Abstract
Numerous studies addressed the diversity of bird Plasmodium and Haemoproteus parasites. However, a few have been carried out in continental avian hotspot regions such as Brazil, a country with markedly different biomes, including Amazon, Brazilian Savanna, Atlantic Forest, Caatinga, Pantanal, and Pampas. We present the first study on hemosporidian (Haemosporida) parasites in free-living birds from an Atlantic Forest fragment where more than 80 avian species have been reported. Within this area, the São Paulo Zoo locates, and it is the fourth largest zoo in the world and the largest in Latin America. A total of 133 free-living bird samples representing 12 species were collected in the zoo, with the overall hemosporidian prevalence of 18 % by PCR-based diagnostics. Twenty-four positive PCR signals were reported from four different bird species, including migratory ones. Columba livia, an urban species, considered nowadays a pest in big cities, showed 100 % prevalence of Haemoproteus spp., mainly Haemoproteus columbae. We discuss the epidemiological importance of new parasites introduced by migratory birds in the São Paulo Zoo area and the risk it poses to the captive species, which are natives or exotics. We also warn about the influence these parasites can have on the biodiversity and the structure of host populations by altering the competitive interaction between the free-living and the captive birds.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Avian hemosporidians (Sporozoa, Haemosporida) are protozoan parasites with over 250 species of the genera Plasmodium, Haemoproteus, Leucocytozoon, and Fallisia, which are described in birds. They are transmitted exclusively by dipteran blood-sucking insects (Garnham 1966; Atkinson et al. 2008; Valkiūnas 2005). Although these parasites are cosmopolitan in distribution, a review of global diversity of Plasmodium and Haemoproteus genetic lineages has shown a substantial geographical bias in studies on continental avian non-hotspot regions in Europe and North America whose focus has been mainly on the Passeriformes species (Clark et al. 2014).
Brazil hosts one of the greatest diversities of fauna and flora in the world, consisting of markedly different biomes (Amazon, Cerrado, Atlantic Forest, Caatinga, Pantanal, and Pampas) and is also considered a hotspot for avian richness (Orme et al. 2005). However, more specifically for the Atlantic Forest, a few studies on the prevalence of blood parasites in birds have been conducted (Ribeiro et al. 2005; Belo et al. 2009; Sebaio et al. 2012; Lacorte et al. 2013; Motta et al. 2013). São Paulo is the second Brazilian state to hold large remaining areas of Atlantic forest (http://mapas.sosma.org.br/dados/) that is one of world’s biodiversity hotspots (Ribeiro et al. 2009). In the county of São Paulo, the state’s capital, there is an area of 26,664 km2 of forest that includes a state park named “Fontes do Ipiranga” (Parque Estadual das Fontes do Ipiranga—PEFI), where over 80 avian species have been reported. Within the PEFI, the São Paulo Zoo locates. This is the fourth largest zoo in the world and the largest in Latin America. This area has a high density and diversity of mosquitoes that can transmit avian malaria (Bueno et al. 2010), and Plasmodium (Novyella) nucleophilum has been identified and characterized molecularly from a captive goose born at the Zoo (Chagas et al. 2013). Biting midges (Ceratopogonidae) and hippoboscid flies (Hippoboscidae), which are vectors of avian Haemoproteus species, have also been reported in the São Paulo State (Felippe-Bauer and Oliveira 2001; Arzua and Valim 2010). In this paper, we present results of the first study on hemosporidian parasites in free-living birds from this area. We also discuss the effects of new parasites introduced by migratory birds to the São Paulo Zoo area and the possible risk of such introduction to the exotic captive bird species.
Materials and methods
Study area
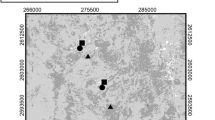
Established in 1958, the São Paulo Zoo is located in an area of 824,529 m2 within one of the last Atlantic Forest remnants within the City of São Paulo, the largest city in Brazil (Fig. 1a). The waters of the Ipiranga stream form a lake that receives several wild species of birds including migratory species (Fig. 1b). Around the lake, the forest shelters wild native animals, forming a diverse wildlife along with the zoo animal collection where more than 3000 animals are on display, representing species of mammals, birds, reptiles, amphibians, and invertebrates. During migration and the wintering of birds from various American countries, as well as from other Brazilian regions, the space destined for zoo animals is shared by migratory species, native and opportunistic species. The latter are behaviorally flexible birds living in variable environmental conditions and sustaining from different food sources.
Collection of blood samples
Samples of different bird species were collected between September 2012 and June 2015. Healthy individual samples were collected primarily for other projects: blood from Dendrocygna viduata was collected for a research project aiming isolation of microorganisms and bacterial and fungal resistance genes in migrating birds (SISBIO 35166-2); blood from Penelope superciliaris was collected for the study of the biology of Penelope sp. (Galliformes: Cracidae) and its ecological importance in the PEFI (SISBIO 41010-1). All other samples were collected for routine activities of the veterinary staff in the park since the birds presented clinical signs of diseases (SISBIO 34605-2).
For capturing Penelope sp., a trap of wooden structure measuring 80.0 × 80.0 × 6.0 cm as a base and covered with mesh braided line fishing net of 1.0 × 1.0 cm was installed on the ground in the most suitable locations in order to minimize stress and injuries to the birds. To attract birds to the trap, cracked corn was used as bait. For capturing the Anatidae species, three traps of 3.0 × 2.4 × 0.3 m were placed on the lake border. These traps were built of wood and fence with doors in the upper side to facilitate the birds’ removal. To attract the birds, we use the same methodology as for catching Penelope sp., but rice and Anatidae species food were added to bait, as established by CEMAVE/IBAMA (1994). The sampled birds were ringed after identification and blood sampling; CEMAVE’s model rings were used.
Blood sampling and microscopic examination
After being caught, the birds were physically restrained and the venous blood was collected. A thin blood smear was prepared without anticoagulant and the remaining blood was placed in a lithium heparin tube and frozen for DNA extraction. The thin blood smear was fixed by 100 % methanol, stained by Giemsa in a work solution prepared with buffered water (10 % stain stock solution was used), and then examined microscopically for 20–25 min: 100 fields were examined at low magnification (×400) and 100 fields were viewed at high magnification (×1000) (Valkiūnas 2005) using an Olympus BX41 light microscope. The intensity of parasitemia was determined by actual counting of the number of parasites per 1000 erythrocytes, as recommended by Godfrey et al. (1987).
Genomic DNA (gDNA) extraction, PCR amplification of Haemoproteus sp. and Plasmodium sp. cytb fragments, sequencing, and the sequence data analysis
DNA from blood samples was extracted with the Wizard® SV 96 Genomic DNA Purification System (Promega) with the following modifications. First, 10 μl of red blood cell pellets was completed to 200 μl with ultrapure water and an initial lysis was performed with Proteinase K (0.8 mg/200 μl sample) for 1 h at 37 °C. Next, 400 μl of Whole Blood Lysis Buffer (Wizard® SV Lysis Buffer with 10 % Triton X-100) was added and incubated overnight at room temperature. The lysates were transferred into the columns and washed according the manufacturer’s instructions. The gDNA was eluted in 100 μl of Nuclease-Free Water and stored at −20 °C. The PCR reactions were conducted using a nested polymerase chain reaction targeting the mitochondrial cytochrome b (cytb) gene of Haemoproteus and Plasmodium (Hellgren et al. 2004). Primers HaemNFI and HaemNR3 and 50 ng gDNA were used in the first reaction and 1-μl aliquot of this product was used as a template for a nested reaction with primers HaemF and HaemR2. Two blood samples with different parasitemias (<0.01 and 6.49 %) of Plasmodium sp. were used as positive control, and ultrapure water was used as a negative control. The PCR product was sequenced by Big Dye Terminator v3.0 Cycle Sequencing Kit in ABI Genetic Analyzer (ABI, USA), using PCR oligonucleotides (HaemF and HaemR2). The sequences of ∼480 bp were obtained and used in this study. Sequences possessing at least one different nucleotide were considered unique lineages. In order to determine already available lineage names, the obtained sequences were aligned with sequences deposited in MalAvi database (available at http://mbio-serv2.mbioekol.lu.se/Malavi/blast.html, see Bensch et al. 2009) using BLASTN (Basic Local Alignment Search Tool, see Altschul et al. 1997). Those absent from the MalAvi database were considered as new and were named according to the MalAvi nomenclature (Bensch et al. 2009) and deposited in GenBank.
Phylogenetic analysis
The phylogenetic relationship among reported parasites was inferred using cytb gene sequences. The phylogenetic reconstruction was performed separately for Haemoproteus and Plasmodium lineages, with 24 sequences used in each analysis. The phylogenetic tree was constructed using the Bayesian inference method implemented in MrBayes v3.2.0 (Huelsenbeck and Ronquist 2001). Bayesian inference was performed with two Markov Chain Monte Carlo searches of 3 million generations, each with sampling of 1 in 300 trees. After a burn-in of 25 %, the remaining 15,002 trees were used to calculate the 50 % majority-rule consensus tree. The phylogeny was visualized using FigTree version 1.4.0 available at http://tree.bio.ed.ac.uk/software/figtree/ (Rambaut 2006).
Results
In total, 133 samples belonging to 124 birds from 12 species were collected and screened for Plasmodium or Haemoproteus infections (Table 1). We detected an overall prevalence of 18 % (24 positive samples) by PCR-based diagnostics. These positive results were found in four different bird species (Table 1), but the prevalence varied significantly among these hosts. Columba livia had highest prevalence (100 %), while other eight sampled bird species were not infected. Of these PCR positive samples, 21 samples were also positive by microscopic examination: one was positive for Plasmodium sp., with a light parasitemia, and 20 samples positive for Haemoproteus, with parasitemias ranging between 0.3 and 15.5 %. No co-infection with hemosporidians belonging to the same or different genera was detected by microscopy. Haemoproteus (Haemoproteus) columbae (Fig. 2) was the only species of hemosporidian parasites morphologically identified. For the first time, we provided morphological support for molecular characterization of several lineages of this parasite (Figs. 2 and 3). Although Plasmodium sp. was seen, the parasites were not identified to species level because of light parasitemia.
Haemoproteus (Haemoproteus) columbae gametocytes from the blood of Columba livia (rock pigeon) in São Paulo Zoo. Bird ID 425 (lineage COLIV03): macrogametocytes, a–b and microgametocytes, c–d; Bird ID 430 (lineage COQUI05): macrogametocytes, e–f and microgametocytes, g–h; Bird ID 478 (lineage COQUI05): macrogametocytes, i–j and microgametocytes, k–l; Bird ID 481 (this parasite was readily seen in blood films, but was not detected by PCR): macrogametocytes, m–n and microgametocytes, o–p. Note large aggregations of volutin and pigment granules readily visible in microgametocytes of all lineages of this parasite. Giemsa-stained thin blood films. Bar = 10 μm
The cytb sequences recovered from the PCR-positive samples showed four birds positive for Plasmodium spp. (3 %) and 20 positive for Haemoproteus spp. (15 %), with seven different cytb lineages (three of Plasmodium and four of Haemoproteus lineages), of which five were obtained from resident birds and two from migratory birds (Table 1). We detected four new lineages: (1) C. livia (sample identification number, ID 481) presented a sequence with 98 % of identity with the lineage AFR119 (Haemoproteus sp.); (2) D. viduata (ID 263) presented a sequence with 98 % of identity with STUSUP02 and VIOLI07 (Plasmodium sp.); (3) D. viduata (ID 249) presented a sequence with 99 % of identity with GRW06 (Plasmodium elongatum); and (4) P. superciliaris (ID 885) and N. nycticorax (ID 854) presented a sequence with 99 % of identity with PESA01 (Plasmodium sp.). Three Haemoproteus lineages had been already deposited in MalAvi: HAECOL1, COQUI05, and COLIV03. However, only the first one has been attributed to certain morphospecies (H. columbae).
The phylogenetic reconstruction of Haemoproteus spp. lineages showed two well-supported clusters (posterior probability is of 100 %), corresponding to each subgenus of this genus (Fig. 3, clades a and b). One included species of the subgenus Haemoproteus (Haemoproteus), with H. columbae lineage HAECOL1, Haemoproteus multipigmentatus and Haemoproteus iwa. The other included species belonging to the subgenus Haemoproteus (Parahaemoproteus). All Haemoproteus sequences obtained from doves in this study clustered with subgenus Haemoproteus (Haemoproteus). The majority of them appeared closely related with H. columbae lineage HAECOL1, while the sequence obtained in C. livia (ID 481) clustered with H. multipigmentatus and H. iwa. No Parahaemoproteus parasite lineages were reported in this study.
Bayesian phylogeny of cytochrome b gene lineages of Haemoproteus species of avian hemosporidian parasites. Lineages recorded in this work are given bolded. Names of the lineages are given after the species names of parasites. GenBank accession numbers of the lineages are provided before the parasite species names. Nodal support values (in percentage) indicate posterior clade probabilities. a Species of the subgenus Haemoproteus (Haemoproteus). b Species belonging to the subgenus Haemoproteus (Parahaemoproteus)
The phylogenetic tree of Plasmodium spp. (Fig. 4) showed low phylogenetic resolution, and morphospecies belonging to different subgenera (Haemamoeba, Giovannolaia, Novyella, Bennettinia, and Huffia) were included in more than one clade. However, the sequences recovered from P. superciliares (ID 885) and N. nycticorax (ID 854) grouped with a well-supported cluster with the majority of P. (Haemamoeba) sequences (94 % of posterior probability); the sequence recovered from D. viduata (ID 263) (DENVID01) grouped with a well-supported cluster containing exclusively P. (Novyella) sequences (96 % of posterior probability); and the sequence recovered from D. viduata (ID 249) (DENVID02) grouped with the only P. (Huffia) sp. sequence available in the tree, with a high posterior probability (100 %).
Bayesian phylogeny of cytochrome b gene lineages of Plasmodium species of avian hemosporidian parasites. Lineages recorded in this work are given bolded. Names of the lineages are given after the species names of parasites. GenBank accession numbers of the lineages are provided before the parasite species names. Nodal support values (in percentage) indicate posterior clade probabilities
Discussion
This study adds to the better understanding of the occurrence and genetic diversity of Plasmodium and Haemoproteus parasites in different groups of birds living freely in the São Paulo Zoo. The majority of examined birds belonged to the Anatidae species, among which two migrant species were investigated and one was found positive. This was Dendrocygna viduata (white-faced whistling duck), a species found in Africa, including Madagascar and Comores Islands, Central America Islands, and South America, including Brazil. Dendrocygna javanica, D. viduata, and D. autunnalis had been previously observed infected with Plasmodium sp. (Beadell et al. 2004; Valkiunas 2005). In this study, we found two white-faced whistling ducks positive for Plasmodium lineages, with two different new lineages (DENVID01 and DENVID02). The lineage DENVID01 is similar (98 % of identity) to two different lineages (STUSUP02 and VIOLI07), which, in turn, have 97 % of identity with each other. The lineage DENVID02 shows high identity (99 % of similarity due to a C-T nucleotide substitution at position 186) to a sequence that has already been characterized as P. elongatum (GRW06), the cosmopolitan and virulent parasite in many bird species (Garnham 1966; Valkiūnas et al. 2008). The lineage DENVID01 has been reported before in other birds (mainly belonging to the Anatidae) in the São Paulo Zoo (K. Kirchgatter, unpublished observation). Inversely, the lineage DENVID02 has not been found in other infected bird species in the Zoo (K. Kirchgatter, unpublished observations). This lineage was confirmed in different assays of PCR and sequencing, indicating a possibility that this bird came to the Zoo being already infected. In response to seasonal fluctuations in food and water availability, the white-faced whistling duck may undertake short migrations (usually less than 500 km) in search of favorable foraging grounds (Madge and Burn 1988; del Hoyo et al. 1992). Their migration is conditioned to the availability of water bodies (Carboneras and Kirwan 2014) and they may use even the polluted ones (Sick 1997). Considering the wide extension of Brazil, the migratory distance of 500 km by D. viduata populations allows us to determine the origin of São Paulo State’s populations only to this country, and probably ranging from South to Southeast, as these birds usually look for sites with warm temperatures. Plasmodium elongatum (here the lineage DENVID02) has a wide range of distribution, and it is of broad specificity having been reported in birds belonging to ten different orders (see MalAvi and Garnham 1966; Valkiūnas 2005), including magellanic penguins from southern Brazil (Vanstreels et al. 2014a).
Birds of the Columbidae family were the second most largely sampled group in this study, with 20 individual rock doves (Columba livia) tested. This bird was introduced to Brazil in XVI century from Europe, and it became well adapted to the urban environment due to its ecological plasticity in the ability to inhabit different environments (Sigrist 2014), particularly those located close to human settlements (Sick 1997; Develey and Endrigo 2011). Despite some studies showing its ability to migrate over hundreds of kilometers, the native populations have been considered sedentary (Baptista et al. 1997). In São Paulo Zoo, the birds usually find food and shelter easily, and that contributes to the spread of the populations. Four different lineages of hemosporidians were found in C. livia from São Paulo Zoo: (1) the lineage HAECOL1 has been formerly found in C. livia in several countries: Botswana, Nigeria, Colombia, and Italy (Waldenstrom et al. 2002; Karamba et al. 2012; Gonzalez et al. 2015; Scaglione et al. 2015); (2) COQUI05 has been found in Coquillettidia (Culicidae, Diptera) mosquitoes from Cameroon (Njabo et al. 2009); (3) COLIV03 has already been found in C. livia from Nigeria (Karamba et al. 2012); (4) COLIV06 is a new sequence, and possibly it belongs to a Haemoproteus species since it showed 10 % of genetic distance with other sequences from HAECOL1 group. However, the bird with COLIV06 was probably co-infected whereas parasites that could present this sequence were not visible during microscopic examination of blood films. Absence of gametocytes in birds PCR positive for hemosporidian parasites has been described and can be explained by light gametocyte parasitemia, amplification of DNA of circulating sporozoites (Valkiūnas et al. 2009), or even the presence of remnants of parasites, which abort development during tissue stage of development (Ferrell et al. 2007; Donovan et al. 2008; Olias et al. 2011; Chagas et al. 2013). COQUI05 and COLIV03 sequences showed 1–2 % of genetic distances with HAECOL1 group sequences and were confirmed by microscopy in this study as H. columbae (Fig. 2), a parasite widespread in at least 14 species of the Columbiformes (Valkiūnas 2005).
Haemoproteus spp. is the most common blood parasite in birds, followed by Leucocytozoon spp. and Plasmodium spp. (Garnham 1966; Valkiūnas 2005). However, in South America, Haemoproteus lineage diversity does not follow the high diversity found among avian and mosquito species (Clark et al. 2014). It is possible that a bias to preferential amplification of Plasmodium cytb sequences exists (Valkiūnas et al. 2006). During this study, we found 83 and 17 % of the positive free-living birds infected with Haemoproteus spp. and Plasmodium spp., respectively, while Leucocytozoon spp. has not been observed in the lowlands of Brazil (Valkiūnas 2005). The high prevalence of Haemoproteus spp. reported in this study is somewhat striking since only 2 % of the positive captive birds of the same region have been found infected with this parasite (K. Kirchgatter, unpublished observation). However, this difference can be attributed to a bias due to high rate of infection found in rock doves. High prevalence (100 %) of Haemoproteus infection is consistent with that found in a study, which examined rock doves in the State of São Paulo (Adriano and Cordeiro 2001).
Columba livia, in its natural habitat, plays an important ecological role, controlling the population of insects and spreading some seeds. As invasive species in the study area and big cities such São Paulo, it is known as a sinantropic animal, competing with local species. However, the parasites that rock doves harbor can be transmitted to other columbiform birds, compromising their fitness. C. livia has a status of least concern (IUCN RED LIST 2015), but in São Paulo county, where PEFI locates, not only rock doves are present but also other species of Columbiformes, such as Columbina talpacoti, Patagioenas picazuro, Patagioenas cayennensis, Patagioenas plumbea, Patagioenas speciose, Zenaida auriculata, Geotrygon montana, Geotrygon violacea, Leptotila verreauxi, and L. rufaxilla (http://www.prefeitura.sp.gov.br/cidade/secretarias/upload/meio_ambiente/arquivos/TRE_EIA.pdf). Although Haemoproteus sp. usually does not cause clinical disease in adapted avian hosts (Valkiūnas 2005), infections with this parasite causing sudden death in non-adapted passerine birds have been reported in San Diego Zoo, USA (Donovan et al. 2008). Haemoproteus (Parahaemoproteus) parasites cause severe disease in some bird species in Europe and Australia (Olias et al. 2011; Cannell et al. 2013) and can be lethal for blood-sucking insects (Valkiūnas et al. 2014). Because rock doves have expanded their range into environmental conservation areas, there is a greater risk of spreading its infections to the native fauna; therefore, population control activities with these birds can be required.
We also found infected Penelope superciliaris and Nycticorax nycticorax. The former bird is the most well-known species of the Cracidae family, which includes 50 species and 20 of them are present in Brazil (Del Hoyo and Kirwan 1994; CBRO 2014). Penelope superciliaris is found from north to south of Brazil, Paraguay, and north of Argentina (Guix 1997). This species eats basically insects, flowers, and fallen fruits, which makes it an important seed disperser in the forest ecosystems (Del Hoyo and Kirwan 1994; Mikich 2002; Sick 1997). Nycticorax nycticorax (Ardeidae) has nightly and twilight habits (CBRO 2014) and can be found living close to water from Canada to south Argentina, including Old World where it has migration habits (Martinez-Vilalta et al. 2014). Its diet consists of fish, crustaceans, amphibians, mollusks, crabs, aquatic insects, polychaetes, small lizards, snakes, rodents, bats, eggs and chicks, and occasionally carcass (Sick 1997). In São Paulo Zoo, it is an opportunistic bird that does not belong to the park, but lives in the neighborhood and feeds using remnants of food of captive animals. Thus, living freely in the area of the zoo, such birds can rapidly take advantage of favorable conditions when they arise. In a report of Plasmodium sp. causing mortality in penguins from São Paulo Zoo, we detected a mosquito engorged with N. nycticorax blood (Bueno et al. 2010). As these birds were frequently found inside the penguin enclosures feeding on the leftover of fish, they might be a source of Plasmodium infections for other bird species in São Paulo Zoo. According to MalAvi database, the lineage found in N. nycticorax (PESA01) has been found in three different hosts of three different orders: Phaeomyias murina, a passeriform species from Brazil (Lacorte et al. 2013); Calidris melanotos, a migratory wader from Alaska (Yohannes et al. 2009); and Leptotila verreauxi, a Columbidae species from Uruguay (Durrant et al. 2006), showing a generalist parasite that may infect a broad range of avian hosts. However, it is impossible to proceed species identification using only the sequence information since a small genetic difference (96 % or less of identity) was reported in readily distinguishable hemosporidian morphospecies, all of Haemamoeba subgenus. Additional morphologic studies are needed to determine Plasmodium species identity of the PESA01 lineage.
We also tested owls that live freely in the Zoo. Although none was found infected in our study, hemosporidian parasites, mainly Haemoproteus parasites, are widespread in owls worldwide and have been found in the USA (Tavernier et al. 2005; Ishak et al. 2008), Europe (Krone et al. 2008; Bukauskaitė et al. 2015), Africa (Ishak et al. 2008), Asia (McClure et al. 1978), and South America (White et al. 1978). The prevalence of infections exceeds 50 % in many owl populations worldwide (Valkiūnas 2005). Owl haemoproteids belong to the subgenus Parahaemoproteus and are transmitted by biting midges of the genus Culicoides (Bukauskaitė et al. 2015). In São Paulo State, one individual of Megascops choliba sampled in 2009 at Carapicuíba was found positive for Haemoproteus syrnii (Vanstreels et al. 2014b). The absence of positives among the owl samples in our study could reflect the low number of birds tested here (nine samples of three birds). However, all the 35 samples tested from captive birds of Strigiformes order from the same area were negative by PCR (K. Kirchgatter, unpublished observation). This result might indicate weak transmission of Parahaemoproteus parasites due to the absence of susceptible species of biting midges at our study site warranting further investigation.
The cost of the infections to the birds depends on many factors, including the age, host immunity, breeding season, migration, and food availability (Sorci and Cornet 2010). In captivity, the animals do not usually need to compete for food, territory, or females, decreasing these stress levels that would affect animals under natural conditions. Wild animals sampled in this study have access to food from the captive animals. Since parasites can influence the biodiversity and the structure of host populations by altering the competitive interaction between the free-living and the captive birds, biodiversity preservation studies should be considered in areas with these two animal communities.
References
Adriano EA, Cordeiro NS (2001) Prevalence and intensity of Haemoproteus columbae in three species of wild doves from Brazil. Mem Inst Oswaldo Cruz 96:175–178
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Arzua M, Valim MP (2010) Bases para o estudo qualitativo e quantitativo em aves. In: Matter SV, Strauber FC, Accordi I, Piacentini V, Cândido JF Jr (eds) Ornitologia e Conservação: Ciência Aplicada, Técnicas de Pesquisa e Levantamento. Technical Books, Rio de Janeiro, pp 347–366
Atkinson CT, Thomas NJ, Hunter BC (2008) Parasitic diseases of wild birds. Wiley-Blackwell, Ames
Baptista LF, Trail PW, Horblit HM (1997) Rock dove (Columba livia). In: del Hoyo J, Elliott A, Sargatal J, Christie DA, de Juana E (eds) (2014). Handbook of the birds of the world alive. Lynx Edicions, Barcelona, Acesso: http://www.hbw.com/node/54097 em 26 de julho de 2015
Beadell JS, Gering E, Austin J, Dumbacher JP, Peirce MA, Pratt TK, Atkinson CT, Fleischer RC (2004) Prevalence and differential host-specificity of two avian blood parasite genera in the Australo-Papuan region. Mol Ecol 13:3829–3844
Belo NO, Passos LF, Júnior LM, Goulart CE, Sherlock TM, Braga EM (2009) Avian malaria in captive psittacine birds: detection by microscopy and 18S rRNA gene amplification. Prev Vet Med 88:220–224
Bensch S, Hellgren O, Pérez-Tris J (2009) MalAvi: a public database of malaria parasites and related haemosporidians in avian hosts based on mitochondrial cytochrome b lineages. Mol Ecol Resour 9:1353–1358
Bueno MG, Lopez RPG, Menezes RMT, Costa-Nascimento MJ, Lima GFMC, Araújo RAS, Guida FJV, Kirchgatter K (2010) Identification of Plasmodium relictum causing mortality in penguins (Spheniscus magellanicus) from São Paulo Zoo, Brazil. Vet Parasitol 173:123–127
Bukauskaitė D, Žiegytė R, Palinauskas V, Iezhova TA, Dimitrov D, Ilgūnas M, Bernotienė R, Markovets MY, Valkiūnas G (2015) Biting midges (Culicoides, Diptera) transmit Haemoproteus parasites of owls: evidence from sporogony and molecular phylogeny. Parasit Vectors 8:303
Cannell BL, Krasnec KV, Campbell K, Jones HI, Miller RD, Stephens N (2013) The pathology and pathogenicity of a novel Haemoproteus spp. infection in wild Little Penguins (Eudyptula minor). Vet Parasitol 197:74–84
Carboneras C, Kirwan GM (2014) White-faced whistling-duck (Dendrocygna viduata). In: del Hoyo J, Elliott A, Sargatal J, Christie DA, de Juana E (eds) 2014. Handbook of the birds of the world alive. Lynx Edicions, Barcelona, Acesso: http://www.hbw.com/node/52799 em 26 de julho de 2015
CBRO (2014) Comitê brasileiro de registros ornitológicos. Listas das aves do Brasil 11ª Edição. http://www.cbro.org.br. Accessed: January 01, 2014
CEMAVE/IBAMA (1994) Manual de anilhamento de aves silvestres, 2nd edn. IBAMA, Brasília, p 146
Chagas CRF, Valkiūnas G, Nery CVC, Henrique PC, Gonzalez IHL, Monteiro EF, Guimarães L, Kirchgatter K (2013) Plasmodium (Novyella) nucleophilum from Egyptian Goose in São Paulo Zoo, Brazil: microscopic confirmation and molecular characterization. Int J Parasitol Parasites Wildl 2:286–291
Clark NJ, Clegg SM, Lima MR (2014) A review of global diversity in avian haemosporidians (Plasmodium and Haemoproteus: Haemosporida): new insights from molecular data. Int J Parasitol 44:329–338
Del Hoyo J, Kirwan GM (1994) Rusty-margined Guan (Penelope superciliaris). In: del Hoyo J, Elliott A, Sargatal J, Christie DA, de Juana E (eds) Handbook of the birds of the world, vol 2, New World vultures to guineafowl. Lynx Edicions, Barcelona, pp 348–350
del Hoyo J, Elliott A, Sargatal J (1992) Handbook of the birds of the world. Volume 1: ostrich to ducks. Lynx Edicions, Barcelona
Develey PF, Endrigo E (2011) Guia de campo: Aves da Grande São Paulo. Aves e Fotos Editora, São Paulo
Donovan TA, Schrenzel M, Tucker TA, Pessier AP, Stalis IH (2008) Hepatic hemorrhage, hemocoelom, and sudden death due to Haemoproteus infection in passerine birds: eleven cases. J Vet Diagn Invest 20:304–313
Durrant KL, Beadell JS, Ishtiaq F, Graves GR, Olson SL, Gering E, Peirce MA, Milensky CM, Schmidt BK, Gebhard C, Fleischer RC (2006) Avian hematozoa in South America: a comparison of temperate and tropical zones. Ornit Monographs 60:98–111
Felippe-Bauer ML, Oliveira SJ (2001) Lista dos exemplares tipos de Ceratopogonidae (Diptera: Nematocera) depositados na Coleção Entomológica do Instituto Oswaldo Cruz, Rio de Janeiro, Brasil. Mem Inst Oswaldo Cruz 96:1109–1119
Ferrell ST, Snowden K, Marlar AB, Garner M, Lung NP (2007) Fatal hemoprotozoal infections in multiple avian species in a zoological park. J Zoo Wildl Med 38:309–316
Garnham PCC (1966) Malaria parasites and other haemosporidia. Blackwell Scientific, Oxford
Godfrey RD, Fedynich AM, Pence DB (1987) Quantification of the hematozoa in blood smears. J Wildl Dis 23:558–565
González AD, Lotta IA, García LF, Moncada LI, Matta NE (2015) Avian haemosporidians from Neotropical highlands: evidence from morphological and molecular data. Parasitol Int 64:48–59
Guix JC (1997) Exclusão geográfica e ecológica de Penelope obscura, Penelope superciliaris e Pipile jacutinga (Galliformes, Cracidae) no estado de São Paulo. Ararajuba 5:195–202
Hellgren O, Waldenström J, Bensch S (2004) A new PCR assay for simultaneous studies of Leucocytozoon, Plasmodium and Haemoproteus from avian blood. J Parasitol 90:797–802
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755
IUCN Red List (2015) [http://www.iucnredlist.org/]
Ishak HD, Dumbacher JP, Anderson NL, Keane JJ, Valkiūnas G, Haig SM, Tell LA, Sehgal RN (2008) Blood parasites in owls with conservation implications for the Spotted Owl (Strix occidentalis). PLoS One 3:e2304
Karamba KI, Kawo AH, Dabo NT, Mukhtar MD (2012) A survey of avian malaria parasite in Kano State, Northern Nigeria. Int J Biotechnol Mol Biol Res 3:8–14
Krone O, Waldenström J, Valkiūnas G, Lessow O, Müller K, Iezhova TA, Fickel J, Bensch S (2008) Haemosporidian blood parasites in European birds of prey and owls. J Parasitol 94:709–715
Lacorte GA, Félix GM, Pinheiro RR, Chaves AV, Almeida-Neto G, Neves FS, Leite LO, Santos FR, Braga EM (2013) Exploring the diversity and distribution of neotropical avian malaria parasites—a molecular survey from Southeast Brazil. PLoS One 8:e57770
Madge S, Burn H (1988) Wildfowl. Christopher Helm, London
Martínez-Vilalta A, Motis A, Kirwan GM (2014) Black-crowned Night-heron (Nycticorax nycticorax). In: del Hoyo J, Elliott A, Sargatal J, Christie DA, de Juana E (eds) Handbook of the birds of the world alive. Barcelona, Lynx Edicions, Acesso: http://www.hbw.com/node/52707 em 30 de julho
McClure HE, Poonswad P, Greiner EC, Laird M (1978) Haematozoa in the birds of Eastern and Southern Asia. St. John’s: Memorial University of Newfoundland.
Mikich AB (2002) A dieta frugívora de Penelope superciliaris (Cracidae) em remanescentes de florestas estacional semidecidual no centro-oeste do Paraná, Brasil e sua relação com Euterpe edulis (Arecaceae). Ararajuba 10:207–217
Motta ROC, Marques MVR, Ferreira JRFC, Andery DA, Horta RS, Peixoto RB, Lacorte GA, Moreira PA, Leme FOP, Melo MM, Martins NRS, Braga EM (2013) Does haemosporidian infection affect hematological and biochemical profiles of the endangered Black-fronted piping-guan (Aburria jacutinga)? PeerJ 1:e45
Njabo KY, Cornel AJ, Sehgal RN, Loiseau C, Buermann W, Harrigan RJ, Pollinger J, Valkiūnas G, Smith TB (2009) Coquillettidia (Culicidae, Diptera) mosquitoes are natural vectors of avian malaria in Africa. Malar J 8:193
Olias P, Wegelin M, Zenker W, Freter S, Gruber AD, Klopfleisch R (2011) Avian malaria deaths in parrots, Europe. Emerg Infect Dis 17:950–952
Orme CD, Davies RG, Burgess M, Eigenbrod F, Pickup N, Olson VA, Webster AJ, Ding TS, Rasmussen PC, Ridgely RS, Stattersfield AJ, Bennett PM, Blackburn TM, Gaston KJ, Owens IP (2005) Global hotspots of species richness are not congruent with endemism or threat. Nature 436:1016–1019
Rambaut A (2006) FigTree tree figure drawing toll version 1.4.0. Institute of Evolutionary Biology, University of Edinburgh [http://tree.bio.ed.ac.uk/software/figtree/).
Ribeiro SF, Sebaio F, Branquinho FC, Marini MA, Vago AR, Braga EM (2005) Avian malaria in Brazilian passerine birds: parasitism detected by nested PCR using DNA from stained blood smears. Parasitology 130:261–267
Ribeiro MC, Metzger JP, Martensen AC, Ponzoni FJ, Hirota MM (2009) The Brazilian Atlantic Forest: how much is left, and how is the remaining forest distributed? Implications for conservation. Biol Conserv 142:1141–1153
Scaglione FE, Pregel P, Cannizzo FT, Pérez-Rodríguez AD, Ferroglio E, Bollo E (2015) Prevalence of new and known species of haemoparasites in feral pigeons in northwest Italy. Malar J 14:99
Sebaio F, Braga EM, Branquinho F, Fecchio A, Marini MÂ (2012) Blood parasites in passerine birds from the Brazilian Atlantic Forest. Rev Bras Parasitol Vet 21:7–15
Sick H (1997) Ornitologia Brasileira, uma introdução. Nova Fronteira, Rio de Janeiro
Sigrist T (2014) Guia de campo Avis Brasilis—Avifauna Brasileira. Avis Brasilis, São Paulo, p 608
Sorci G, Cornet S (2010) Immunity and virulence in bird-parasite interactions. Open Ornithol J 3:33–40
Tavernier P, Saggese M, Van Wettere A, Redig P (2005) Malaria in an eastern screech owl (Otus asio). Avian Dis 49:433–435
Valkiūnas G (2005) Avian malaria parasites and other haemosporidia. CRC Press, New York, p 935
Valkiūnas G, Bensch S, Iezhova TA, Krizanauskiené A, Hellgren O, Bolshakov CV (2006) Nested cytochrome b polymerase chain reaction diagnostics underestimate mixed infections of avian blood haemosporidian parasites: microscopy is still essential. J Parasitol 92:418–422
Valkiūnas G, Zehtindjiev P, Dimitrov D, Krizanauskiene A, Iezhova TA, Bensch S (2008) Polymerase chain reaction-based identification of Plasmodium (Huffia) elongatum, with remarks on species identity of haemosporidian lineages deposited in GenBank. Parasitol Res 102:1185–1193
Valkiūnas G, Iezhova T, Loiseau C, Thomas S, Sehgal RNM (2009) Nested cytochrome B polymerase chain reaction diagnostics detect sporozoites of hemosporidian parasites in peripheral blood of naturally infected birds. J Parasitol 95:1512–1525
Valkiūnas G, Kazlauskienė R, Bernotienė R, Bukauskaitė D, Palinauskas V, Iezhova TA (2014) Haemoproteus infections (Haemosporida, Haemoproteidae) kill bird-biting mosquitoes. Parasitol Res 113:1011–1018
Vanstreels RET, Kolesnikovas CKM, Sandri S, Silveira P, Belo NO, Ferreira Junior FC, Epiphanio S, Steindel M, Braga EM, Catão-Dias JL (2014a) Outbreak of avian malaria associated to multiple species of Plasmodium in magellanic penguins undergoing rehabilitation in southern Brazil. PLoS One 9:e94994
Vanstreels RET, Kolesnikovas CKM, Sandri S, Silveira P, Belo NO, Ferreira Junior FC, Epiphanio S, Steindel M, Braga EM, Catão-Dias JL (2014b) Correction: Outbreak of avian malaria associated to multiple species of Plasmodium in magellanic penguins undergoing rehabilitation in southern Brazil. PLoS One 9:e116554
Waldenström J, Bensch S, Kiboi S, Hasselquist D, Ottosson U (2002) Cross-species infection of blood parasites between resident and migratory songbirds in Africa. Mol Ecol 11:1545–1554
White EM, Greiner EC, Bennett GF, Herman CM (1978) Distribution of the hematozoa of Neotropical birds. Rev Biol Trop 26:43–102
Yohannes E, Križanauskiene A, Valcu M, Bensch S, Kempenaers B (2009) Prevalence of malaria and related haemosporidian parasites in two shorebird species with different winter habitat distribution. J Ornithol 150:287–291
Acknowledgments
We thank Prof. Dr. Claudio Marinho for kindly providing the use of Zeiss Axio Imager M2 light microscope equipped with a Zeiss Axio Cam HRc in which the photographs of this paper were produced. We thank the São Paulo Zoological Park Foundation (Fundação Parque Zoológico de São Paulo) for the support provided to this research. This research was funded by Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP 2012/51427-1). All procedures were approved by the Ethical Principles in Animal Research, of the Ethics Committee of Institute of Tropical Medicine, University of São Paulo (CPE-IMT/193), and were in full compliance with federal permits issued by the Brazilian Ministry of the Environment (SISBIO 35166-2, 41010-1, and 34605-2).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chagas, C.R.F., Guimarães, L.d.O., Monteiro, E.F. et al. Hemosporidian parasites of free-living birds in the São Paulo Zoo, Brazil. Parasitol Res 115, 1443–1452 (2016). https://doi.org/10.1007/s00436-015-4878-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-015-4878-0