identified as T. scolopax and three species were recognized as new forms: T. acanthophora, T. dugesiae, and T. nigricans. Thus, 25% of the examined planarian taxa are positive for Tetrahymena species and three of them represent new taxa, indicating a large undescribed ciliate diversity in freshwater planarians. According to phylogenetic analyses, histophagous tetrahymenas show a low phylogenetic host specificity. Although T. acanthophora, T. dugesiae, and T. scolopax clustered together within the “borealis” clade, the former species has been detected exclusively in G. tigrina, while the two latter species only in D. gonocephala. Tetrahymena nigricans, which has been isolated only from G. tigrina, was classified within the “paravorax” clade along with T. glochidiophila which feeds on glochidia. The present phylogenetic reconstruction of ancestral life strategies suggested that the last common ancestor of the family Tetrahymenidae was free-living, unlike the progenitor of the subclass Hymenostomatia which was very likely parasitic. Consequently, there were at least seven independent shifts back to parasitism/histophagy within Tetrahymena: one each in the “paravorax” and “australis” clades and at least five transfers back to parasitism in the “borealis” clade.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The hymenostome genus Tetrahymena Furgason, 1940 belongs together with Paramecium Müller, 1773 and Oxytricha Bory, 1824 among the best studied genera of the phylum Ciliophora Doflein, 1901 (Lynn 2008). The most explored Tetrahymena species is undoubtedly T. thermophila Nanney & McCoy, 1976, formerly classified in the pyriformis complex as T. pyriformis syngen 1 (Nanney and McCoy 1976). Its successful cultivation on sterile medium by Lwoff (1923) initiated a new chapter of research in the field of biochemistry and physiology of unicellular organisms. The easy cultivability of T. thermophila also contributed to several important discoveries, ranging from the existence of cell-cycle regulation mechanisms (Frankel 1999), through the activity of enzymes indispensable for DNA-matrix reproduction (Brownell et al. 1996; Greider and Blackburn 1985) to even a more significant finding from the medical perspective, the presence of tandem repeats of hexanucleotide units protecting chromosome ends (Blackburn and Gall 1978; Yao and Yao 1981; Yao et al. 1981).
The common morphological feature of this genus, from which its name also originated, is the structure of the oral ciliature. Tetrahymena possesses three oral polykinetids on the left and a single paroral membrane on the right of the oral cavity (Furgason 1940). Although this structure remains practically the same among different species, the size of the oral apparatus varies considerably, depending on the feeding strategy and life style.
Corliss (1969, 1970) provided first insights into the intrageneric taxonomy of Tetrahymena based on the following categories of characters: ciliature and cortex, life cycle, ecology, and physiologic as well as biochemical properties. As a result, Corliss (1970) proposed dividing tetrahymenas into three groups: (1) the pyriformis complex, containing bacteria-feeding microphagous ciliates with tendencies to become parasites; (2) the rostrata complex, gathering facultative histophagous and/or parasitic species often dividing inside of a cyst; and (3) the patula complex of non-parasitic, weakly histophagous but occasionally cannibalistic macrostome forms. However, these three complexes very likely do not reflect the evolutionary history of the genus, since they do not form distinct clusters in phylogenetic trees (Chantangsi et al. 2007; Doerder 2019; Lynn and Doerder 2012; Lynn et al. 2018; Quintela-Alonso et al. 2013; Pitsch et al. 2017; Rataj and Vďačný 2019; Strüder-Kypke et al. 2001; Zahid et al. 2014). On the other hand, the analysis of the D2 domain of the 23S rRNA gene (now designated as 28S rRNA gene) by Nanney et al. (1998) has suggested a parallel evolutionary origin of macrostomy and histophagy within the genus Tetrahymena, thus supporting Corliss’ (1972) tentative evolutionary scenario. Likewise, the 18S rRNA gene has indicated that histophagy evolved among tetrahymenas several times convergently (Rataj and Vďačný 2019; Strüder-Kypke et al. 2001). This ribosomal gene has also suggested division of the genus Tetrahymena into two fundamental lineages—one leading to the borealis group and the second gathering species of the australis group (Jerome et al. 1996; Strüder-Kypke et al. 2001). In spite of a small portion of today known species involved in the early analyses, this image remained almost unchanged when further species have been added in 18S rRNA gene phylogenies. However, a slightly different branching pattern has been offered by the mitochondrial gene coding for cytochrome c oxidase subunit 1 (COI), placing several taxa outside the australis/borealis dichotomy (Kher et al. 2011; Lynn et al. 2018; Quintela-Alonso et al. 2013).
Until 2018, more than 40 species have been recognized in Tetrahymena (Lynn et al. 2018). An actual breakthrough is represented by the study of Doerder (2019), which introduced 37 new names for molecularly unique species, considering genetic distances in the 18S rDNA and COI sequences. Still, it is hard to determine the overall species richness within the genus Tetrahymena due to the uncovered hidden diversity of cryptic species or even lost isolates (Doerder 2019), not to mention the need for examination of all sorts of habitats or potential host organisms. Indeed, invertebrates represent an insufficiently explored habitat of parasitic and potentially new tetrahymenas, as indicated in our previous study (Rataj and Vďačný 2019). Although Tetrahymena infections of invertebrates have been only seldomly studied, it has been recognized that multiple Tetrahymena species can invade a broad variety of invertebrates. For instance, black flies can be parasitized by T. dimorpha Batson, 1983 (Batson 1983) or T. rotunda Lynn et al., 1981 (Lynn et al. 1981), alderflies by T. sialidos Batson, 1985 (Batson 1985), mosquitoes by T. empidokyrea Jerome et al., 1996 (Jerome et al. 1996), midges by T. chironomi Corliss, 1960 (Corliss 1960), slugs by T. limacis (Warren 1932) Kozloff, 1946 (Kozloff 1946) or T. rostrata (Kahl 1926) Corliss, 1952 (Brooks 1968), glochidia of freshwater mussels by T. glochidiophila Lynn et al., 2018 (Lynn et al. 2018), and freshwater planarians by T. pyriformis (Ehrenberg 1830) Lwoff, 1947 and T. corlissi Thompson, 1955 (Wright 1981) as well as by some other so far undescribed species (Rataj and Vďačný 2019).
In the present paper, we focused on the prevalence of tetrahymenas in eight freshwater planarian species. Associations of ciliates with planarians are an interesting emerging but still very insufficiently explored topic (Rataj and Vďačný 2018, 2019). Our extensive sampling yielded three new morphologically distinguishable histophagous Tetrahymena species, inhabiting the gut and mesenchyme tissues of freshwater planarians. We were also successful in amplifying three molecular markers (the nuclear 18S rDNA and the mitochondrial 16S rDNA and COI) of these parasitic tetrahymenas, which corroborated their distinctness and enabled us to study the evolution of histophagy in hymenostomes from the phylogenetic perspective. And, finally, the three Tetrahymena species parasitizing planarians are formally described here as new taxa.
Material and methods
Material collection and processing
Collection, identification, storage, and consecutive dissection of host organisms were carried out as described in our previous publications (Rataj and Vďačný 2018, 2019). During the course of this study, samples were obtained together from 27 collection sites between October 2016 and May 2019. These sites were represented by the coastal zones of lakes, slowly running streams as well as by the shallow sections of bigger rivers in Slovakia. Their characterization with GPS coordinates and corresponding planarian species is summarized in Table 1.
Altogether, 784 planarians were inspected for the presence of tetrahymenas. Representative specimens of each planarian species are shown in Supplementary Fig. S1. Living Tetrahymena individuals were isolated from the mesenchyme and dissected gastrointestinal tissues of host organisms using an adjusted Pasteur micropipette (Foissner 2014). They were consequently washed through several drops of physiological saline solution (0.6% NaCl). Some sufficiently washed ciliates with no visible surrounding tissue particles were then put into the lysis buffer for later DNA extraction. Individual samples contained one, usually two, and rarely up to five Tetrahymena cells. Sequencing chromatograms of samples containing from one to five cells gave no evidence of multiple peaks, documenting that a single Tetrahymena species was analyzed each time. Other successfully isolated cells were subjected to a thorough morphological examination under an optical microscope Zeiss Axio Imager 2 equipped with differential interference contrast optics. Cell shape, ciliary pattern, locomotory behavior, contractile vacuole cycle together with other observable characteristics were captured on microphotographs and videos by a Canon EOS 70D camera with a format size of 1920 × 1080 pixels and a rate of 25 frames per second. All size measurements were conducted in ImageJ ver. 1.49. Illustrations consist of free-hand drawings and sketches made in Inkscape ver. 0.92.4, all based on microphotographs.
ZooBank registration number of the present work (Recommendation 8A of the International Commission on Zoological Nomenclature 2012) is urn:lsid:zoobank.org:pub:900AC676-1F58-49E6-95D2-7BC98E64C4FE.
Molecular methods
Thoroughly washed cells of Tetrahymena spp. intended for DNA extraction were lysed in 180 μl of cell lysis buffer (Promega, Fitchburg, Wisconsin, USA) and stored in a refrigerator at 8 °C until processed. DNA extraction followed the protocol of the Relia Prep™ Blood gDNA Miniprep System (Promega). Amplification of the nuclear 18S rRNA gene was performed using universal forward and reverse eukaryotic primers EukA (5′-AAC CTG GTT GAT CCT GCC AGT-3′) and EukB (5′-TGA TCC TTC TGC AGG TTC AC-3′) (Medlin et al. 1988). The respective thermo cycler program consisted of three stages: (i) initial hot start incubation of 15 min at 95 °C; (ii) 30 identical amplification cycles, each composed of 45 s denaturation at 95 °C, 1 min primer annealing at 55 °C, and 2.5 min extension at 72 °C; and (iii) final extension of 10 min at 72 °C (Vďačný et al. 2011). In the case of the mitochondrial 16S rRNA gene, amplification was carried out with 16S-mtSSU-F (5′-TGT GCC AGC AGC CGC GGT AA-3′) and 16S-mtSSU-R (5′-CCC MTA CCR GTA CCT TGT GT-3′) (van Hoek et al. 2000) primers. The respective PCR reaction was composed of four stages: (i) initial hot start incubation of 3 min at 94 °C; (ii) 5 amplification cycles (denaturing at 94 °C for 30 s, annealing at 50 °C for 1 min, and extension at 68 °C for 75 s); (iii) 30 amplification cycles (denaturing at 94 °C for 30 s, annealing at 60 °C for 1 min, and extension at 68 °C for 75 s); and (iv) final extension at 68 °C for 10 min (Lynn and Strüder-Kypke 2006). Finally, the mitochondrial COI gene was amplified using COI-FW-mod (5′-ATG TGA GTT GAT TTT ATA GA-3′) (Chantangsi et al. 2007) and COI-689-RW (5′-CTC TTC TAT GTC TTA AAC CAG GCA-3′) (Doerder 2014) primers and a slight modification of the previous PCR program. Thus, the primer annealing temperature of the stages (ii) and (iii) was lowered from 50 to 45 °C and from 60 to 55 °C, respectively, and the number of cycles in the stage (iii) was increased from 30 to 35. Quality check of the amplified DNA was constantly performed by agarose gel electrophoresis. PCR products of high quality were purified using the NucleoSpin Gel and PCR clean-up Kit (Macherey-Nagel) and consequently sequenced on an ABI 3730 automatic sequencer (Macrogen) with primers used for DNA amplification. The newly obtained sequences were inspected for quality in Chromas ver. 2.33 (Technelysium Pty Ltd) and the noise-free sequence fragments with strong signal were trimmed and assembled into contigs using BioEdit ver. 7.2.5 (Hall 1999).
Phylogenetic methods
Datasets and alignment procedures
Four sets of alignments were assembled to assess the phylogenetic position of three new Tetrahymena species. The first dataset contained concatenated sequences of the nuclear 18S rRNA gene and the mitochondrial COI gene, coming from 98 Tetrahymena spp. and some related hymenostomes (Supplementary Table S1). All Tetrahymena strains having sequences of both molecular markers have been selected for this dataset (Chantangsi et al. 2007; Doerder 2014, 2019; MacColl et al. 2015). The second dataset consisted of two concatenated mitochondrial markers, the 16S rRNA gene and COI, belonging to 22 Tetrahymena taxa (Supplementary Table S2). This dataset was distinctly smaller, since only comparatively few strains have available sequences from both markers. Because there were much more Tetrahymena GenBank entries with only one molecular marker available, two further alignments were constructed to cover the known diversity of the genus Tetrahymena. One contained 106 COI sequences and the second one consisted of 84 sequences coding for the 16S rRNA molecule. Sampling in these two alignments followed mainly Doerder (2019).
All sequences, except for those obtained in the present study, were downloaded from GenBank (https://www.ncbi.nlm.nih.gov/genbank/). The 16S rRNA gene and COI sequences used in the second alignment were extracted from the whole mitochondrial genomes (Brunk et al. 2003; Burger et al. 2000; Edqvist et al. 2000; Moradian et al. 2007). Ribosomal RNA genes were aligned with the ClustalW algorithm as implemented in BioEdit. The protein-coding COI gene sequences were aligned based on the predicted amino acid sequences with MEGA-X (Kumar et al. 2018), using the protozoan mitochondrial genetic code and the Muscle codon algorithm. No masking strategy was employed, since all columns were aligned unambiguously.
Tree-building methods
The evolutionary history of tetrahymenas was reconstructed in the maximum likelihood (ML) and Bayesian frameworks. Both analyses were run on the CIPRES portal ver. 3.1 (http:// www.phylo.org/) (Miller et al. 2010). ML trees were calculated with IQ-TREE on XSEDE ver. 1.6.10 (Nguyen et al. 2015). The model tester implemented in the IQ-TREE package was used to select the best model for each gene, according to the Bayesian Information Criterion. To assess the reliability of the branching pattern of the ML trees, 1000 non-parametric bootstrap replicates were conducted. Bayesian inferences (BI) were carried out in the program MrBayes on XSEDE ver. 3.2.7 (Ronquist et al. 2012) under the best evolutionary models selected for each molecular marker separately in jModelTest ver. 2.1.10 on the basis of the Akaike Information Criterion (Darriba et al. 2012). The settings of the MCMC analysis were as follows: (i) four simultaneously running chains, one cold and three heated; (ii) five million generations and a sampling frequency of one hundred; and (iii) the burn-in fraction of 25%.
Reconstruction of ancestral character states
Reconstruction of ancestral life strategies of the subclass Hymenostomatia Delage and Herouard, 1896 was conducted in SIMMAP ver. 1.5.2 (Bollback 2006). Two character states were considered: a parasitic (histophagic) and a free-living life style. Data were mostly obtained from Lynn et al. (2018), Doerder (2019), and references cited therein. Priors for ancestral state analysis were estimated from the 50% majority-rule Bayesian consensus tree inferred from the first dataset, using an MCMC analysis implemented in SIMMAP. The R script provided with the SIMMAP package was used to find the best fitting distribution for samples from the posterior distributions of the MCMC analyses. Priors of the ancestral states reconstruction analysis were (i) α = 1.00 and k = 31 for the beta distribution of state frequencies; (ii) equal bias prior 1/k for the beta distribution; and (iii) α = 2.399, β = 0.036, and k = 60 for the gamma distribution of the overall rate of character change. A set of 1000 trees was generated from the posterior distribution of the Bayesian analysis of the first concatenated alignment, using a custom Python script containing a random sampling function. The randomly selected post burn-in trees served to incorporate phylogenetic uncertainty. Ten samples were analyzed per each tree with 20 priors drawn from the prior distributions. Branch lengths were re-scaled that the overall length of trees is one. Results were plotted as pie charts and mapped onto the 50% majority-rule Bayesian consensus tree using the R script “PlotSimMap.R” (https://github.com/nylander/PlotSimMap).
Diversification analyses
The effect of life styles on diversification of tetrahymenas over their evolutionary history was assessed using the binary-state speciation and extinction (BiSSE) method (Maddison et al. 2007), as implemented in the program BayesRate ver. 1.3.41 (Silvestro et al. 2011). The BiSSE model has six free parameters: speciation (λ0, λ1) and extinction (μ0, μ1) rate for each character state as well as a transition rate of change between the two states (q01, q10). These six parameters were variously constrained to specify nested diversification models. Bayesian analyses were performed for each model over a sample of 1000 randomly selected trees from the posterior distribution of the Bayesian analysis of the first concatenated dataset. Trees were mid-point rooted and ultrametrized using a custom R script and the “force.ultrametric” function with the “extend” method, as implemented in the R-package phylotools (Revell 2012). MCMC simulations included 200 iterations of slice sampling per tree. The first 20 samples per tree were discarded as burn-in. Diffuse priors were set for the diversification-rate parameters, following Johnson et al. (2011). Finally, log marginal likelihoods were estimated for each model with the thermodynamic integration option. Nested models (M0) were compared with the full model (M1), using the Bayes factor test which is defined as the ratio between their marginal likelihoods. Thus, the log Bayes factor (BF) between pairs of models M0 and M1 can be calculated as BF = 2(M1 − M0). According to Kass and Raftery (1995), BF = 2–6 is interpreted as positive evidence against M0, BF = 6–10 as strong evidence, and BF > 10 as very strong evidence.
Since Bayes factor tests provided positive to very strong evidence against all nested models, posterior distributions of all diversification parameters (speciation and extinction rate for each character state and transition rates of change between the two states) were calculated under the full model. The MCMC simulations with exponential prior were conducted on the ultrametrized 50% majority-rule Bayesian consensus tree inferred from the first dataset, using the R-package diversitree (FitzJohn 2012). The step argument w for the MCMC sampler was determined by running a 100-step long chain. The range of observed samples was then used as a measure of the “step size” in a 10,000-step long MCMC chain. The 95% credibility intervals for marginal distributions of diversification parameters were calculated and plotted using the R “profiles.plot” function.
Results
Distribution and prevalence of Tetrahymena in planarians
Altogether eight freshwater planarian species, belonging to two ecological groups, were collected in the territory of Slovakia, Central Europe, and investigated for the presence of ciliates during the years 2016 and 2019. The first ecological group contained planarians sampled in a variety of running waters: 165 specimens of Dugesia gonocephala (Dugès, 1830) Girard, 1850; 169 individuals of Polycelis felina (Dalyell, 1814) Ehrenberg, 1831; and four exemplars of Crenobia alpina (Dana, 1766) Kenk, 1930. The second group included planarians from stagnant waters: 424 specimens of Girardia tigrina (Girard, 1850) Ball, 1974; eight individuals of Dendrocoelum lacteum (Müller, 1774) Ørsted, 1844; seven exemplars of Schmidtea lugubris (Schmidt, 1861) Ball, 1974; six specimens of Schmidtea polychroa (Schmidt, 1861) Ball, 1974; and a single individual of Polycelis nigra (Müller, 1774) Ehrenberg, 1831.
Three new histophagous species of the genus Tetrahymena were discovered after dissection of 784 planarian exemplars. However, only individuals of D. gonocephala and G. tigrina, the most abundant planarian species in each type of the aquatic environments, were found to be parasitized (Table 1). Interestingly, D. gonocephala was parasitized only by a single Tetrahymena species, described here as T. dugesiae sp. n. The cells of T. dugesiae were found only in 17 out of the 165 examined D. gonocephala individuals, which corresponds to a prevalence of approximately 10.3%. As concerns the two other Tetrahymena species, T. acanthophora sp. n., and T. nigricans sp. n., they were detected solely inside of G. tigrina. Simultaneous infections by both Tetrahymena species were noted only at a single collection site (Table 1). Tetrahymena infections in G. tigrina were rarer than in D. gonocephala. Specifically, T. acanthophora was detected in 14 out of the 424 examined G. tigrina individuals, which corresponds to a prevalence of ca. 3.3%. The prevalence of T. nigricans was even lower than 1%, since this ciliate appeared only in four planarians at two collection sites and always only a single cell per planarian was recorded. This makes T. nigricans the rarest among the three new tetrahymenas described in this paper. In addition to the three new Tetrahymena species, a single already described species, T. scolopax Doerder, 2019, was detected in D. gonocephala at one sampling site (Table 1). This species was identified based on the COI sequence (100% identity). The Slovak isolate of T. scolopax co-occurred with T. dugesiae.
No signs of pathological changes were detected in G. tigrina specimens infected by T. acanthophora and T. nigricans. The only indication of parasitism included the presence of planarian tissues in food vacuoles of both ciliate species. On the other hand, some D. gonocephala exemplars infected by T. dugesiae had open wounds. Possibly this Tetrahymena species enters planarians through wounds and then begins to feed on their mesenchyme. The numbers of T. acanthophora in G. tigrina ranged from one to about 50 cells (on average 11 cells, n = 14), while only a single cell of T. nigricans per host was detected each time (n = 4). The abundance of T. dugesiae per planaria varied from one to 15 cells (on average four cells, n = 17). All our attempts to cultivate the two former Tetrahymena species on the mesenchyme tissues failed, while one temporary culture of T. dugesiae was established in a Petri dish, using the D. gonocephala tissues.
Description of Tetrahymena acanthophora sp. n.
ZooBank registration number: urn:lsid:zoobank.org:act:30865328-6B2A-44FD-AD5F-918E31AB8B76.
Diagnosis: Size about 110 × 55 μm. Body elliptical, ovate or clavate with anterior end apiculate and posterior end broadly rounded. Macronucleus oval with a single large nucleolus. Contractile vacuole dorsal and subterminal. About 28 to 30 sigmoidal ciliary rows on each body side, no caudal cilia. Oral apparatus about 10 μm long.
Type locality: Veľkobielske jazero lake, recreation area in the village of Veľký Biel, Podunajská rovina plain (48° 12′ 26.4″ N, 17° 21′ 28.3″ E).
Type host: Girardia tigrina (Girard, 1850) Ball, 1974.
Type material: A DNA sample of holotype specimen has been deposited in Natural History Museum, Vajanského nábrežie 2, 810 06 Bratislava, Slovakia (ID Collection Code 1427132).
Gene sequences: The nuclear 18S rRNA gene sequence of holotype specimen has been deposited in GenBank (accession no. MN994469). The sequence is 1712 nucleotides long and has a GC content of 43.11%. The mitochondrial 16S rRNA gene sequence of holotype specimen has been deposited in GenBank (accession no. MN994474). The sequence is 996 nucleotides long and has a GC content of 29.92%. The COI gene sequence of holotype specimen has been deposited in GenBank (accession no. MN991314). The sequence is 946 nucleotides long and has a GC content of 25.58%.
Etymology: The specific epithet acanthophora is a composite of the stem of the Greek noun ákantha (ἄκανθα, thorn), the thematic vowel ·o-, and the Latinized Greek adjective phor·us, –a, –um [m, f, n] (bearing), referring to the apiculate anterior pole of the body.
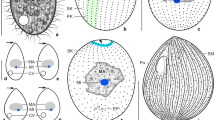
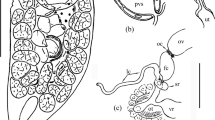
Description: The body size was approximately 83–133 × 32–73 μm (n = 10) in free-swimming individuals. The shape was elliptical, broadly to narrowly ovate or clavate but might have become fusiform after manipulation with a micropipette. The anterior body end was differentiated into a conspicuous ca. 4 μm long thorn-like structure, whereas the posterior cell pole was narrowly to broadly rounded (Figs. 1a–e, g–o; 2a–f, i, j; and 3a, c). There was a single oval macronucleus situated roughly in the central part of the cell and recognizable only after application of methyl green (Fig. 3a–e). The stained macronucleus measured 11–16 × 9–13 μm and contained a single nucleolus being about 6 × 4 μm in size. Micronucleus was not observed either after methyl green or in differential interference contrast optics. One contractile vacuole was located subterminally on the dorsal body side (Fig. 1a). The pulsation cycle of the contractile vacuole took around 18 s. At the beginning of diastole, there were several smaller oval to elliptical vesicles that fused to form a single compact circular vacuole reaching a diameter of about 12 μm at the end of diastole. The cytoplasm was colorless and packed with granules, lipid droplets about 1.3–2.0 μm across, and food vacuoles being up to 7 μm in diameter and containing pieces of host tissues and pigment host cells (Figs. 1a and 2k).
Tetrahymena acanthophora sp. n. from life. a Lateral view, showing the ovoid body. The posterior body end is broadly rounded, while the anterior end is notable apiculate (arrow). A single oval macronucleus is located in the central part of the cell. The globular contractile vacuole is subterminal and situated on the dorsal side. The oral apparatus is situated in the anterior body fourth. b, c Lateral and ventrolateral views, showing the sigmoidal course of somatic kineties. Arrows denote the apiculate anterior cell pole. d Schematic sequence of rotating cell during swimming motion. e Ventral view of the anterior body third, showing the course of somatic kineties right and left of the oral apparatus. Arrow marks the apiculate anterior body end. f Surface view, showing the arrangement of cortical granules between somatic kineties (arrowheads). g–o Variability of body shape and size of free-swimming specimens. Drawn to scale. Explanations = contractile vacuole (CV), cortical granules (CG), macronucleus (MA), oral apparatus (OA), somatic kineties (SK), undulating membrane (UM). Scale bars = 5 μm (f), 10 μm (e), and 50 μm (a–d, g–o)
Tetrahymena acanthophora sp. n. from life. a–d Overviews, showing free-swimming cells. Arrows indicate the apiculate anterior cell pole. e, f Lateral and ventrolateral views, showing the sigmoidal course of somatic kineties. Arrows point to the apiculate anterior body end. g Ventral view of a late divider. Division occurs in freely motile (non-encysted) conditions and binary fission is homothetogenic. Opposed arrowheads mark the division furrow, arrow denotes the apiculate cell pole of the proter. h Detail of the oral apparatus whose right side is bordered by an undulating membrane composed of a single row of cilia. i Detail of the apiculate anterior body end (arrow). j Ventral view of the anterior body third, showing the course of somatic kineties in the area between the apex of the body (arrow) and the oral apparatus. k Detail of the cytoplasm, showing several larger food vacuoles and many globular lipid droplets. Explanations = food vacuoles (FV), lipid droplets (LD), oral apparatus (OA), somatic kineties (SK). Scale bars = 5 μm (h, i), 10 μm (j, k), and 50 μm (a–g)
Tetrahymena acanthophora sp. n. after methyl green staining. a–c, e Dorsolateral views, showing the position as well as the shape of the macronucleus. Arrows denote the apiculate anterior cell pole. d Detail of the elliptical macronucleus with a single large nucleolus. Scale bars = 5 μm (d) and 50 μm (a–c, e)
Somatic cilia were about 8.5–10.5 μm long and arranged in 28–30 rows on each side of the cell. Individual rows were approximately 3 μm distant from each other in the mid-body, while only about 1.5 μm at the anterior and posterior cell poles. The course of somatic kineties appeared slightly sigmoidal in lateral view (Figs. 1b, c and 2e). Ventral ciliary rows formed a suture extending from the apical body end to the beginning of the oral apparatus (Figs. 1e and 2j). Between each two somatic kineties, there were cortical granules arranged in two to four staggered and densely spaced rows (Fig. 1f). The movement was elegant by swimming and rotating about the longitudinal body axis.
The oral apparatus was about 20 μm away from the anterior body end. It was tear-shaped and measured only approximately 10 × 6 μm. The right side of the buccal cavity was lined by the paroral membrane whose cilia were ca. 10 μm long and covered the buccal cavity, causing that the adoral organelles could not be recognized in vivo (Figs. 1b–e and 2f–j).
Division occurred in freely motile (non-encysted) conditions and it was homothetogenic (Fig. 2g).
Description of Tetrahymena nigricans sp. n.
ZooBank registration number: urn:lsid:zoobank.org:act:B31A3E54-70BF-4A2D-8865-8FDA231DE02B.
Diagnosis: Size about 85 × 65 μm. Body oval with both ends broadly rounded. Contractile vacuole dorsal and in third fourth of body length. Cytoplasm packed with lipid droplets and food vacuoles, causing a dark appearance of cell at low magnifications. About 18 meridional ciliary rows on each body side, no caudal cilia.
Type locality: Veľkobielske jazero lake, recreation area in the village of Veľký Biel, Podunajská rovina plain (48° 12′ 26.4″ N, 17° 21′ 28.3″ E).
Type host: Girardia tigrina (Girard, 1850) Ball, 1974.
Type material: A DNA sample of holotype specimen has been deposited in Natural History Museum, Vajanského nábrežie 2, 810 06 Bratislava, Slovakia (ID Collection Code 1427127).
Gene sequences: The nuclear 18S rRNA gene sequence of holotype specimen has been deposited in GenBank (accession no. MN994472). The sequence is 1707 nucleotides long and has a GC content of 43.06%. The mitochondrial 16S rRNA gene sequence of holotype specimen has been deposited in GenBank (accession no. MN994483). The sequence is 991 nucleotides long and has a GC content of 31.08%. The COI gene sequence of holotype specimen has been deposited in GenBank (accession no. MN991323). The sequence is 915 nucleotides long and has a GC content of 25.35%.
Etymology: The specific epithet nigrican·s, –s, –s [m, f, n] (blackish) is a Latin adjective referring to the dark appearance of the species caused by the densely packed cytoplasm with food vacuoles.
Description: Morphological description of this species is considerably limited due to its extremely rare occurrence in planarians. Specifically, only four specimens of T. nigricans were detected, two were sampled for molecular analyses and two for in vivo observations. Thus, all measurements were based only on two cells. Unfortunately, the densely packed cytoplasm made observations of most taxonomically important features almost impossible.
The body size was about 75–90 × 55–70 μm in vivo (n = 2). The shape was oval with both poles broadly rounded and did not distinctly change under the coverslip pressure (Fig. 4a–f). The only clearly visible organelle was the contractile vacuole. It was spherical, 12–13 μm in diameter and located dorsally in the third fourth of the body length (Fig. 4b, d). One pulsation cycle from the beginning of systole until the contractile vacuole was fully formed from smaller vesicles took approximately 13 s. The cytoplasm was colorless and packed with a huge amount of food vacuoles being 3.0–7.8 μm in diameter and 1.0–1.7 μm-sized lipid droplets (Fig. 4g). Somatic cilia were about 8.5–9.5 μm long. Based on the spacing of some visible ciliary rows and the width of the anterior body region, it was estimated that there might have been ca. 18 meridional ciliary rows on each side of the cell.
Tetrahymena nigricans sp. n. from life. a, b, e, f Lateral overviews, showing the oval body with cytoplasm densely packed with food vacuoles. Note that the contractile vacuole is situated slightly below the mid-body on dorsal body side. c Detail of dorsal body margin. Opposed arrows denote the comparatively thick cortex. d Detail of globular contractile vacuole during diastole. g Detail of cytoplasm, showing larger food vacuoles and smaller lipid droplets. Explanations = contractile vacuole (CV), food vacuoles (FV), lipid droplets (LD), somatic kineties (SK). Scale bars = 5 μm (g), 10 μm (c, d), and 50 μm (a, b, e, f)
Description of Tetrahymena dugesiae sp. n.
2019 Tetrahymena sp. – Rataj and Vďačný, Dis Aquat Org 134: 157 (Figs. 3A–H and 4A–K)
ZooBank registration number: urn:lsid:zoobank.org:act:F5F67F43-1FE7-4E98-8BC0-EDD4F1812274.
Diagnosis: Size about 115 × 50 μm. Body narrowly to broadly fusiform with anterior end differentiated into a plate-like rostrum and posterior end broadly rounded. Macronucleus oval. Contractile vacuole dorsal and subterminal. About 10 to 12 meridional ciliary rows on each body side, no caudal cilia. Oral apparatus about 10 μm long.
Type locality: Milošovanský potok stream, inundation area in the vicinity of the village of Milošová, Turzovská vrchovina highlands (49° 28′ 17.1″ N, 18° 45′ 18.4″ E).
Type host: Dugesia gonocephala (Dugès, 1830) Girard, 1850.
Type material: A DNA sample of holotype specimen has been deposited in Natural History Museum, Vajanského nábrežie 2, 810 06 Bratislava, Slovakia (ID Collection Code 1425919).
Gene sequences: The nuclear 18S rRNA gene sequence of holotype specimen has been deposited in GenBank (accession no. MK454732). The sequence is 1749 nucleotides long and has a GC content of 43.28%. The mitochondrial 16S rRNA gene sequence of holotype specimen has been deposited in GenBank (accession no. MN994480). The sequence is 987 nucleotides long and has a GC content of 31.12%. The COI gene sequence of holotype specimen has been deposited in GenBank (accession no. MN991320). The sequence is 967 nucleotides long and has a GC content of 25.23%.
Etymology: The specific epithet dugesiae (from Dugesia) is a singular genitive case of the Latin noun Dugesi·a, –ae [f]. It refers to the presence of this ciliate exclusively in Dugesia specimens. According to Article 11.9.1.4. of the International Commission on Zoological Nomenclature (1999), the species-group name is to be treated as an adjective used as a substantive in the genitive case, because of its derivation from the host’s generic name.
Description: Morphological description is provided in Rataj and Vďačný (2019).
Phylogenetic analyses
In the present study, we obtained altogether 28 new sequences of three molecular markers belonging to the three new Tetrahymena species and T. scolopax. Their length, guanine–cytosine content, and GenBank accession numbers are summarized in Table 2. Intraspecies sequence similarities in the 16S and 18S rRNA genes as well as in the COI gene are 100%, except for the single T. dugesiae M17 isolate which differs by 0.2% in the 16S rRNA gene and by 2.3% in the COI sequences from the four other conspecific isolates. The COI sequences of Slovak and American isolates of T. scolopax are 100% identical.
To assess the phylogenetic position of the newly described tetrahymenas, Bayesian and ML analyses were conducted (Figs. 5, 6, and 7 and Supplementary Fig. S1). Tetrahymenas isolated from planarians were consistently classified within the same clusters across all phylogenetic analyses, even when the overall tree topologies slightly differed depending on the molecular marker(s) and the dataset analyzed. Specifically, T. acanthophora, T. dugesiae, and T. scolopax always formed a group together with T. bergeri and T. corlissi within the variably supported “borealis” clade. Tetrahymena dugesiae was consistently depicted as sister to T. scolopax with usually full statistical support across all analyses. These two species were shown in a sister relationship with T. acanthophora when T. bergeri and T. corlissi were not included into the analyses (Fig. 7). In datasets, where all three latter species were present, T. dugesiae and T. scolopax formed their sister clade (Figs. 5 and 6). Relationships among T. acanthophora, T. bergeri, and T. corlissi could not be, however, reliably resolved. On the other hand, T. nigricans was in every scenario a part of the “paravorax” clade. This cluster was nested within the “australis” clade although without any statistical support in COI trees (Fig. 6), while it was placed outside the “core” of the genus Tetrahymena in analyses of the concatenated 18S rRNA gene + COI dataset (Fig. 5) and the mitochondrial 16S rRNA gene (Fig. 7). Nevertheless, the internal structure of the paravorax clade was consistent across all analyses. Tetrahymena paravorax Corliss, 1957 was sister to the branch leading to T. glochidiophila, T. nigricans, and T. pennsylvaniensis Doerder, 2019 (Figs. 5, 6, and 7). Moreover, phylogenetic analyses suggested a closer relationship of T. nigricans to T. pennsylvaniensis than to T. glochidiophila but only with poor statistical support (Figs. 5 and 6).
Phylogeny based on the concatenated nuclear 18S rRNA gene and mitochondrial COI sequences, showing the systematic position of tetrahymenas isolated from freshwater planarians. Bootstrap values for maximum likelihood (ML) and posterior probabilities for Bayesian inference (BI) were mapped onto the best scoring IQ tree. Dashes indicate mismatch between ML and BI trees. Sequences in bold face were obtained during this study. The strain MP80 was mislabeled as T. elliotti by Chantangsi et al. (2007). According to Doerder (2019), T. elliotti is related to T. gruchyi. For accession numbers, see Supplementary Table S1. The scale bar denotes one substitution per ten nucleotide positions
Phylogeny based on the mitochondrial COI sequences, showing the systematic position of tetrahymenas isolated from freshwater planarians. Bootstrap values for maximum likelihood (ML) and posterior probabilities for Bayesian inference (BI) were mapped onto the best scoring IQ tree. Dashes indicate mismatch between ML and BI trees. Sequences in bold face were obtained during this study. The strain MP80 was mislabeled as T. elliotti by Chantangsi et al. (2007). The scale bar denotes 35 substitutions per hundred nucleotide positions
Phylogeny based on the mitochondrial 16S rRNA gene sequences, showing the systematic position of tetrahymenas isolated from freshwater planarians. Bootstrap values for maximum likelihood (ML) and posterior probabilities for Bayesian inference (BI) were mapped onto the best scoring IQ tree. Dashes indicate mismatch between ML and BI trees. Sequences in bold face were obtained during this study. The scale bar denotes eight substitutions per hundred nucleotide positions
Reconstructions of ancestral life strategies indicated that the last common progenitor of hymenostomes led a parasitic way of life (Fig. 8). Very likely there were two independent shifts to a free-living strategy, i.e., at the base of the “paravorax” clade and in the last common ancestor of Colpidium Stein, 1860, Dexiostoma Jankowski, 1968, Glaucoma Ehrenberg, 1830, and the “core” tetrahymenas (“australis” + “borealis” clade). Or this transfer might have happened only once at the base of these two clades (Fig. 8, arrowhead), depending on the tree topology, especially, on the phylogenetic position of the histophagous Agolohymena aspidocauda Bourland and Strüder-Kypke, 2010. The SIMMAP analyses suggested at least seven independent shifts back to parasitism: (i) in T. glochidiophila and T. nigricans within the “paravorax” clade; (ii) in T. empidokyrea within the “australis” clade; and (iii–vii) in T. mobilis (Kahl, 1926) Lynn and Doerder, 2012, T. farleyi Lynn et al., 2000, T. limacis, T. rostrata as well as in the T. acanthophora + T. bergeri + T. corlissi + T. dugesiae + T. scolopax cluster within the “borealis” clade (Fig. 8, arrows).
SIMMAP reconstruction of ancestral life strategies based on a set of 1000 randomly selected trees from the posterior distribution of the Bayesian analysis of the concatenated nuclear 18S rRNA gene and mitochondrial COI dataset. Relative proportions of character states were mapped onto the Bayesian 50% majority-rule consensus tree
The comparison of log marginal likelihoods of various binary-state speciation and extinction (BiSSE) models for diversification of parasitic and free-living hymenostomes indicated that the model with all parameters free is to be preferred over models where speciation, extinction, and transition rates were constrained (Table 3). The subsequent MCMC simulations suggested that the free-living hymenostome lineages have a distinctly higher speciation rate than the parasitic lineages (Fig. 9, left upper panel). Although the extinction rate between the two types of lineages overlapped significantly, the parasitic branches appear to be subjected to even a much higher risk of becoming extinct (Fig. 9, right upper panel). Consequently, the net diversification rate of the free-living lineages was revealed to be distinctly higher than in the parasitic ones (Fig. 9, left lower panel). As concerns the transition rates, the estimated values distinctly overlapped, although the q01 rate seemed to be higher (Fig. 9, right lower panel). This conspicuous overlap was very likely the reason why the best fitting model did not receive strong evidence (BF < 6.0) against the model with q01 and q10 constrained to be equal (Table 3). In turn, this indicates that both transition rates might have not been different and hence transfers from the free-living to the parasitic strategy and vice versa are equally probable in hymenostome ciliates.
Discussion
The three new Tetrahymena species
In the present study, we have introduced three species of Tetrahymena which undoubtedly represent new taxa also with respect to the ciliate-description recommendations proposed by Warren et al. (2017). However, because most species in this genus lack morphologically unique features, erection of new forms based solely on morphological characteristics is impossible (Lynn et al. 2018). Therefore, the distinctness of Slovak isolates was grounded, especially, on three molecular markers including the 16S and 18S rRNA genes as well as the barcoding COI gene, following Doerder (2019). In their detailed tribute to ciliate taxonomy, Warren et al. (2017) also stated that the genetic divergence cannot serve as an indisputable tool for species identification only by itself due to the absence of one generally accepted threshold value for every molecular marker utilized. Nevertheless, it was already shown that tetrahymenas exhibit an average of about 10.5% interspecific genetic divergence in the barcoding region of their COI sequences, while less than 1% intraspecific variability in this region (Chantangsi et al. 2007). Nevertheless, Doerder (2014, 2019) used a 4% interspecific divergence threshold and mentioned exceptions to the 1% intraspecific variability rule. The threshold proposed by Chantangsi et al. (2007) holds for tetrahymenas infecting planarians and belonging to the “borealis” clade. Specifically, the COI pairwise differences between T. acanthophora, T. dugesiae, and their closest relatives, T. corlissi, T. bergeri, and T. scolopax, were consistently ~ 11%. Tetrahymena acanthophora and T. dugesiae were also depicted as very distinct branches in all phylogenetic trees (Figs. 5, 6, and 7 and Supplementary Fig. S2). Moreover, the conspicuous thorn situated at the anterior body pole together with the uniquely sigmoidally arranged somatic kineties make T. acanthophora to be comparatively easily morphologically distinguishable already by detailed in vivo examination. The only other Tetrahymena species with a distinctly apiculate anterior end is the parasitic stage of T. limacis (Brooks 1968; Kozloff 1946). However, its ciliary rows are arranged meridionally, as usual in the genus Tetrahymena. In addition, our phylogenetic analyses also document that T. acanthophora and T. limacis are phylogenetically distant (Figs. 5 and 6), whereby their COI sequences share only 85% identity. The detailed morphological comparison of T. dugesiae with its closest relatives is provided in Rataj and Vďačný (2019).
As concerns the third new species proposed here, T. nigricans undoubtedly belongs to the “paravorax” clade. Interestingly, the degree of 18S rRNA and COI sequence identities differs considerably among the members of this group. Unlike the cluster containing T. acanthophora and T. dugesiae whose 18S rRNA gene sequences differ by 0.3–1.2%, T. glochidiophila, T. pennsylvaniensis, and T. nigricans, all belonging to the “paravorax” clade, share completely identical 18S rRNA gene sequences. Even though T. nigricans appears in a closer relationship with the free-living T. pennsylvaniensis than with the parasitic T. glochidiophila (Figs. 5 and 6), the COI sequence of T. nigricans is more similar to that of T. glochidiophila (5.4% divergence) than to that of T. pennsylvaniensis (7.2% divergence). This along with the identical 18S rRNA gene sequences indicates a rather recent origin and rapid radiation of T. glochidiophila, T. pennsylvaniensis, and T. nigricans driven by adaptive changes in new ecological situations. The former two species have been so far reported only from North America, T. glochidiophila is a parasite of glochidia and T. pennsylvaniensis is free-living (Doerder 2019; Lynn et al. 2018). We have detected T. nigricans in the planarian G. tigrina which is native to North America (Ball 1974) and was introduced into many European countries and Japan with aquatic plants (Dahm 1958; Kawakatsu et al. 1985), possibly also with its ciliate parasite. Although detailed morphological data are available only for T. glochidiophila (Lynn et al. 2018) and the morphological description of T. nigricans is considerably limited, both species are very dissimilar already at first glance. Tetrahymena nigricans is much bigger and has an oval body with both poles broadly rounded, while T. glochidiophila is only about half of its size and has an ovoid body with anterior pole notably tapered. Moreover, T. glochidiophila possesses a prominent oral apparatus and its bacterivorous form bears a single caudal cilium. None of these features were recognized in our isolate. Both species differ also by the location of the contractile vacuole which is situated subterminally in T. glochidiophila, while slightly below the mid-body in T. nigricans. Moreover, T. nigricans is a parasite of planarians, while T. glochidiophila attacks glochidia of bivalves. Even though the COI divergence is below the 10% threshold, T. glochidiophila and T. nigricans are highly morphologically and ecologically dissimilar, which justifies their species status.
As mentioned above, the three new species can be differentiated from their nearest relatives also by detailed in vivo observations. Nevertheless, it is important to mention some problems that might arise during the morphological identification. First, the morphological properties of cultured cells might differ from those freshly isolated from the host, as it has been documented for the parasitic T. limacis (for a review, see Corliss 1973). Second, it cannot be excluded that the three new species might have also free-living forms, as indicated by the detection of T. scolopax both in D. gonocephala (present study) and in freshwater (Doerder 2019). Because there is no guarantee that morphologies of parasitic and free-living forms are similar, the identification needs to be proven also by molecular data. Finally, since our morphological data are based only on freshly isolated living cells, we had difficulties to recognize the micronucleus, which represents an important taxonomic character (Doerder 2014). Whether the tree new species are amicronucleates needs to be therefore confirmed by protargol impregnation, DAPI staining, and/or detailed observations of mid-dividers in which the division spindle of the micronucleus is well recognizable.
Diversity of tetrahymenas infecting planarians
Despite our extensive sampling effort, we isolated altogether only four Tetrahymena species and they were found only in two out of the eight freshwater planarian species studied, namely, in Dugesia gonocephala and Girardia tigrina. On the other hand, Wright (1981) recorded Tetrahymena infections in populations of Crenobia alpina, Polycelis felina, and Phagocata vitta (Dugès, 1830) Leidy, 1847 in North Wales. The two former planarian species were Tetrahymena-free in our samples and the latter species was not found during material collection in Slovakia. Wright (1981) mentioned that ciliates from C. alpina were identified as T. pyriformis based on silver preparations. He further stated that T. corlissi was also detected in some C. alpina individuals from East Anglia. Unfortunately, no DNA of these tetrahymenas was isolated and, therefore, their specificity cannot be tested with modern molecular phylogenetic methods. Apart from Wright’s (1981) reports, it seems that T. pyriformis was also responsible for some infections of Dendrocoelum lacteum (Wright 1969). Considering that this Tetrahymena species was present not only in freshwater planarians (e.g., Antipa and Small 1971; Nekuie Fard et al. 2011), we assume that it does not prefer one specific type of triclads but a rather wide spectrum of accessible hosts. Indeed, tetrahymenas very likely show a weak structural/phylogenetic host specificity (Rataj and Vďačný 2019; Strüder-Kypke et al. 2001). However, this needs to be also confirmed by much more extensive molecular data, because all new tetrahymenas described in this study turned out to be confined to a certain planarian species. Even though the prevalence of tetrahymenas in D. gonocephala and G. tigrina was very low (ranging from ~ 1 to 10.3%), the infections were never coincidental. Specifically, all three new tetrahymenas were recorded at multiple collection sites and tetrahymenas isolated from G. tigrina were never found to feed on D. gonocephala and vice versa. Both planarians are also ecologically different and very likely do not meet in nature. The former species lives in stagnant water bodies, while the latter is restricted to running waters.
Whether the presence of T. scolopax in a single planarian sample was accidental, remains to be further studied as we have detected it only once. The type locality of T. scolopax is the Woodcock Creek in Pennsylvania, USA (Doerder 2019). Unfortunately, we have not examined water samples from the site where infected planarians were detected. Since Doerder (2019) found free-living forms of species also reported as parasites, further investigations are needed to corroborate whether T. scolopax is an obligate or a facultative parasite of aquatic animals, having both free-living and parasitic stages. In the present study, T. scolopax co-occurred with T. dugesiae in D. gonocephala. This is not very surprising, since mixed Tetrahymena infections were already observed in gray garden slug (Brooks 1968).
To summarize, the present study and literature data (Reynoldson and Bellamy 1973; Wright 1981) indicate that 25% of planarian species are positive for Tetrahymena. The prevalence of Tetrahymena infections ranges from about 1 to ~ 10%. Out of the four Tetrahymena species found during the present study, three were recognized to be new taxa. This indicates a large undescribed ciliate diversitylinking histone acetylation to gene activation in freshwater planarians. Nevertheless, six species have been so far reported to infect planarians: T. acanthophora, T. corlissi, T. dugesiae, T. nigricans, T. pyriformis, and T. scolopax. The identity of T. corlissi and T. pyriformis needs to be supported by molecular data. We believe that intensive research on insufficiently explored or completely neglected planarians might still reveal new Tetrahymena species.
Evolution of life strategies in Tetrahymena
The ancestral way of life in hymenostome ciliates has been analyzed in two previous studies, using the parsimony framework (Rataj and Vďačný 2019; Strüder-Kypke et al. 2001). However, both reconstructions were not unequivocal for the node of the “core” Tetrahymenidae Corliss, 1952 and, therefore, it was not possible to state whether the life style of tetrahymenas was ancestrally free-living or parasitic. Here, we applied a different statistical strategy, the stochastic character mapping in a combination with the Bayesian approach that accounts also for uncertainty in the inferred phylogeny. The present reconstruction of the ancestral way of life was unambiguous and suggested that the last common ancestor of the subclass Hymenostomatia was parasitic, while that of tetrahymenas was free-living. Consequently, there were at least seven independent shifts back to parasitism/histophagy: one each in the “paravorax” and “australis” clades, and at a minimum five reversals to parasitism in the “borealis” clade (Fig. 8). Interestingly, the BiSSE analyses suggested that the parasitic lineages have slower speciation and net diversification rates than the free-living lineages. In addition, according to the MCMC simulations, the parasitic lineages face a higher risk to become extinct than the free-living lineages (Fig. 9). This contrasts with two other ciliate classes, the Armophorea Lynn, 2004 and the Litostomatea Small and Lynn, 1981, where both free-living and endosymbiotic lineages occur. Specifically, our previous diversification analyses suggested that the endobiotic armophorean lineages have several times higher speciation and net diversification rates than the free-living armophorean lineages. This is indirectly corroborated also in that there are only about 80 recognized free-living taxa, while almost 200 endosymbiotic forms (Vďačný et al. 2019). A similar picture can be found in litostomateans which include about 300 free-living species (Foissner et al. 1999), while about 1000 endosymbiotic taxa (Cedrola et al. 2020). By contrast, there are about dozen of obligate or facultative parasitic Tetrahymena species, while about 60 free-living taxa (Fig. 8). Also this fact along with phylogenetic trees suggests that tetrahymenas were ancestrally very likely free-living and there were multiple independent shifts to parasitism. On the other hand, there was only a single shift to endobiotic life style in case of litostomateans (Vďačný 2018) and two transfers in case of armophoreans (Vďačný et al. 2019). When obligate endobiosis in the digestive tract of both invertebrates and vertebrates was established, then there was a burst of endobiotic armophorean and litostomatean lineages. Obviously, tetrahymenas prefer different evolutionary strategies than armophoreans and litostomateans, and hence their diversification patterns are also different.
References
Antipa GA, Small EB (1971) The occurrence of thigmotrichous ciliated protozoa inhabiting the mantle cavity of unionid molluscs of Illinois. Trans Am Microsc Soc 90:463–472. https://doi.org/10.2307/3225461
Ball IR (1974) A contribution to the phylogeny and biogeography of the freshwater triclads (Platyhelminthes, Turbellaria). In: Riser W, Morse MP (eds) Biology of the Turbellaria. McGraw-Hill, New York, pp 339–401
Batson BS (1983) Tetrahymena dimorpha sp. nov. (Hymenostomatida: Tetrahymenidae), a new ciliate parasite of Simuliidae (Diptera) with potential as a model for the study of ciliate morphogenesis. Philos Trans R Soc Lond B 301:345–363. https://doi.org/10.1098/rstb.1983.0027
Batson BS (1985) A paradigm for the study of insect-ciliate relationships: Tetrahymena sialidos sp. nov. (Hymenostomatida: Tetrahymenidae), parasite of larval Sialis lutaria (Linn.) (Megaloptera: Sialidae). Philos Trans R Soc Lond B 310:123–144. https://doi.org/10.1098/rstb.1985.0102
Blackburn EH, Gall JG (1978) A tandemly repeated sequence at the termini of the extrachromosomal ribosomal RNA genes in Tetrahymena. J Mol Biol 120:33–53. https://doi.org/10.1016/0022-2836(78)90294-2
Bollback JP (2006) SIMMAP: stochastic character mapping of discrete traits on phylogenies. BMC Bioinformatics 7:88. https://doi.org/10.1186/1471-2105-7-88
Brooks W (1968) Tetrahymenid ciliates as parasites of the gray garden slug. Hilgardia 39:205–276. https://doi.org/10.3733/hilg.v39n08p205
Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD (1996) Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84:843–851. https://doi.org/10.1016/s0092-8674(00)81063-6
Brunk CF, Lee LC, Tran AB, Li J (2003) Complete sequence of the mitochondrial genome of Tetrahymena thermophila and comparative methods for identifying highly divergent genes. Nucleic Acids Res 31:1673–1682. https://doi.org/10.1093/nar/gkg270
Burger G, Zhu Y, Littlejohn TG, Greenwood SJ, Schnare MN, Lang BF, Gray MW (2000) Complete sequence of the mitochondrial genome of Tetrahymena pyriformis and comparison with Paramecium aurelia mitochondrial DNA. J Mol Biol 297:365–380. https://doi.org/10.1006/jmbi.2000.3529
Cedrola F, Senra MVX, Rossi MF, Fregulia P, D’Agosto M, Dias RJP (2020) Trichostomatid ciliates (Alveolata, Ciliophora, Trichostomatia) systematics and diversity: past, present and future. Front Microbiol 10:2967. https://doi.org/10.3389/fmicb.2019.02967
Chantangsi C, Lynn DH, Brandl MT, Cole JC, Hetrick N, Ikonomi P (2007) Barcoding ciliates: a comprehensive study of 75 isolates of the genus Tetrahymena. Int J Syst Evol Microbiol 57:2412–2425. https://doi.org/10.1099/ijs.0.64865-0
Corliss JO (1960) Tetrahymena chironomi sp. nov., a ciliate from midge larvae, and the current status of facultative parasitism in the genus Tetrahymena. Parasitology 50:111–153. https://doi.org/10.1017/s0031182000025245
Corliss JO (1969) An up-to-date analysis of the systematics of the ciliate genus Tetrahymena. J Protozool 16:6–7. https://doi.org/10.1111/j.1550-7408.1969.tb04358.x
Corliss JO (1970) The comparative systematics of species comprising the hymenostome ciliate genus Tetrahymena. J Protozool 17:198–209. https://doi.org/10.1111/j.1550-7408.1970.tb02356.x
Corliss JO (1972) Tetrahymena and some thoughts on the evolutionary origin of endoparasitism. Trans Am Microsc Soc 91:566–573. https://doi.org/10.2307/3225485
Corliss JO (1973) History, taxonomy, ecology, and evolution of species of Tetrahymena. In: Elliott AM (ed) Biology of Tetrahymena. Dowden, Hutchinson & Ross, Stroudsburg, pp 1–55
Dahm AG (1958) Taxonomy and ecology of five species groups in the family Planariidae (Turbellaria Tricladida Paludicola). Nya Litografen, Malmö
Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9:772. https://doi.org/10.1038/nmeth.2109
Doerder FP (2014) Abandoning sex: multiple origins of asexuality in the ciliate Tetrahymena. BMC Evol Biol 14:112. https://doi.org/10.1186/1471-2148-14-112
Doerder FP (2019) Barcodes reveal 48 new species of Tetrahymena, Dexiostoma, and Glaucoma: phylogeny, ecology, and biogeography of new and established species. J Eukaryot Microbiol 66:182–208. https://doi.org/10.1111/jeu.12642
Edqvist J, Burger G, Gray MW (2000) Expression of mitochondrial protein-coding genes in Tetrahymena pyriformis. J Mol Biol 297:381–393. https://doi.org/10.1006/jmbi.2000.3530
FitzJohn RG (2012) Diversitree: comparative phylogenetic analyses of diversification in R. Methods Ecol Evol 3:1084–1092. https://doi.org/10.1111/j.2041-210X.2012.00234.x
Foissner W (2014) An update of ‘basic light and scanning electron microscopic methods for taxonomic studies of ciliated protozoa’. Int J Syst Evol Microbiol 64:271–292. https://doi.org/10.1099/ijs.0.057893-0
Foissner W, Berger H, Schaumburg J (1999) Identification and ecology of limnetic plankton ciliates. Informationsberichte des Bayer Landesamtes für Wasserwirtschaft 3(99):1–793
Frankel J (1999) Cell biology of Tetrahymena thermophila. Methods Cell Biol 125:27–125
Furgason WH (1940) The significant cytostomal pattern of the “Glaucoma-Colpidium group,” and a proposed new genus and species, Tetrahymena geleii. Arch Protistenkd 94:224–266
Greider CW, Blackburn EH (1985) Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43:405–413. https://doi.org/10.1016/0092-8674(85)90170-9
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
International Commission on Zoological Nomenclature (1999) International code of zoological nomenclature, 4th edn. Tipografia La Garangola, Padova
International Commission on Zoological Nomenclature (2012) Amendment of Articles 8, 9, 10, 21 and 78 of the International Code of Zoological Nomenclature to expand and refine methods of publication. Bull Zool Nomencl 69:161–169
Jerome CA, Simon EM, Lynn DH (1996) Description of Tetrahymena empidokyrea n.sp., a new species in the Tetrahymena pyriformis sibling species complex (Ciliophora, Oligohymenophorea), and an assessment of its phylogenetic position using small-subunit rRNA sequences. Can J Zool 74:1898–1906. https://doi.org/10.1139/z96-214
Johnson MTJ, FitzJohn RG, Smith SD, Rausher MD, Otto SP (2011) Loss of sexual recombination and segregation is associated with increased diversification in evening primroses. Evolution 65:3230–3240. https://doi.org/10.1111/j.1558-5646.2011.01378.x
Kass RE, Raftery AE (1995) Bayes factors. J Am Stat Assoc 90:773–795. https://doi.org/10.1080/01621459.1995.10476572
Kawakatsu M, Oki I, Tamura S, Yamayoshi T (1985) Reexamination of freshwater planarians found in tanks of tropical fishes in Japan, with a description of a new species, Dugesia austroasiatica sp. nov. (Turbellaria; Tricladida; Paludicola). Bull Biogeogr Soc Jpn 40:1–19
Kher CP, Doerder FP, Cooper J, Ikonomi P, Achilles-Day U, Küpper FC, Lynn DH (2011) Barcoding Tetrahymena: discriminating species and identifying unknowns using the cytochrome c oxidase subunit I (cox-1) barcode. Protist 162:2–13. https://doi.org/10.1016/j.protis.2010.03.004
Kozloff EN (1946) The morphology and systematic position of a holotrichous ciliate parasitizing Deroceras agreste (L.). J Morphol 79:445–465. https://doi.org/10.1002/jmor.1050790305
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Lwoff A (1923) Sur la nutrition des infusoires. C R Acad Sci 176:928–930
Lynn DH (2008) The ciliated protozoa. Characterization, classification and guide to the literature. Springer, Dordrecht
Lynn DH, Doerder FP (2012) The life and times of Tetrahymena. Methods Cell Biol 109:9–27. https://doi.org/10.1016/B978-0-12-385967-9.00002-5
Lynn DH, Doerder FP, Gillis PL, Prosser PS (2018) Tetrahymena glochidiophila n. sp., a new species of Tetrahymena (Ciliophora) that causes mortality to glochidia larvae of freshwater mussels (Bivalvia). Dis Aquat Org 127:125–136. https://doi.org/10.3354/dao03188
Lynn DH, Molloy D, LeBrun R (1981) Tetrahymena rotunda n. sp. (Hymenostomatida: Tetrahymenidae), a ciliate parasite of the hemolymph of Simulium (Diptera: Simuliidae). Trans Am Microsc Soc 100:134–141. https://doi.org/10.2307/3225796
Lynn DH, Strüder-Kypke MC (2006) Species of Tetrahymena identical by small subunit rRNA gene sequences are discriminated by mitochondrial cytochrome c oxidase I gene sequences. J Eukaryot Microbiol 53:385–387. https://doi.org/10.1111/j.1550-7408.2006.00116.x
MacColl E, Therkelsen MD, Sherpa T, Ellerbrock H, Johnston LA, Jariwala RH, Chang WS, Gurtowski J, Schatz MC, Mozammal Hossain M, Cassidy-Hanley DM, Clark TG, Chang WJ (2015) Molecular genetic diversity and characterization of conjugation genes in the fish parasite Ichthyophthirius multifiliis. Mol Phylogenet Evol 86:1–7. https://doi.org/10.1016/j.ympev.2015.02.017
Maddison WP, Midford PE, Otto SP (2007) Estimating a binary character’s effect on speciation and extinction. Syst Biol 56:701–710. https://doi.org/10.1080/10635150701607033
Medlin L, Elwood HJ, Stickel S, Sogin ML (1988) The characterization of enzymatically amplified eukaryotic 16S-like rRNA coding regions. Gene 71:491–499. https://doi.org/10.1016/0378-1119(88)90066-2
Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE). Piscataway, N.J., New Orleans, Louisiana, pp 1–8. https://doi.org/10.1109/GCE.2010.5676129
Moradian MM, Beglaryan D, Skozylas JM, Kerikorian V (2007) Complete mitochondrial genome sequence of three Tetrahymena species reveals mutation hot spots and accelerated nonsynonymous substitutions in Ymf genes. PLoS One 2:e650. https://doi.org/10.1371/journal.pone.0000650
Nanney DL, McCoy JW (1976) Characterization of the species of the Tetrahymena pyriformis complex. Trans Am Microsc Soc 95:664–682. https://doi.org/10.2307/3225391
Nanney DL, Park C, Preparata R, Simon EM (1998) Comparison of sequence differences in a variable 23S rRNA domain among sets of cryptic species of ciliated protozoa. J Eukaryot Microbiol 45:91–100. https://doi.org/10.1111/j.1550-7408.1998.tb05075.x
Nekuie Fard A, Motalebi AA, Jalali Jafari B, Aghazadeh Meshgi M, Azadikhah D, Afsharnasab M (2011) Survey on fungal, parasites and epibionts infestation on the Astacus leptodactylus (Eschscholtz, 1823), in Aras reservoir West Azarbaijan, Iran. Iran J Fish Sci 10:266–275
Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. https://doi.org/10.1093/molbev/msu300
Pitsch G, Adamec L, Dirren S, Nitsche F, Šimek K, Sirová D, Posch T (2017) The green Tetrahymena utriculariae n. sp. (Ciliophora, Oligohymenophorea) with its endosymbiotic algae (Micractinium sp.), living in traps of a carnivorous aquatic plant. J Eukaryot Microbiol 64:322–335. https://doi.org/10.1111/jeu.12369
Quintela-Alonso P, Nitsche F, Wylezich C, Arndt H, Foissner W (2013) A new Tetrahymena (Ciliophora, Oligohymenophorea) from groundwater of Cape Town, South Africa. J Eukaryot Microbiol 60:235–246. https://doi.org/10.1111/jeu.12021
Rataj M, Vďačný P (2018) Dawn of astome ciliates in light of morphology and time-calibrated phylogeny of Haptophrya planariarum, an obligate endosymbiont of freshwater turbellarians. Eur J Protistol 64:54–71. https://doi.org/10.1016/j.ejop.2018.03.004
Rataj M, Vďačný P (2019) Living morphology and molecular phylogeny of oligohymenophorean ciliates associated with freshwater turbellarians. Dis Aquat Org 134:147–166. https://doi.org/10.3354/dao03366
Revell LJ (2012) Phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol Evol 3:217–223. https://doi.org/10.1111/j.2041-210X.2011.00169.x
Reynoldson TB, Bellamy LS (1973) Interspecific competition in lake-dwelling triclads. A laboratory study. Oikos 24:301–313
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542. https://doi.org/10.1093/sysbio/sys029
Silvestro D, Schnitzler J, Zizka G (2011) A Bayesian framework to estimate diversification rates and their variation through time and space. BMC Evol Biol 11:311. https://doi.org/10.1186/1471-2148-11-311
Strüder-Kypke MC, Wright ADG, Jerome CA, Lynn DH (2001) Parallel evolution of histophagy in ciliates of the genus Tetrahymena. BMC Evol Biol 1:5. https://doi.org/10.1186/1471-2148-1-5
van Hoek AHAM, Akhmanova AS, Huynen MA, Hackstein JHP (2000) A mitochondrial ancestry of the hydrogenosomes of Nyctotherus ovalis. Mol Biol Evol 17:202–206. https://doi.org/10.1093/oxfordjournals.molbev.a026234
Vďačný P (2018) Evolutionary associations of endosymbiotic ciliates shed light on the timing of the marsupial-placental split. Mol Biol Evol 35:1757–1769. https://doi.org/10.1093/molbev/msy071
Vďačný P, Bourland WA, Orsi W, Epstein SS, Foissner W (2011) Phylogeny and classification of the Litostomatea (Protista, Ciliophora), with emphasis on free-living taxa and the 18S rRNA gene. Mol Phylogenet Evol 59:510–522. https://doi.org/10.1016/j.ympev.2011.02.016
Vďačný P, Rajter Ľ, Stoeck T, Foissner W (2019) A proposed timescale for the evolution of armophorean ciliates: clevelandellids diversify more rapidly than metopids. J Eukaryot Microbiol 66:167–181. https://doi.org/10.1111/jeu.12641
Warren A, Patterson DJ, Dunthorn M, Clamp JC, Achilles-Day UEM, Aescht E, al-Farraj SA, al-Quraishy S, al-Rasheid K, Carr M, Day JG, Dellinger M, el-Serehy HA, Fan Y, Gao F, Gao S, Gong J, Gupta R, Hu X, Kamra K, Langlois G, Lin X, Lipscomb D, Lobban CS, Luporini P, Lynn DH, Ma H, Macek M, Mackenzie-Dodds J, Makhija S, Mansergh RI, Martín-Cereceda M, McMiller N, Montagnes DJS, Nikolaeva S, Ong'ondo G, Pérez-Uz B, Purushothaman J, Quintela-Alonso P, Rotterová J, Santoferrara L, Shao C, Shen Z, Shi X, Song W, Stoeck T, la Terza A, Vallesi A, Wang M, Weisse T, Wiackowski K, Wu L, Xu K, Yi Z, Zufall R, Agatha S (2017) Beyond the ‘code’: a guide to the description and documentation of biodiversity in ciliated protists (Alveolata, Ciliophora). J Eukaryot Microbiol 64:539–544. https://doi.org/10.1111/jeu.12391
Wright JF (1969) The ecology of stream-dwelling triclads. Dissertation, University of Wales
Wright JF (1981) Tetrahymena pyriformis (Ehrenberg) and T. corlissi Thompson parasitic in stream-dwelling triclads (Platyhelminthes: Turbellaria). J Parasitol 67:131–133. https://doi.org/10.2307/3280799
Yao MC, Blackburn EH, Gall JG (1981) Tandemly repeated C-C-C-C-A-A hexanucleotide of Tetrahymena rDNA is present elsewhere in the genome and may be related to the alteration of the somatic genome. J Cell Biol 90:515–520. https://doi.org/10.1083/jcb.90.2.515
Yao MC, Yao CH (1981) Repeated hexanucleotide C-C-C-C-A-A is present near free ends of macronuclear DNA of Tetrahymena. Proc Natl Acad Sci U S A 78:7436–7439. https://doi.org/10.1073/pnas.78.12.7436
Zahid MT, Shakoori FR, Zulifqar S, Jahan N, Shakoori AR (2014) A new ciliate species, Tetrahymena farahensis, isolated from the industrial wastewater and its phylogenetic relationship with other members of the genus Tetrahymena. Pakistan J Zool 46:1433–1445
Acknowledgments
We thank two anonymous reviewers for their valuable comments. We are also grateful to Ján Kočišek, Erik Mravec, Ivan Rúrik, Martin Sečanský, Eva Tirjaková, and Michal Veselický for their help with sampling, and to Ivan Rúrik for his help with some bioinformatics tools.
Funding
This work was supported by the Slovak Research and Development Agency under the contract no. APVV-15-0147, by the Grant Agency of the Ministry of Education, Science, Research and Sport of the Slovak Republic and Slovak Academy of Sciences under the Grant VEGA 1/0041/17, and by the Comenius University in Bratislava under the Grant UK/160/2020.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
All applicable institutional, national, and international guidelines for the care and use of animals were followed.
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Handling Editor: Julia Walochnik
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 2998 kb)
Rights and permissions
About this article
Cite this article
Rataj, M., Vďačný, P. Multi-gene phylogeny of Tetrahymena refreshed with three new histophagous species invading freshwater planarians. Parasitol Res 119, 1523–1545 (2020). https://doi.org/10.1007/s00436-020-06628-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-020-06628-0