Abstract
Several rhoptry proteins (ROPs) have been confirmed to be critical virulence factors of Toxoplasma gondii strains from North America and Europe. The two active kinases ROP17 and ROP18, and pseudokinase ROP5 were thought to be the key determinants of parasites’ virulence in laboratory mice. Given the genetic diversity of Toxoplasma strains from different geographical regions, the virulence determinants in other strains, particularly the ones that are phylogenetically distant to the North American and European strains, are yet to be elucidated. In this study, we sought to examine the contribution of three known virulence factors to the virulence of a type I strain (T.gHB1) isolated from Central China. We deleted ROP17 and ROP18 individually, as well as in combination with GRA7 by the CRISPR-Cas9 system in this local isolate. Subsequent virulence tests in mice indicated that deletion of GRA7, ROP17, or ROP18 in T.gHB1showed similar attenuation in mice as the type I RH strain lacking the corresponding proteins. However, in contrast to the reported double knockouts in RH, double deletions of GRA7 plus ROP17 or GRA7 plus ROP18 in T.gHB1 did not show significant further virulence attenuation compared to the ROP17 or ROP18 single knockouts. These results indicated that GRA7, ROP18 and ROP17 may play different roles in virulence determination in genetically diverse strains of Toxoplasma.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Toxoplasma gondii is a widely prevalent intracellular parasite, which infects almost all warm-blooded animals and leads to the zoonotic toxoplasmosis (Dubey 2008; Dubey 2014). In addition, Toxoplasma gondii has been developed as a model organism for Apicomplexan parasites, which contain important pathogens such as Plasmodium and Cryptosporidium (Sibley 2003). T. gondii has an interesting population structure, with three main clonal types (Type I, II, and III) of strains prevalent in North America and Europe. For these clonal lineages, the strain types correlate well with their acute virulence in mice. Type I strains such as RH and GT1 are the most virulent, one parasite is enough to kill a mouse. Type III strains such as VEG are almost avirulent, whereas type II strains such as ME49 have intermediate virulence (Sibley and Ajioka 2008).
However, the genetic diversity of Toxoplasma strains was immediately seen after the isolation and characterization of strains from regions outside North American and Europe (Sibley et al. 2002; Ajzenberg et al. 2004; Lehmann et al. 2006; Khan et al. 2007; Chen et al. 2011; Su et al. 2012; Yang et al. 2013; Ajioka and Sibley 2014). In addition, the limited loci used for strain genotyping are not enough to distinguish virulence differences in different strains. For example, most (about 78 %) strains isolated from China belong to the Chinese I or ToxoDB#9 group, yet different isolates from this group may display very different acute virulence in laboratory mice, suggesting that the strain genotype may not accurately predict its virulence (Dubey et al. 2007; Chen et al. 2011; Jiang et al. 2013; Wang et al. 2013; Cheng et al. 2015). The T.gHB1 strain isolated from central China was determined to be a type I strain, and it displayed similarly acute virulence as RH and GT1 in mice, but the factors that determine its acute virulence have not yet been identified. Considering the geographical distance of T.gHB1 to classic type I strains such as RH, we were interested in the virulence determination mechanisms in this local strain.
When the host detects T. gondii infection, activated dendritic cells and macrophages secrete proinflammatory cytokines such as IL-12 to stimulate the secretion of IFN-γ from NK cells and CD4/CD8 T cells (Yap and Sher 1999). IFN-γ is the key factor for the host to control Toxoplasma infection. It induces the immunity-related GTPases (IRG) to attack the parasites by disrupting the integrity of the parasitiphorous vacuolar membrane (PVM) (Butcher et al. 2005; Hunn et al. 2008; Zhao et al. 2009a, b). These immunity-related GTPases include Irga6, Irgb6, Irgd, Irgm1, Irgm2, and Irgm3. They play different roles in defending Toxoplasma infection. Irgb6 and Irgb10 are assembled on the PVM once infection is detected, followed by Irga6 and other IRGs (Papic et al. 2008; Khaminets et al. 2010). However, loading of these IRG proteins to the PVM is significantly affected by Toxoplasma strain types. Avirulent strains accumulate IRGs on their PVM robustly, and as a consequence, their tachyzoites are efficiently eliminated by the host. In contrast, virulent strains secrete defense factors on the PVM that resist the clearance mechanisms from host cells; therefore, they are able to multiply and lead to acute toxoplasmosis (Taylor 2007; Steinfeldt et al. 2010).
The strategies used by Toxoplasma cells to resist the host’s immune clearance were extensively studied over the last 10 years. A set of proteins were identified to be involved in this process; these include rhoptry kinases ROP17 and ROP18, the pseudokinase ROP5, and the dense granule protein GRA7(Bradley et al. 2005; EI Hajj et al. 2007; Alaganan et al. 2014; Etheridge et al. 2014; Grzybowski et al. 2015). These proteins display significant polymorphisms among strains and are thought to be the main determinants of acute virulence in mice. Although the detailed mechanisms are still under investigation, these proteins lead to the phosphorylation of IRGs and disassemble them on the PVM (Etheridge et al. 2014). ROP18 directly phosphorylates Irga6 under the regulation of ROP5 and GRA7 (Hermanns et al. 2015). ROP17 phosphorylates Irgb6 and works independently of ROP5 (Etheridge et al. 2014). Individual inactivation of GRA7, ROP17, or ROP18 resulted in a modest attenuation of virulence in mice. However, double deletions of GRA7 plus ROP18 or ROP17 plus ROP18 in RH strain led to almost complete loss of virulence (Alaganan et al. 2014; Etheridge et al. 2014), suggesting that they work synergistically. On the other hand, disruption of ROP18 in RH and exotic South American strains resulted in different degrees of virulence attenuation, indicating that the same protein may have different contributions to virulence in genetically diverse strains (Behnke et al. 2015).
Given these differences in virulence determination in different strains, we sought to check the contribution of known virulence factors to the acute virulence of a local strain (T.gHB1) isolated from central China. GRA7, ROP17, and ROP18 were deleted individually or in combination by the CRISPR-Cas9 system, tested their growth in vitro, and assessed their virulence in mice. The results suggested that these factors do play roles in virulence determination in T.gHB1, but their degree of contribution and mode of action may be different than that in the RH strain.
Materials and methods
Propagation of T. gondii
All the genetically modified strains produced in this study were based on the parental strain T.gHB1 isolated from central China. Tachyzoites were cultured in human foreskin fibroblasts (HFF, ATCC SCRC-1041) cells grown with Dulbecco’s modified Eagle medium (DMEM) supplemented with 10 % fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin.
Plasmid construction
All the primers and plasmids used in this study are listed in Table S1 and Table S2 in the Supplemental Material. The strategies used for plasmid construction are described in Fig. 1a. The UPRT targeting guide RNA (gRNA) in pSAG1::CAS9-U6::sgUPRT (Addgene #54467) (Shen et al. 2014) was replaced with GRA7, ROP17, or ROP18 targeting gRNAs by PCR mutagenesis to generate gene-specific CRISPR plasmids. To make the homologous templates for gene replacements, left (L) and right (R) homologous arms of GRA7, ROP17, and ROP18 were amplified from the genomic DNA of the T.gHB1 strain respectively. Subsequently, the L and R arms of each gene, along with the selectable markers DHFR*-Ts or CAT amplified from the plasmids pUPRT-DHFR-D or pTubYFP, were cloned into pUC19 by multi-fragment cloning. All the plasmids were verified by enzyme digestion and DNA sequencing before use.
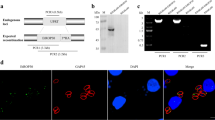
Schematic illustration of plasmids construction and gene disruption using CRISPR-Cas9. a Construct the homologous template plasmid containing homologous arms and selectable marker by one-step cloning (upper panel). Amplify the fragments with appropriate primers (arrows with color tails) and then clone them into a linearized vector by recombinase-mediated multi-fragments cloning. Change the sgRNA sequence in the CRISPR plasmid by PCR; the target-specific gRNA sequence (pink bar) was incorporated into both forward and reverse primers (lower panel). b CRISPR-Cas9-mediated gene disruption by homologous recombination in T. gondii. CRISPR-Cas9 is used to introduce double-strand DNA break in the target gene and L-DHFR*/CAT-R is used as templates for homologous recombination. PCR1/2/3 are used for identification of knockout clones
Production of GRA7 and ROP17 antisera
The coding sequences of GRA7 and ROP17 lacking signal peptides were amplified from the T.gHB1 cDNA (primers are listed in Table S1) and cloned into the pGEX-KG and pE-SUMO, respectively, by homologous recombination-mediated cloning, to make the expression plasmids pGEX-KG-GRA7 and pE-SUMO-ROP17. Subsequently, these two plasmids were introduced into E. coli BL21(DE3) pLysS (Transgen Biotech, China) competent cells for recombinant GST-GRA7 and HIS-SUMO-ROP17 expression and purification. After purification, 0.5 mg of each protein (GRA7 or ROP17, diluted to 1 mg/ml) emulsified with equal volume of Freund’s adjuvant (Sigma) was injected subcutaneously into 2.5-kg female rabbits to produce rabbit anti-GRA7 and ROP17 antisera (Khan et al. 2014).
Parasite transfection and selection
Freshly egressed tachyzoites were collected, purified by filtration through polycarbonate membranes with a pore size of 3 μm, washed with incomplete cytomix (120 mM KCl, 0.15 mM CaCl2, 10 mM K2HPO4/KH2PO4, 25 mM HEPES, 2 mM EGTA, 5 mM MgCl2, pH = 7.6 as the incomplete cytomix; addition of 2 mM ATP and 5 mM glutathione (final concentrations) makes complete cytomix (van den Hoff et al. 1992)), counted with a hemocytometer, and re-suspended in complete cytomix to achieve a concentration of 3.3 × 107 parasites/ml. For transfection, 300-μl parasites were mixed with 10-μg CRISPR/Cas9 plasmid DNA and 1-μg homologous templates (in the form of purified PCR products amplified from homologous template plasmids generated in Fig. 1a) and then electroporated in 4-mm gap cuvettes using a Bio-Rad electroporator (2 pulses of 1.5 kv, 50 Ω, and 25 μF). After transfection, tachyzoites were immediately transferred into T25 flasks seeded with confluent HFF cells and selected with corresponding drugs (pyrimethamine for DHFR-TS and chloramphenicol for CAT) (Jacot et al. 2014).
Single clones were obtained by limiting dilution in 96-well plates seeded with HFF host cells and expanded in T25 flasks. Diagnostic PCRs (PCR1, PCR2, and PCR3 as illustrated in Fig. 1b, the primers used are listed in Table S1) were used to screen individual clones. PCR reactions were performed using Taq or Pfu DNA polymerase in a 25-μl reaction mixture containing 0.5-μl genomic DNA extracted from single clones as templates. Subsequently, the products were examined by agarose gel electrophoresis.
Plaque assay
Two hundred freshly egressed tachyzoites were used to infect HFF monolayer seeded in 6-well plates and grown for 7 days. Then, the medium was aspirated, the wells rinsed with PBS, the cells fixed in 4 % paraformaldehyde for 10 min, stained with 2 % crystal violet for 10 min, and subsequently washed with water (Jacot et al. 2014). The number and sizes of plaques formed by each parasite strain were recorded as indications of parasite growth. All parasite strains were assayed three times independently, each with triplicates.
Immunofluorescence microscopy
Rabbit antisera (against GRA7 or ROP17) were used to examine the expression of target genes. HFF cells seeded on coverslips were infected with tachyzoites for 24 h. Subsequently, the samples were fixed with 4 % paraformaldehyde (in PBS), permeabilized with 0.25 % Triton X100 (in PBS), blocked with 1 % BSA, and then incubated with primary antibodies (rabbit anti-TgGRA7 or rabbit anti-TgROP17; mouse anti TgSAG1 was used as positive control) for 30 min. After washing the coverslips five times with PBS (3 min each time), secondary antibodies (Alexa Fluor 488-conjugated goat anti-mouse IgG, and Alexa Fluor 594-conjugated goat anti-rabbit IgG) were added and incubated for 15 min, counter-stained the nuclei with DAPI solution, and finally washed the coverslips with PBS four times. Images were obtained using the Axio Scope A1 fluorescence microscope (Zeiss, Germany) equipped with an AxioCam 503 mono camera (Zeiss, Germany).
Western blotting
Purified parasites were lysed in SDS sample buffer and boiled for 10 min. The protein samples were then separated on 8 % SDS-PAGE gels and transferred to PVDF membranes for Western blotting analysis. Rabbit anti TgGRA7 or TgROP17 was used to detect GRA7 and ROP17, respectively (rabbit anti TgALD was used as loading control). HRP-conjugated goat anti-rabbit IgG was used as the secondary antibody, and the blots were imaged on the imaging system (MF-Chemi BIS 3.2, Israel).
Virulence test in laboratory mice
Seven-week-old female ICR mice were purchased from Hubei Provincial Center for Disease Control and Prevention and maintained in the laboratory animal center of Huazhong Agricultural University. One hundred purified tachyzoites were injected intraperitoneally into mice (10 mice for each parasite strain), and the animals were monitored daily for signs of infection. All the statistical analyses and graphics were done with GraphPad prism 5.
Results
Deletion of GRA7 and ROP18 in T.gHB1
To check the contribution of GRA7 and ROP18 to the virulence of T.gHB1, they were individually knocked out using the CRISPR-Cas9 technology. The coding sequence of GRA7 was replaced with DHFR*-TS and selected with pyrimethamine, whereas ROP18 was substituted by the chloramphenicol acetyltransferase (CAT) mini gene and selected with chlorampenicol. Diagnostic PCR1 and PCR2 with expected bands of 1498 and 1335 bp in T.gHB1 △GRA7, and 1170 bp and 1117 bp in T.gHB1 △ROP18 (Fig. 2a) confirmed the correct integration of selectable markers to replace GRA7 and ROP18, respectively. Loss of the 346 bp band in GRA7 mutant and the 1665 bp band in ROP18 mutant in PCR3 (Fig. 2a) further confirmed the corresponding gene deletion. These results demonstrated that GRA7 and ROP18 in T.gHB1 were successfully knocked out.
Phenotypic analysis of the T.gHB1 △GRA7 and T.gHB1 △ROP18 mutants. a Diagnostic PCR to verify the homologous replacement of GRA7 by DHFR* in clone F7 and ROP18 by CAT in clone B3. PCR1 and PCR2 examined the integration of DHFR* or CAT into corresponding genes, whereas PCR3 checked the deletion of GRA7 or ROP18 sequences. b Plaque assay checking the growth of T.gHB1 and deletion mutants in vitro. c Survival curves of ICR mice infected with 100 purified tachyzoites (by i.p. inoculation) of indicated strains (n = 10 per group). ***P < 0.0001, Gehan-Breslow-Wilcoxon test
Characterization of the GRA7 and ROP18 knockouts in vitro and in vivo
In order to test the growth of deletion mutants in vitro, plaque assay was performed to compare the growth of mutants to WT parasites. After plaque development for 7 days, the number of plaques and the size of individual plaques formed by each strain were recorded (Fig. 2b). Comparison of these strains indicated that there is no obvious difference in plaque formation between WT and T.gHB1△GRA7 or T.gHB1△ROP18 mutants, suggesting that GRA7 or ROP18 deletion did not affect parasite growth in vitro.
To check the impact of GRA7 and ROP18 deletions on parasite growth and virulence in vivo, 100 freshly egressed tachyzoites of WT or mutant strains were injected into mice and their survival monitored daily. The results indicated that both GRA7 and ROP18 knockouts exhibited a slight virulence attenuation (Fig. 2c). Compared to wild-type parasite infection, mice injected with T.gHB1△GRA7 delayed animal death for 1–2 days, whereas T.gHB1△ROP18-infected mice survived three to four more days.
GRA7 and ROP18 double knockout strain construction
GRA7 was thought to regulate the function of ROP18 in the process of defending the host’s immune clearance. Combined deletion of GRA7 and ROP18 in type I strain RH increased the virulence defects of the single mutants (Alaganan et al. 2014; Hermanns et al. 2015). We were interested in whether deleting both GRA7 and ROP18 in T.gHB1 would affect its virulence in a similar way as in RH. To this end, a GRA7 ROP18 double knockout strain in T.gHB1 was constructed by deleting ROP18 in T.gHB1 △GRA7. After selection with pyrimethamine and chloramphenicol, diagnostic PCRs were used to identify positive clones that had DHFR*-Ts and CAT integrated at the correct place and the complete removal of GRA7 and ROP18 coding sequences (Fig. 3a). Such double knockouts were obtained with regular frequency.
Confirmation and analysis of the GRA7 and ROP18 double deletion mutant in T.gHB1. The GRA7 ROP18 double knockout strain was constructed by deleting ROP18 in T.gHB1 ΔGRA7. a Diagnostic PCR checking the homologous replacement of ROP18 by CAT four randomly selected clones (1–4). Clones #1 and #4 were clean double knockouts. b Plaque assay on the T.gHB1 and T.gHB1△GRA7△ROP18 strains. c Survival curves of ICR mice challenged with 100 purified tachyzoites of T.gHB1 or T.gHB1△GRA7△ ROP18 (n = 10 per group) by i.p injection. ***P < 0.0001; **P < 0.01, ns not significant, Gehan-Breslow-Wilcoxon test
Characterization of the GRA7 and ROP18 double knockout in vitro and in vivo
As mentioned above, individual deletion of GRA7 or ROP18 did not affect parasite growth in vitro (Fig. 2b). To test whether deletion of both genes has any effect on parasite growth, a plaque assay was done as described above. Plaquing results (Fig. 3b) indicated that the double knockout strain displayed similar growth as WT and single knockout strains, further confirming that these two genes do not contribute to parasite growth in vitro. To examine the virulence of the double knockout in vivo, ICR mice were infected with WT or knockout mutants through intraperitoneal injection and animal survival monitored. Surprisingly, we found that double deletion of GRA7 and ROP18 did not cause further virulence attenuation than the single knockouts (Fig. 3c); this is different than what was observed in the RH strain (Alaganan et al. 2014) but is consistent with the fact that GRA7 and ROP18 function in the same pathway to resist host clearance (Hermanns et al. 2015).
Constructing ROP17 single and GRA7 ROP17 double knockouts in T.gHB1
ROP17 plays a role similar to ROP18 to phosphorylate IRGs and to protect the integrity of the PVM. Deletion of ROP17 in the RH strain led to a slight virulence attenuation, indicating that ROP17 is another virulence factor. To test the role of ROP17 in T.gHB1, we generated a mutant lacking ROP17 using the CRISPR-Cas9 strategy mentioned above to replace the ROP17 coding sequence with CAT. We also deleted ROP17 in T.gHB1△GRA7 to generate the double knockout T.gHB1 △GRA7 △ROP17. Using similar diagnostic PCRs described above, we identified positive knockout clones and confirmed the correct gene replacements (Fig. 4a). In addition, we further confirmed the deletion of GRA7 and ROP17 at protein levels by immunofluorescence assay (IFA) and Western blotting. As shown in Fig. 4b–d, both Western blotting and IFA analysis showed the disappearance of target proteins in corresponding mutants, confirming the successful construction of these knockout strains.
Confirmation of ROP17 deletion mutants. a Diagnostic PCR on single clones of T.gHB1 △ROP17 and T.gHB1 △GRA7 △ROP17 (three clones for each strain) to check the replacement of ROP17 by CAT. b, c Immunofluorescent staining checking the expression of GRA7 and ROP17 in WT vs mutant strains; SAG1 was used as a positive control. d Western blotting to detect GRA7 and ROP17 expression in wild-type and mutant strains; TgALD was used as a loading control
Characterization of GRA7 and ROP17 knockouts in vitro and in vivo
Plaque assay was used to estimate the growth of knockout strains in vitro, as described above. The plaquing results (Fig. 5a) indicated that both △ROP17 single and △GRA7 △ROP17 double mutants produced a similar number of plaques as the WT strain, but the plaques were smaller in size, suggesting that ROP17 may play a role to support robust parasite growth in vitro. When challenged with 100 parasites, ICR mice infected with T.gHB1 △ROP17 survived a few more days than mice infected with WT parasites (Fig. 5b). Deleting GRA7 in addition to ROP17 knockout did not lead to further attenuation of the virulence. These results suggested that GRA7 and ROP17 are indeed important virulence factors in T.gHB1, but their contribution is partial and there must be other factors involved in determining the virulence of T.gHB1 in mice, which deserve further investigation.
Discussion
In this study, the roles of selected virulence factors in a local Toxoplasma strain isolated from central China were examined. The focus was on the dense granule protein GRA7 (Coppens et al. 2006) and two rhoptry kinases ROP17 and ROP18 (Etheridge et al. 2014). Using the CRISPR-Cas9 genome editing tool (Shen et al. 2014), GRA7, ROP17, and ROP18 were knocked out individually or in combination. The results suggested that ROP17 might be important for optimal parasite growth in vitro, but ROP18 and GRA7 did not play significant roles in this regard. Virulence tests in ICR mice indicated that all three proteins contributed to the acute virulence of parasites in mice. However, compared with previous studies in RH or South American strains, deletion of GRA7, ROP17, or ROP18 in T.gHB1 seemed to attenuate the virulence to a lesser degree. This difference is even more dramatic in double knockout mutants. Double deletion of GRA7 and ROP18 in the RH strain caused significantly greater virulence attenuation than each single mutant (Alaganan et al. 2014). In contrast, GRA7 and ROP18 double deletion in T.gHB1 resulted in very similar virulence attenuation as the ΔGRA7 or ΔROP18 single mutants, suggesting that the virulence determination mechanisms may be different in T.gHB1 than in RH and other Toxoplasma strains.
One contribution of ROP5 as a major virulence determinant was thought to be its interactions with ROP17 and ROP18, which are active serine/threonine kinases known to target and phosphorylate mouse IRG proteins and protect the integrity of the PVM (Etheridge et al. 2014). In addition, GRA7 was also found to be in complex with ROP5/ROP18 and play a supporting role to reduce the loading of IRG on the PVM (Hermanns et al. 2015). Previous studies showed that deleting ROP5 in RH resulted in almost complete loss of virulence, but △GRA7, △ROP17, and △ROP18 single mutants displayed only modest virulence attenuation in mice (Alaganan et al. 2014; Etheridge et al. 2014; Zhao and Yap 2014). However, double deletions of GRA7 plus ROP18 or ROP17 plus ROP18 resulted in much greater virulence attenuation. According to these findings, it was proposed that GRA7, ROP17, and ROP18 function together in a synergistic manner to defend IRG loading. Nonetheless, the contribution of each factor in resisting IRG and virulence determination may be different in different strain backgrounds. For example, recent studies showed that knocking out ROP18 in South American strains led to much greater virulence attenuation than that in RH (Behnke et al. 2015).
From the perspective of innate immunity, IRGs induced by IFN-γ provided cell-autonomous resistance to inhibit the proliferation of various avirulent T. gondii strains, but not virulent ones. The accumulation of IRGs at the PVM differs significantly between avirulent and virulent strains. Polymorphisms in ROP18 and ROP5 are thought to be largely responsible for these differences (Jensen et al. 2015). ROP18 is an important inhibitor of the recruitment of Irgb6 and Irga6 (Fentress et al. 2010; Fentress et al. 2012; Hermanns et al. 2015). When infected with avirulent strains, Irgb6 accumulates on the PVM as an early sentinel to clear the parasite. If the parasite expresses a type I ROP18, less Irgb6, as well as other IRGs such as Irga6 and Irgb10, would be observed (Zhao et al. 2009a, b; Fentress et al. 2010). Association of GRA7 with ROP5 has a direct effect on ROP18 to phosphorylate Irga6 (Hermanns et al. 2015). ROP17 phosphorylates Irgb6 and does so independent of ROP5. ROP18, with the assistance of GRA7 and ROP5, phosphorylates Irga6 and Irgb10 (Fentress et al. 2010; Steinfeldt et al. 2010). Upon phosphorylation, these IRG complexes are disassembled from the PVM. Besides these factors, there might be other effectors involved to resist the host’s immune clearance.
In this study, checking the contribution of GRA7, ROP17, and ROP18 to the acute virulence of the local strain T.gHB1, we found that although these factors do affect parasite virulence more or less, the degree of their contribution and the mode of action may be different than in other strains, such as RH. This is consistent with a recent study on South American strains (Behnke et al. 2015). Particularly, double deletion of GRA7 and ROP18 did not result in further attenuation than the single knockout mutants. This is different than in RH, in which deletion of both GRA7 and ROP18 caused much greater virulence attenuation than the corresponding GRA7 or ROP18 single knockouts. These results may imply that although all these proteins are involved in resisting the host’s immune clearance, the detailed working mechanisms may be different in genetically diverse strains. For the T.gHB1 isolate, since disruption of GRA7, ROP18, and ROP17, individually or in combination, did not attenuate the virulence completely, it suggests that other factors or mechanisms might be involved, which deserve further investigation.
References
Ajioka JW, Sibley LD (2014) Chapter 16—Development and application of classical genetics in Toxoplasma gondii. Toxoplasma Gondii:551-576
Ajzenberg D, Bañuls AL, Su C, Dumètre A, Demar M, Carme B, Dardé ML (2004) Genetic diversity, clonality and sexuality in Toxoplasma gondii. Int J Parasitol 34(10):1185–1196
Alaganan A, Fentress SJ, Tang K, Wang Q, Sibley LD (2014) Toxoplasma GRA7 effector increases turnover of immunity-related GTPases and contributes to acute virulence in the mouse. Proc Natl Acad Sci U S A 111(3):1126–1131
Behnke MS, Khan A, Lauron EJ, Jimah JR, Wang Q, Tolia NH, Sibley LD (2015) Rhoptry proteins ROP5 and ROP18 are major murine virulence factors in genetically divergent South American strains of Toxoplasma gondii. PLoS Genet 11(8), e1005434
Bradley PJ, Ward C, Cheng SJ, Alexander DL, Coller S, Coombs GH, Dunn JD, Ferguson DJ, Sanderson SJ, Wastling JM, Boothroyd JC (2005) Proteomic analysis of rhoptry organelles reveals many novel constituents for host-parasite interactions in Toxoplasma gondii. J Biol Chem 280(40):34245–34258
Butcher BA, Greene RI, Henry SC, Annecharico KL, Weinberg JB, Denkers EY, Sher A, Taylor GA (2005) p47 GTPases regulate Toxoplasma gondii survival in activated macrophages. Infect Immun 73(6):3278–3286
Chen ZW, Gao JM, Huo XX, Wang L, Yu L, Halm-Lai F, Xu YH, Song WJ, Hide G, Shen JL, Lun ZR (2011) Genotyping of Toxoplasma gondii isolates from cats in different geographic regions of China. Vet Parasitol 183(1-2):166–170
Cheng W, Liu F, Li M, Hu X, Chen H, Pappoe F, Luo Q, Wen H, Xing T, Xu Y, Shen J (2015) Variation detection based on next-generation sequencing of type Chinese 1 strains of Toxoplasma gondii with different virulence from China. BMC Genomics 16:888
Coppens I, Dunn JD, Romano JD, Pypaert M, Zhang H, Boothroyd JC, Joiner KA (2006) Toxoplasma gondii sequesters lysosomes from mammalian hosts in the vacuolar space. Cell 125(2):261–274
Dubey JP (2008) The history of Toxoplasma gondii-the first 100 years. J Eukaryot Microbiol 55(6):467–475
Dubey JP (2014) Chapter 1-the history and life cycle of toxoplasma gondii. Toxoplasma Gondii 1-17
Dubey JP, Zhu XQ, Sundar N, Zhang H, Kwok OC, Su C (2007) Genetic and biologic characterization of Toxoplasma gondii isolates of cats from China. Vet Parasitol 145(3-4):352–356
EI Hajj H, Lebrun M, Arold ST, Vial H, Labesse G, Dubremetz JF (2007) Rop18 is a rhoptry kinase controlling the intracellular proliferation of toxoplasma gondii. PLoS Pathog 3(2), e14
Etheridge RD, Alaganan A, Tang K, Lou HJ, Turk BE, Sibley LD (2014) The Toxoplasma pseudokinase ROP5 forms complexes with ROP18 and ROP17 kinases that synergize to control acute virulence in mice. Cell Host Microbe 15(5):537–550
Fentress SJ, Behnke MS, Dunay IR, Mashayekhi M, Rommereim LM, Fox BA, Bzik DJ, Taylor GA, Turk BE, Lichti CF, Townsend RR, Qiu W, Hui R, Beatty WL, Sibley LD (2010) Phosphorylation of immunity-related GTPases by a Toxoplasma gondii-secreted kinase promotes macrophage survival and virulence. Cell Host Microbe 8(6):484–95
Fentress SJ, Steinfeldt T, Howard JC, Sibley LD (2012) The arginine-rich N-terminal domain of ROP18 is necessary for vacuole targeting and virulence of Toxoplasma gondii. Cell Microbiol 14(12):1921–1933
Grzybowski MM, Dziadek B, Gatkowska JM, Dzitko K, Długońska H (2015) Towards vaccine against toxoplasmosis: evaluation of the immunogenic and protective activity of recombinant ROP5 and ROP18 Toxoplasma gondii proteins. Parasitol Res 114(12):4553–4563
Hermanns T, Müller UB, Könen-Waisman S, Howard JC, Steinfeldt T (2015) The Toxoplasma gondii rhoptry protein ROP18 is an Irga6-specific kinase and regulated by the dense granule protein GRA7. Cell Microbiol 18(2):244–59
Hunn JP, Koenen-Waisman S, Papic N, Schroeder N, Pawlowski N, Lange R, Kaiser F, Zerrahn J, Martens S, Howard JC (2008) Regulatory interactions between IRG resistance GTPases in the cellular response to Toxoplasma gondii. EMBO J 27(19):2495–2509
Jacot D, Meissner M, Sheiner L, Soldati-Favre D, Striepen B (2014) Genetic manipulation of Toxoplasma gondii - Toxoplasma Gondii (Second Edition) - Chapter 17. Toxoplasma Gondii:577–611
Jensen KD, Camejo A, Melo MB, Cordeiro C, Julien L, Grotenbreg GM, Frickel EM, Ploegh HL, Young L, Saeij JP (2015) Toxoplasma gondii superinfection and virulence during secondary infection correlate with the exact ROP5/ROP18 allelic combination. MBio 6(2), e02280
Jiang HH, Huang SY, Zhou DH, Zhang XX, Su C, Deng SZ, Zhu XQ (2013) Genetic characterization of Toxoplasma gondii from pigs from different localities in China by PCR-RFLP. Parasitol Vectors 6(2):207–215
Khaminets A, Hunn JP, Könen-Waisman S, Zhao YO, Preukschat D, Coers J, Boyle JP, Ong YC, Boothroyd JC, Reichmann G, Howard JC (2010) Coordinated loading of IRG resistance GTPases on to the Toxoplasma gondii parasitophorous vacuole. Cell Microbiol 12(7):939–961
Khan A, Fux B, Su C, Dubey JP, Darde ML, Ajioka JW, Rosenthal BM, Sibley LD (2007) Recent transcontinental sweep of Toxoplasma gondii driven by a single monomorphic chromosome. Proc Natl Acad Sci U S A A104(37):14872–14877
Khan MK, He L, Zhang W, Wang Y, Tao Q, Song Q, Sajid MS, Yu Q, Hu J, Fang R, Hu M, Zhou Y, Zhao J (2014) Identification of two novel HSP90 proteins in Babesia orientalis: molecular characterization, and computational analyses of their structure, function, antigenicity and inhibitor interaction. Parasitol Vectors 7(1):1–15
Lehmann T, Marcet PL, Graham DH, Dahl ER, Dubey JP (2006) Globalization and the population structure of Toxoplasma gondii. Proc Natl Acad Sci U S A 103(30):11423–11428
Papic N, Hunn JP, Pawlowski N, Zerrahn J, Howard JC (2008) Inactive and active states of the interferon-inducible resistance GTPase, Irga6, in vivo. J Biol Chem 283(46):32143–32151
Shen B, Brown KM, Lee TD, Sibley LD (2014) Efficient gene disruption in diverse strains of Toxoplasma gondii using CRISPR/CAS9. MBio 5(3):e01114–14
Sibley LD (2003) Recent origins among ancient parasites. Vet Parasitol 115(2):185–198
Sibley LD, Ajioka JW (2008) Population structure of Toxoplasma gondii: clonal expansion driven by infrequent recombination and selective sweeps. Annu Rev Microbiol 62:329–351
Sibley LD, Mordue DG, Su C, Robben PM, Howe DK (2002) Genetic approaches to studying virulence and pathogenesis in Toxoplasma gondii. Philos Trans R Soc Lond B Biol Sci 357(1417):81–88
Steinfeldt T, Könen-Waisman S, Tong L, Pawlowski N, Lamkemeyer T, Sibley LD, Hunn JP, Howard JC (2010) Phosphorylation of mouse immunity-related GTPase (IRG) resistance proteins is an evasion strategy for virulent Toxoplasma gondii. PLoS Biol 8(12), e1000576
Su C, Khan A, Zhou P, Majumdar D, Ajzenberg D, Dardé ML, Zhu XQ, Ajioka JW, Rosenthal BM, Dubey JP, Sibley LD (2012) Globally diverse Toxoplasma gondii isolates comprise six major clades originating from a small number of distinct ancestral lineages. Proc Natl Acad Sci U S A 109(15):5844–5849
Taylor GA (2007) IRG proteins: key mediators of interferon-regulated host resistance to intracellular pathogens. Cell Microbiol 9(5):1099–1107
van den Hoff MJ, Moorman AF, Lamers WH (1992) Electroporation in ‘intracellular’ buffer increases cell survival. Nucleic Acids Res 20(11):2902–2902
Wang L, Chen H, Liu D, Huo X, Gao J, Song X, Xu X, Huang K, Liu W, Wang Y, Lu F, Lun ZR, Luo Q, Wang X, Shen J (2013) Genotypes and mouse virulence of Toxoplasma gondii isolates from animals and humans in China. PLoS One 8(5), e53483
Yang N, Farrell A, Niedelman W, Melo M, Lu D, Julien L, Marth GT, Gubbels MJ, Saeij JP (2013) Genetic basis for phenotypic differences between different Toxoplasma gondii type I strains. BMC Genomics 14(2):1–19
Yap GS, Sher A (1999) Effector cells of both nonhemopoietic and hemopoietic origin are required for interferon (IFN)-gamma- and tumor necrosis factor (TNF)-alpha-dependent host resistance to the intracellular pathogen, Toxoplasma gondii. J Exp Med 189(7):1083–1092
Zhao Y, Yap GS (2014) Toxoplasma’s arms race with the host interferon response: a Ménage à Trois of ROPs. Cell Host Microbe 15(5):517–518
Zhao Y, Ferguson DJ, Wilson DC, Howard JC, Sibley LD, Yap GS (2009a) Virulent Toxoplasma gondii evade immunity-related GTPase-mediated parasite vacuole disruption within primed macrophages. J Immunol 182(6):3775–3781
Zhao YO, Khaminets A, Hunn JP, Howard JC (2009b) Disruption of the Toxoplasma gondii parasitophorous vacuole by IFNgamma-inducible immunity-related GTPases (IRG proteins) triggers necrotic cell death. PLoS Pathog 5(2), e1000288
Acknowledgments
The authors thanked Dr. Aditi Alaganan (Pasteur Institute, Paris, France) for her critical reading and valuable suggestions for the improvement of this manuscript. This work was supported by the National Natural Science Foundation of China (Grants No. 31372429 and No. 31572510), National Key Basic Research Program (973program) of China (Grant No. 2015CB150302), and Project 2662015PY048 from the Fundamental Research Funds for the Central Universities in China.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Human and animals rights informed consent
All the ICR mice and rabbits used in all experiments were purchased from Laboratory Animals Research Centre of Hubei Province (permit number SCXK (E)-2015-0018) and raised under the standard conditions according to the Regulations for the Administration of Affairs Concerning Experimental Animals. The animal experiments were approved by the ethical committee of Huazhong Agricultural University according to the Regulations of the Care and Use of Laboratory Animals in China.
Rights and permissions
About this article
Cite this article
Zhang, W., Li, L., Xia, N. et al. Analysis of the virulence determination mechanisms in a local Toxoplasma strain (T.gHB1) isolated from central China. Parasitol Res 115, 3807–3815 (2016). https://doi.org/10.1007/s00436-016-5141-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-016-5141-z