Abstract
The protistan pathogens Cryptosporidium and Enterocytozoon bieneusi can cause significant intestinal diseases in animals and humans. However, limited information is available regarding prevalence and molecular characterization of Cryptosporidium and E. bieneusi in ruminants in Northern China. In this study, the overall prevalence of Cryptosporidium and E. bieneusi was 19.3% (62/321) and 28.97% (93/321) in dairy calves and 1.10% (9/818) and 13.57% (111/818) in sika deer (Cervus nippon) in four provinces in Northern China, respectively. The prevalence of Cryptosporidium and E. bieneusi in different factor groups was various. Five Cryptosporidium species/genotypes were identified, of which C. parvum, C. ryanae, C. bovis, and C. andersoni were only found in dairy calves, and only Cryptosporidium deer genotype was found in sika deer. Moreover, J, I, and BEB4 ITS genotypes of E. bieneusi were found in dairy calves, and six known genotypes (JLD-III, JLD-IX, JLD-VII, EbpC, BEB6, and I) and ten novel genotypes (namely LND-I and JLD-XV to JLD-XXIII) were found in sika deer in this study. Cryptosporidium parvum and E. bieneusi genotype J were identified as the predominant species/genotypes in dairy calves, whereas the predominance of Cryptosporidium spp. and E. bieneusi in sika deer was Cryptosporidium deer genotype and BEB6, respectively. The present study reported the prevalence and genotypes of Cryptosporidium and E. bieneusi in dairy calves and sika deer in four provinces in northern China. The present findings also suggest that investigated dairy calves and sika deer may play an important role in the transmission of E. bieneusi and Cryptosporidium to humans and other animals, and also in an effort to better understand the epidemiology of these enteric pathogens in China.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cryptosporidium and Enterocytozoon bieneusi are the most important enteropathogens, which can infect a wide range of animals including cattle and sika deer (Cervus nippon) (Huang et al. 2014, 2018; Parsons et al. 2015; Ma et al. 2015; Naguib et al. 2015; Zhang et al. 2016; Baroudi et al. 2018; Wu et al. 2018; Amer et al. 2019; Lombardelli et al. 2019). Transmission of Cryptosporidium and E. bieneusi were mainly through ingestion of water and food contaminated by infective spores or oocysts, respectively (Wang et al. 2017). In general, the two pathogens infection in humans may cause acute or chronic diarrhea. However, their co-infection with immune-compromised individuals could cause death (Checkley et al. 2015). Moreover, they also have negative effects on the growth and cognitive functions of children (Pawlowic et al. 2017).
More than 96 Cryptosporidium species/genotypes and 240 distinct ITS E. bieneusi genotypes have been identified worldwide (Zhang et al. 2015a, 2016). C. parvum, C. andersoni, C. ryanae, and C. bovis are the four commonest species which are responsible for infections in cattle (Chalmers and Katzer 2013), and C. parvum mostly infects pre-weaned calves and causes diarrhea (Santín et al. 2004; Mercado et al. 2015). Only 12 Cryptosporidium species/genotypes (including Cryptosporidium deer genotype) have also been found in cervids around the world (Huang et al. 2018). E. bieneusi genotype group 1 were the zoonotic genotypes and group 2 were bovine-specific genotypes (Ma et al. 2015), but some genotypes (I, J, and BEB4) from group 2 also have recently been reported in humans (Li et al. 2016a). Of these, C. hominis and C. parvum are the commonest species for human cryptosporidiosis and group 1 genotypes of E. bieneusi are responsible for the majority of human microsporidiosis (Naguib et al. 2018; Zhang et al. 2018a). Sika deer and cattle are the important reservoir hosts of Cryptosporidium and E. bieneusi (Hu et al. 2017; Huang et al. 2018; Santin and Fayer 2015). To date, more than 10 Cryptosporidium species/genotypes and 35 distinct E. bieneusi genotypes have been found in cattle and sika deer (Wang et al. 2011a; Del Coco et al. 2014; Ma et al. 2014; Zhao et al. 2015; Jiang et al. 2015; Li et al. 2016b; Huang et al. 2017, 2018). Although some information regarding prevalence and molecular characterization of Cryptosporidium and E. bieneusi in dairy cattle and sika deer have been reported, the information regarding the two pathogens in dairy cattle and sika deer in China is also limited.
In this study, we further investigated the prevalence of Cryptosporidium and E. bieneusi and infection in dairy cattle and sika deer (Cervus nippon) in northern China and identified species/genotypes/subtypes of these pathogens. Moreover, the present study also aims to estimate their zoonotic potential for transmission to humans.
Materials and methods
Specimen collection and preparation
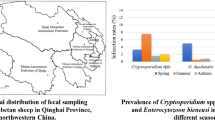
One thousand one hundred and thirty-nine fecal samples of dairy calves and sika deer (Cervus nippon) were randomly collected from farms from Heilongjiang Province (43° 26′~53° 33′ N, 121° 11′~135° 05′ E), Jilin Province (40° 50′~46° 19′ N, 121° 38′~131° 19′ E), Inner Mongolia (37° 24′~53° 33′ N, 97° 12′~126° 04′ E), Liaoning Province (38° 43′~43° 26′ N, 118° 53′~125° 46′ E), Northern China, between September 2017 and December 2018. All the sika deer are the same species. The pre-weaned sika deer was considered the young deer, the post-weaned sika deer was the adults. Every fresh fecal sample was collected from the rectum of each animal, and then placed into sterile gloves and transported back to the laboratory. The Stool DNA kit (OMEGA, USA) was used to extract the genomic DNAs of the fresh fecal samples. All the operations were strictly performed according to the manufacturer’s instructions. Then, DNA were stored at − 20 °C and analyzed by PCR. Information regarding geographic origin, breed, and age was recorded and listed in Tables 1, 2, 3, and 4.
PCR amplification
The PCR based on the SSU rRNA gene was performed to determine the Cryptosporidium spp. species (Zhang et al. 2015a). The nested PCR of the ITS region of the ribosomal RNA (rRNA) gene was used to identify the E. bieneusi genotype (Wang et al. 2016). PCR reaction (25 μl) composed of 1× Ex Taq buffer (Mg2+ free), 2 mM MgCl2, 200 μM each deoxy-ribonucleoside triphosphate (dNTP), 0.4 μM of each primer, 0.625 U of Ex Taq DNA polymerase (TAKARA, Japan), and 2 μl of DNA template. The cycling conditions were 5 min at 95 °C, 35 cycles of 45 s at 94 °C, 45 s at suitable temperature, and 1 min at 72 °C, followed by final extension at 72 °C for 10 min. Both positive and negative controls were included in each test. Amplification products were observed under UV light after electrophoresis in 1.5% agarose gel containing GoldView (Solarbio, China).
Sequencing and phylogenetic analyses
Positive secondary PCR products were sent to Sangon Biotech Company (Shanghai, China) for sequencing. The Cryptosporidium and E. bieneusi species/subtypes were determined by alignments with known reference sequences available in GenBank using the BLAST (http://www.ncbi.nlm.nih.gov/BLAST/), and the computer program ClustalX 1.83. The phylogenetic trees were re-constructed by using neighbor-joining (NJ) method (Kimura two-parameter model) in Mega 5.0 (http://www.megasoftware.net/), and bootstrapping was performed using 1000 replicates.
Statistical analysis
The variation in Cryptosporidium and E. bieneusi prevalence (y) of dairy calves and sika deer of different geographical location (x1), season (x2), and age (x3) were analyzed by Chi-square in Statistical Analysis System (SAS, Version 9.0), respectively. Results were considered statistically significant when P < 0.05. Odds ratios (ORs) and their 95% confidence intervals (95% CIs) were also provided.
Data availability statement
Representative nucleotide sequences were submitted to GenBank under accession numbers: MH754165-MH754181 and MN056193-MN056201 for Cryptosporidium and MH732748-MH732751 and MN056202-MN056217 for E. bieneusi.
Committee of Ethics and Animal Welfare
This study was approved by the Animal Ethics Committee of Heilongjiang Bayi Agricultural University (registration protocol Mar 2017). The dairy calves and sika deer, which the feces were collected, were handled in accordance with good animal practices required by the Animal Ethics Procedures and Guidelines of the People’s Republic of China.
Results
A total of 62 (19.31%) out of 321 examined fecal samples of dairy calves were test positive for Cryptosporidium (Table 1), whereas only 9 (1.10%) samples were Cryptosporidium positive among 858 sika deer samples (Table 2). All the Cryptosporidium-positive samples in sika deer were detected from farm 18 (n = 8) from Jilin Province and farm 25 (n = 1) from Heilongjiang Province. Pre-weaned dairy calves (22.75%, 38/167) had a higher Cryptosporidium infection rate than post-weaned dairy calves (15.58%, 24/154), but prevalence of Cryptosporidium in young sika deer (0/31) was lower than adults (9/787) (Table 3). The prevalence of Cryptosporidium in dairy calves collected in different seasons ranging from 1.03% (1/97) in summer to 33.67% (33/98) in spring, and the difference was statistically significant (P < 0.0001) (Table 3). Moreover, no Cryptosporidium-positive samples were found in female sika deer; whereas, 9 (1.29%) male sika deer were detected as Cryptosporidium positive. Sequencing and phylogenetic analysis suggested five Cryptosporidium species/genotypes were detected in the present study. Of which, C. parvum (n = 46), C. ryanae (n = 5), C. bovis (n = 7), and C. andersoni (n = 4) were only found in dairy calves (Table 1), and Cryptosporidium deer genotype (n = 9; 8 from farm 18 in Jilin Province and 1 from farm 25 in Heilongjiang Province) was only found in sika deer (Table 2; Fig. 1).
Of the 818 sika deer specimens analyzed, 111 (13.57%) were positive for E. bieneusi infection (Table 3). By sequence analysis, 6 known genotypes, namely JLD-III, JLD-IX, JLD-VII, EbpC, BEB6, and I) and 10 novel genotypes (namely LND-I and JLD-XV to JLD-XXIII) were identified (Table 2; Fig. 2). Prevalence of E. bieneusi in sika deer in different provinces ranges from 0% (0/106) in Inner Mongolia to 17.84% (96/538) in Jilin; the difference was statistically significant (P < 0.0001) (Table 3). Pre-weaned dairy calves (38.92%, 65/167) had a significant higher E. bieneusi infection rate than post-weaned dairy calves (18.18%, 28/154, P<0.0001), but prevalence of E. bieneusi in young sika deer (0%, 0/31) was lower than adults (14.10%, 111/787) (Table 3). Significant differences in infection rate were also observed among dairy calves collected during different seasons (Spring, 25.51%, 25/98; Summer, 20.62%, 20/97; Autumn, 45.35%, 39/86; Winter, 15.00%, 9/60; P = 0.0001; Table 3). In this study, prevalence of E. bieneusi in dairy calves was 28.97% (93/321), which was much higher than that in investigated sika deer (13.57%, 111/699), although only J, I, and BEB4 were identified in dairy calves (Table 1). Phylogenetic analysis revealed that EbpC was clustered into group 1d, and JLD-III, JLD-XIX, JLD-XVII, JLD-XXII, and JLD-XVI were classified in group 1a, and other 13 genotypes (JLD-IX, JLD-VII, BEB4, BEB6, I, J, LND-I, JLD-XV, JLD-IX, JLD-XXIII, JLD-XX, JLD-XVII, and JLD-XXI) were grouped in group 2 (Fig. 2).
In this study, 4 (0.49%, n = 818) samples were detected as co-infection of E. bieneusi and Cryptosporidium in sika deer. All the samples were adult sika deer collected from Shuangyang City. Moreover, 18 (5.61%) out of 321 dairy calves were positive to E. bieneusi and Cryptosporodium (Table 4).
Discussion
In this study, the overall prevalence of Cryptosporidium infection in sika deer was 1.1%, which was lower than that in sika deer in Henan (2.41%, 2/83) and Jilin (Wang et al. 2008) but higher than the 0% in red deer and Pere David’s deer in Zhengzhou, China (Wang et al. 2008). It is also lower than that of the 1.3–80% prevalence of Cryptosporidium in other deer species in the USA, Nepal, Spain, Japan, Czech Republic, UK, Australia, and some regions of China (Deng and Cliver 1999; Feng et al. 2012; García-Presedo et al. 2013; Santin and Fayer 2015; Wells et al. 2015; Koehler et al. 2016; Kotkova et al. 2016; Kato et al. 2016; Huang et al. 2018). The overall 23.01% Cryptosporidium prevalence was found in dairy calves in this study, which was higher than the reported infection rates in Heilongjiang (19.15%, 158/825) (Zhao et al. 2014a), Anhui (14.9%, 52/350), Shanghai (12.5%, 55/440), Jiangsu (20.7%, 251/1215) (Chen and Qiu 2012), Henan (21.5%, 172/801) (Wang et al. 2011b), Ningxia (5.5%, 92/1688; 1.69%, 23/1366) (Huang et al. 2014; Zhang et al. 2015a), Gansu (4.6%, 58/1257) (Zhang et al. 2015a), and Shaanxi (20.2%, 52/258) (Qi et al. 2015) but lower than that in Heilongjiang (47.62%, 72/151) (Zhang et al. 2013) and Shanghai (37%, 303/818) (Cai et al. 2017). The differences in prevalence might partially be related to the timing of specimen collection, the different susceptibility of different animal species, different sample sizes, the ecological conditions, the different susceptibility of domesticated deer and wild deer, and host health status. In addition, geographical and seasonal differences can also affect the prevalence of Cryptosporidium.
Since Cryptosporidium infection in dairy cattle and sika deer were recorded, 13 genotypes/genotypes (C. parvum, C. andersoni, C. suis-like, C. hominis, C. bovis, C. ryanae, C. ubiquitum, C. muris, Cryptosporidium deer genotype, Cryptosporidium muskrat II genotype, Cryptosporidium suis-like genotype, Cryptosporidium hominis-like genotype, Cryptosporidium caribou genotype) have been found in sika deer worldwide (Jellison et al. 2009; Perz and Le Blancq 2001; García-Presedo et al. 2013; Kváč et al. 2014; Wells et al. 2015; Koehler et al. 2016; Kotkova et al. 2016), and at least 9 Cryptosporidium species/genotypes, namely C. andersoni, C. bovis, C. parvum, C. ryanae, C. ubiquitum, C. meleagridis, C. xiaoi, C. serpentis, and C. suis-like (Liu et al. 2009; Wang et al. 2011b; Chen and Huang 2012; Zhang et al. 2013; Cui et al. 2014; Ma et al. 2014, 2015) have been detected in cattle in China. However, only Cryptosporidium deer genotype was found in sika deer (Table 2) and only four Cryptosporidium spp., namely C. parvum (n = 71), C. ryanae (n = 7), C. bovis (n = 7), and C. andersoni (n = 4) were found in dairy calves in the present study (Table 1), which indicated that the five Cryptosporidium spp. were endemic in sika deer and dairy calves in investigated areas, respectively. The results of the present study also indicated that C. parvum (in 6 farms) was the commonest species on dairy cattle farms, which were similar with previous observations in pre-weaned calves in Ningxia (Huang et al. 2014), Henan (Wang et al. 2011b), and Xinjiang. Because C. parvum was frequently found in humans (Naguib et al. 2018), dairy calf has been indicated as one of the most important potential resources for human cryptosporidiosis. Cryptosporidium deer genotype was only identified in deer, including Red deer (UK), Roe deer (UK), White-tailed deer (USA, Czech Republic), and sika deer (Japan, China) (Robinson et al. 2011; Santin and Fayer 2015; Wells et al. 2015; Kotkova et al. 2016; Kato et al. 2016; Huang et al. 2018), which suggested that it seems to be a host-adapted genotype. Other deer-derived Cryptosporidium species/genotypes (including C. parvum) were not found in this study, which suggested that more sample size should be collected for further studies.
Presently, the prevalence of E. bieneusi in sika deer was 13.57% (111/818), which was higher than that in white-tailed deer in New York, USA (12.2%, 6/49) (Guo et al. 2014), sika deer in Jilin Province (7.1%, 23/326), China (Zhang et al. 2016), sambar deer (Rusa unicolor) (0%, 0/3), fallow deer (Dama dama) (0%, 0/7), and Pere David’s deer (Elaphurus davidianus) (0%, 0/3) in Sichuan, China (Li et al. 2016b), and sambar deer (Rusa unicolor) (4.1%, 25/516), red deer (Cervus elaphus) (0%, 0/77), and fallow deer (Dama dama) (0%, 0/17) in Australia (Zhang et al. 2018b) but lower than the 17.04–75% prevalence of E. bieneusi in various species of deer in the USA (Santin and Fayer 2015), Korea (Amer et al. 2019), and China (Zhao et al. 2014b; Zhang et al. 2015b; Li et al. 2016b; Huang et al. 2017; Song et al. 2018). In the present study, only four farms were positive for E. bieneusi, with infection rates ranging between 5.71 and 47.96%. The highest infection rate in sika deer was found in farm 18 (47.96%, 94/196), while the lowest (0%) was in farms 17 (0/34), 19 (0/59), 20 (0/58), 21 (0/60), 22 (0/58), 24 (0/44), 27 (0/58), and 28 (0/106) (Table 2). The overall infection rate of E. bieneusi was 30.16% (122/386) in dairy calves, which was lower than that in cattle in Jilin (37.6%, 35/93) (Zhang et al. 2011), but it was higher than that in cattle in Shandong (2.0%, 3/148) (Ma et al. 2015) and Heilongjiang (30.1%, 40/133 and 5.89%, 31/526) (Zhao et al. 2015; Jiang et al. 2015) and calves in Shanghai (26.5%, 214/809) (Tang et al. 2018) and Shaanxi 19.68% (73/371) (Wang et al. 2016). The infection rate in this study was higher than in most parts of China, which may be due to the difference in detection methods, the different susceptibility of different animal species, diagnostic methods, study design, and climate. E. bieneusi-positive samples were detected in 13 dairy cattle farms (all in Heilongjiang) and 4 sika deer farms (2 from Jilin, 1 from Heilongjiang, 1 from Liaoning) in this study, which may be due to worse livestock management, host health status, and animal welfare in these farms. Moreover, all the investigated sika deer were collected in farms in the present study which differ from most samples of deer reported previously which are collected from wild deer. In addition, the breeding periods of these deer in each farm may also affect the pathogen transmission in the farm, which will be investigated in further studies.
Sequence analysis revealed that three known genotypes were detected in dairy calves, including J (n = 63), I (n = 36), and BEB4 (n = 23) (Table 1). We found genotype J was predominant in dairy calves in this study. Genotype J was also the dominant genotype found in dairy cattle in Xinjiang (Qi et al. 2017), Henan, and Ningxia (Li et al. 2016a). Meanwhile, similar results have been reported in pre-weaned calves in Shanghai (Tang et al. 2018). In contrast, other results reported genotype I was the dominant genotype in dairy cattle in Henan and Shandong (Ma et al. 2015) and in beef and dairy cattle in Shaanxi (Wang et al. 2016), genotype O was the dominant genotype in dairy cattle in Heilongjiang (Zhao et al. 2015), and genotype CHN3 was the dominant genotype in cows in Jilin (Zhang et al. 2011). The differences in the distribution of E. bieneusi genotypes in cattle from different areas in China may be because of geographical and host segregation. More than 60 E. bieneusi genotypes have been found in deer worldwide (Guo et al. 2014; Zhao et al. 2014b; Santin and Fayer 2015; Zhang et al. 2015b, 2016, 2018b; Li et al. 2016b; Huang et al. 2017; Song et al. 2018; Amer et al. 2019). However, only 16 genotypes, including 6 known genotypes (JLD-III, JLD-IX, JLD-VII, EbpC, BEB6, and I) and 10 novel genotypes (namely LND-I and JLD-XV to JLD-XXIII), were identified in this study. Genotypes BEB4, J, I, and BEB6, clustered in group 2, were widely found in lots of animals; however, all of them have also been found in humans (Huang et al. 2017; Yang et al. 2018). Genotype EbpC, known to cause human microsporidiosis (Liu et al. 2017; Huang et al. 2017), and genotypes JLD-III, JLD-XIX, JLD-XVII, JLD-XXII, and JLD-XVI clustered in group 1, have zoonotic potential. The findings suggested that the investigated dairy calves and sika deer may play an important role in the transmission of E. bieneusi to humans and other animals. Further studies will investigate the molecular characterization of E. bieneusi in these animals’ feeders in order to further prove the evidence of transmission of E. bieneusi between humans and dairy calves or sika deer.
Conclusion
Cryptosporidium and E. bieneusi infection frequently occurred in dairy cattle and sika deer in Northern China. Co-infection of Cryptosporidium and E. bieneusi was present in investigated dairy calves and sika deer. Five Cryptosporidium species/genotypes were identified, of which C. parvum, C. ryanae, C. bovis, and C. andersoni were only found in dairy calves, and only Cryptosporidium deer genotype was found in sika deer. Moreover, J, I, and BEB4 ITS genotypes of E. bieneusi were found in dairy calves; 6 known genotypes (JLD-III, JLD-IX, JLD-VII, EbpC, BEB6, and I) and 10 novel genotypes (namely LND-I, JLD-XV to JLD-XXIII) were found in sika deer in this study. C. parvum and E. bieneusi genotype J were identified as the predominant species/genotypes in dairy calves, whereas the predominance of Cryptosporidium spp. and E. bieneusi in sika deer was Cryptosporidium deer genotype and BEB6, respectively. Genotypes EbpC, JLD-III, JLD-XIX, JLD-XVII, JLD-XXII, and JLD-XVI were clustered in group 1; the members of which have zoonotic potential, which also suggested that the investigated dairy calves and sika deer may play an important role in the transmission of Cryptosporidium and E. bieneusi to humans.
References
Amer S, Kim S, Han JI, Na KJ (2019) Prevalence and genotypes of Enterocytozoon bieneusi in wildlife in Korea: a public health concern. Parasit Vectors 12:160
Baroudi D, Zhang H, Amer S, Khelef D, Roellig DM, Wang Y, Feng Y, Xiao L (2018) Divergent Cryptosporidium parvum subtype and Enterocytozoon bieneusi genotypes in dromedary camels in Algeria. Parasitol Res 117:905–910
Cai M, Guo Y, Pan B, Li N, Wang X, Tang C, Feng Y, Xiao L (2017) Longitudinal monitoring of Cryptosporidium species in pre-weaned dairy calves on five farms in Shanghai, China. Vet Parasitol 241:14–19
Chalmers RM, Katzer F (2013) Looking for Cryptosporidium: the application of advances in detection and diagnosis. Trends Parasitol 9:237–251
Checkley W, White AC Jr, Jaganath D, Arrowood MJ, Chalmers RM, Chen XM, Fayer R, Griffiths JK, Guerrant RL, Hedstrom L, Huston CD, Kotloff KL, Kang G, Mead JR, Miller M, Petri WA Jr, Priest JW, Roos DS, Striepen B, Thompson RC, Ward HD, Van Voorhis WA, Xiao L, Zhu G, Houpt ER (2015) A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for Cryptosporidium. Lancet Infect Dis 15:85–94
Chen F, Qiu H (2012) Identification and characterization of a chinese isolate of Cryptosporidium serpentis from dairy cattle. Parasitol Res 111:1785–1791
Chen F, Huang K (2012) Prevalence and molecular characterization of Cryptosporidium spp. in dairy cattle from farms in China. J Vet Sci 13:15–22
Cui Z, Wang R, Huang J, Wang H, Zhao J, Luo N, Li J, Zhang Z, Zhang L (2014) Cryptosporidiosis caused by Cryptosporidium parvum subtype IIdA15G1 at a dairy farm in Northwestern China. Parasit Vectors 7:529
Del Coco VF, Córdoba MA, Bilbao G, de Almeida CP, Basualdo JA, Santín M (2014) First report of Enterocytozoon bieneusi from dairy cattle in Argentina. Vet Parasitol 199:112–115
Deng MQ, Cliver DO (1999) Improved immunofluorescence assay for detection of Giardia and Cryptosporidium from asymptomatic adult cervine animals. Parasitol Res 85:733–736
Feng Y, Karna SR, Dearen TK, Singh DK, Adhikari LN, Shrestha A, Xiao L (2012) Common occurrence of a unique Cryptosporidium ryanae variant in zebu cattle and water buffaloes in the buffer zone of the Chitwan National Park, Nepal. Vet Parasitol 185:309–314
García-Presedo I, Pedraza-Díaz S, González-Warleta M, Mezo M, Gómez-Bautista M, Ortega-Mora LM, Castro-Hermida JA (2013) The first report of Cryptosporidium bovis, C. ryanae and Giardia duodenalis sub-assemblage A-II in roe deer (Capreolus capreolus) in Spain. Vet Parasitol 197:658–664
Guo Y, Alderisio KA, Yang W, Cama V, Feng Y, Xiao L (2014) Host specificity and source of Enterocytozoon bieneusi genotypes in a drinking source watershed. Appl Environ Microbiol 80:218–225
Hu S, Liu Z, Yan F, Zhang Z, Zhang G, Zhang L, Jian F, Zhang S, Ning C, Wang R (2017) Zoonotic and host-adapted genotypes of Cryptosporidium spp., Giardia duodenalis and Enterocytozoon bieneusi in dairy cattle in Hebei and Tianjin, China. Vet Parasitol 248:68–73
Huang J, Yue D, Qi M, Wang R, Zhao J, Li J, Shi K, Wang M, Zhang L (2014) Prevalence and molecular characterization of Cryptosporidium spp. and Giardia duodenalis in dairy cattle in Ningxia, northwestern China. BMC Vet Res 10:292
Huang J, Zhang Z, Yang Y, Wang R, Zhao J, Jian F, Ning C, Zhang L (2017) New genotypes of Enterocytozoon bieneusi isolated from sika deer and red deer in China. Front Microbiol 8:879
Huang J, Zhang Z, Zhang Y, Yang Y, Zhao J, Wang R, Jian F, Ning C, Zhang W, Zhang L (2018) Prevalence and molecular characterization of Cryptosporidium spp. and Giardia duodenalis in deer in Henan and Jilin, China. Parasit Vectors 11:239
Jellison KL, Lynch AE, Ziemann JM (2009) Source tracking identifies deer and geese as vectors of human-infectious Cryptosporidium genotypes in an urban/suburban watershed. Environ Sci Technol 43:4267–4272
Jiang Y, Tao W, Wan Q, Li Q, Yang Y, Lin Y, Zhang S, Li W (2015) Zoonotic and potentially host-adapted Enterocytozoon bieneusi genotypes in sheep and cattle in Northeast China and an increasing concern about the zoonotic importance of previously considered ruminant-adapted genotypes. Appl Environ Microbiol 81:3326–3335
Kato S, Yanagawa Y, Matsuyama R, Suzuki M, Sugimoto C (2016) Molecular identification of the Cryptosporidium deer genotype in the Hokkaido sika deer (Cervus nippon yesoensis) in Hokkaido, Japan. Parasitol Res 115:1463–1471
Koehler AV, Haydon SR, Jex AR, Gasser RB (2016) Cryptosporidium and Giardia taxa in faecal samples from animals in catchments supplying the city of Melbourne with drinking water (2011 to 2015). Parasit Vectors 9:315
Kotkova M, Nemejc K, Sak B, Hanzal V, Kvetonova D, Hlaskova L, Condlova S, McEvoy J, Kvac M (2016) Cryptosporidium ubiquitum, C. muris and Cryptosporidium deer genotype in wild cervids and caprines in the Czech Republic. Folia Parasitol (Praha) 25:63
Kváč M, McEvoy J, Stenger B, Clark M (2014) Cryptosporidiosis in other vertebrates. In: Cacciò SM, Widmer G (eds) Cryptosporidium: parasite and disease. Springer, Vienna, pp 237–323
Li J, Luo N, Wang C, Qi M, Cao J, Cui Z, Huang J, Wang R, Zhang L (2016a) Occurrence, molecular characterization and predominant genotypes of Enterocytozoon bieneusi in dairy cattle in Henan and Ningxia, China. Parasit Vectors 9:142
Li W, Deng L, Yu X, Zhong Z, Wang Q, Liu X, Niu L, Xie N, Deng J, Lei S, Wang L, Gong C, Zhou Z, Hu Y, Fu H, Xu H, Geng Y, Peng G (2016b) Multilocus genotypes and broad host-range of Enterocytozoon bieneusi in captive wildlife at zoological gardens in China. Parasit Vectors 9:395
Liu A, Wang R, Li Y, Zhang L, Shu J, Zhang W, Feng Y, Xiao L, Ling H (2009) Prevalence and distribution of Cryptosporidium spp. in dairy cattle in Heilongjiang Province, China. Parasitol Res 105:797–802
Liu H, Jiang Z, Yuan Z, Yin J, Wang Z, Yu B, Zhou D, Shen Y, Cao J (2017) Infection by and genotype characteristics of Enterocytozoon bieneusi in HIV/AIDS patients from Guangxi Zhuang autonomous region, China. BMC Infect Dis 17:684
Lombardelli JA, Tomazic ML, Schnittger L, Tiranti KI (2019) Prevalence of Cryptosporidium parvum in dairy calves and GP60 subtyping of diarrheic calves in central Argentina. Parasitol Res 118:2079–2086
Ma J, Cai J, Ma J, Feng Y, Xiao L (2014) Occurrence and molecular characterization of Cryptosporidium spp. in yaks (Bos grunniens) in China. Vet Parasitol 202:113–118
Ma J, Li P, Zhao X, Xu H, Wu W, Wang Y, Guo Y, Wang L, Feng Y, Xiao L (2015) Occurrence and molecular characterization of Cryptosporidium spp. and Enterocytozoon bieneusi in dairy cattle, beef cattle and water buffaloes in China. Vet Parasitol 207:220–227
Mercado R, Peña S, Ozaki LS, Fredes F, Godoy J (2015) Multiple Cryptosporidium parvum subtypes detected in a unique isolate of a Chilean neonatal calf with diarrhea. Parasitol Res 114:1985–1988
Naguib D, El-Gohary AH, Roellig D, Mohamed AA, Arafat N, Wang Y, Feng Y, Xiao L (2018) Molecular characterization of Cryptosporidium spp. and Giardia duodenalis in children in Egypt. Parasit Vectors 11:403
Naguib D, El-Gohary AH, Roellig D, Mohamed AA, Arafat N, Wang Y, Feng Y, Xiao L (2015) Epidemiology and molecular characterization of Cryptosporidium spp. in humans, wild primates, and domesticated animals in the Greater Gombe Ecosystem, Tanzania. PLoS Negl Trop Dis 9:e0003529
Parsons MB, Travis D, Lonsdorf EV, Lipende I, Roellig DM, Collins A, Kamenya S, Zhang H, Xiao L, Gillespie TR (2015) Epidemiology and molecular characterization of Cryptosporidium spp. in humans, wild primates, and domesticated animals in the Greater Gombe Ecosystem, Tanzania. PLoS Negl Trop Dis 9:e0003529
Pawlowic MC, Vinayak S, Sateriale A, Brooks CF, Striepen B (2017) Generating and maintaining transgenic Cryptosporidium parvum parasites. Curr Protoc Microbiol 46:20B.2.1–20B.2.32
Perz JF, Le Blancq SM (2001) Cryptosporidium parvum infection involving novel genotypes in wildlife from lower New York State. Appl Environ Microbiol 67:1154–1162
Qi MZ, Fang YQ, Wang XT, Zhang LX, Wang RJ, Du SZ, Guo YX, Jia YQ, Yao L, Liu QD, Zhao GH (2015) Molecular characterization of Cryptosporidium spp. in pre-weaned calves in Shaanxi Province, north-western China. J Med Microbiol 64:111–116
Qi M, Jing B, Jian F, Wang R, Zhang S, Wang H, Ning C, Zhang L (2017) Dominance of Enterocytozoon bieneusi genotype J in dairy calves in Xinjiang, Northwest China. Parasitol Int 66:960–963
Robinson G, Chalmers RM, Stapleton C, Palmer SR, Watkins J, Francis C, Kay D (2011) A whole water catchment approach to investigating the origin and distribution of Cryptosporidium species. J Appl Microbiol 111:717–730
Santín M, Trout JM, Xiao L, Zhou L, Greiner E, Fayer R (2004) Prevalence and age-related variation of Cryptosporidium species and genotypes in dairy calves. Vet Parasitol 122:103–117
Santin M, Fayer R (2015) Enterocytozoon bieneusi, Giardia, and Cryptosporidium infecting white-tailed deer. J Eukaryot Microbiol 62:34–43
Song Y, Li W, Liu H, Zhong Z, Luo Y, Wei Y, Fu W, Ren Z, Zhou Z, Deng L, Cheng J, Peng G (2018) First report of Giardia duodenalis and Enterocytozoon bieneusi in forest musk deer (Moschus berezovskii) in China. Parasit Vectors 11:204
Tang C, Cai M, Wang L, Guo Y, Li N, Feng Y, Xiao L (2018) Genetic diversity within dominant Enterocytozoon bieneusi genotypes in pre-weaned calves. Parasit Vectors 11:170
Wang R, Wang J, Sun M, Dang H, Feng Y, Ning C, Jian F, Zhang L, Xiao L (2008) Molecular characterization of the Cryptosporidium cervine genotype from a sika deer (Cervus nippon Temminck) in Zhengzhou, China and literature review. Parasitol Res 103:865–869
Wang R, Wang H, Sun Y, Zhang L, Jian F, Qi M, Ning C, Xiao L (2011a) Characteristics of Cryptosporidium transmission in preweaned dairy cattle in Henan, China. J Clin Microbiol 49:1077–1082
Wang R, Ma G, Zhao J, Lu Q, Wang H, Zhang L, Jian F, Ning C, Xiao L (2011b) Cryptosporidium andersoni is the predominant species in post-weaned and adult dairy cattle in China. Parasitol Int 60:1–4
Wang R, Zhao G, Gong Y, Zhang L (2017) Advances and perspectives on the epidemiology of bovine Cryptosporidium in China in the past 30 years. Front Microbiol 8:1823
Wang XT, Wang RJ, Ren GJ, Yu ZQ, Zhang LX, Zhang SY, Lu H, Peng XQ, Zhao GH (2016) Multilocus genotyping of Giardia duodenalis and Enterocytozoon bieneusi in dairy and native beef (Qinchuan) calves in Shaanxi province, northwestern China. Parasitol Res 115:1355–1361
Wells B, Shaw H, Hotchkiss E, Gilray J, Ayton R, Green J, Katzer F, Wells A, Innes E (2015) Prevalence, species identification and genotyping Cryptosporidium from livestock and deer in a catchment in the Cairngorms with a history of a contaminated public water supply. Parasit Vectors 8:66
Wu J, Han JQ, Shi LQ, Zou Y, Li Z, Yang JF, Huang CQ, Zou FC (2018) Prevalence, genotypes, and risk factors of Enterocytozoon bieneusi in Asiatic black bear (Ursus thibetanus) in Yunnan Province, Southwestern China. Parasitol Res 117:1139–1145
Yang H, Mi R, Cheng L, Huang Y, An R, Zhang Y, Jia H, Zhang X, Wang X, Han X, Chen Z (2018) Prevalence and genetic diversity of Enterocytozoon bieneusi in sheep in China. Parasit Vectors 11:587
Zhang X, Wang Z, Su Y, Liang X, Sun X, Peng S, Lu H, Jiang N, Yin J, Xiang M, Chen Q (2011) Identification and genotyping of Enterocytozoon bieneusi in China. J Clin Microbiol 49:2006–2008
Zhang XX, Tan QD, Zhou DH, Ni XT, Liu GX, Yang YC, Zhu XQ (2015a) Prevalence and molecular characterization of Cryptosporidium spp. in dairy cattle, northwest China. Parasitol Res 114:2781–2787
Zhang Z, Huang J, Karim MR, Zhao J, Dong H, Ai W, Li F, Zhang L, Wang R (2015b) Zoonotic Enterocytozoon bieneusi genotypes in Pere David’s deer (Elaphurus davidianus) in Henan, China. Exp Parasitol 155:46–48
Zhang XX, Cong W, Liu GH, Ni XT, Ma JG, Zheng WB, Zhao Q, Zhu XQ (2016) Prevalence and genotypes of Enterocytozoon bieneusi in sika deer in Jilin province, Northeastern China. Acta Parasitol 61:382–388
Zhang Y, Koehler AV, Wang T, Robertson GJ, Bradbury RS, Gasser RB (2018a) Enterocytozoon bieneusi genotypes in people with gastrointestinal disorders in Queensland and Western Australia. Infect Genet Evol 65:293–299
Zhang Y, Koehler AV, Wang T, Haydon SR, Gasser RB (2018b) First detection and genetic characterisation of Enterocytozoon bieneusi in wild deer in Melbourne’s water catchments in Australia. Parasit Vectors 11:2
Zhang W, Wang R, Yang F, Zhang L, Cao J, Zhang X, Ling H, Liu A, Shen Y (2013) Distribution and genetic characterizations of Cryptosporidium spp. in pre-weaned dairy calves in Northeastern China's Heilongjiang Province. PLoS ONE 8:e54857
Zhao W, Wang R, Zhang W, Liu A, Cao J, Shen Y, Yang F, Zhang L (2014a) MLST subtypes and population genetic structure of Cryptosporidium andersoni from dairy cattle and beef cattle in northeastern China’s Heilongjiang Province. PLoS ONE 9:e102006
Zhao W, Zhang W, Wang R, Liu W, Liu A, Yang D, Yang F, Karim MR, Zhang L (2014b) Enterocytozoon bieneusi in sika deer (Cervus nippon) and red deer (Cervus elaphus): deer specificity and zoonotic potential of ITS genotypes. Parasitol Res 113:4243–4250
Zhao W, Zhang W, Yang F, Zhang L, Wang R, Cao J, Shen Y, Liu A (2015) Enterocytozoon bieneusi in dairy cattle in the Northeast of China: genetic diversity of ITS gene and evaluation of zoonotic transmission potential. J Eukaryot Microbiol 62:553–560
Funding
Project support was provided by the Heilongjiang Provincial Key Laboratory of Prevention and Control of Bovine Diseases (PCBD201706). No competing financial interests exist. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the funding agencies.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Section Editor: Panagiotis Karanis
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Tao, WF., Ni, HB., Du, HF. et al. Molecular detection of Cryptosporidium and Enterocytozoon bieneusi in dairy calves and sika deer in four provinces in Northern China. Parasitol Res 119, 105–114 (2020). https://doi.org/10.1007/s00436-019-06498-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-019-06498-1