Abstract
Ghrelin is a stomach-derived hormone that regulates a variety of biological functions such as food intake, gastrointestinal function and blood glucose metabolism, among others. Ghrelin acts via the growth hormone secretagogue receptor (GHSR), a G-protein-coupled receptor located in key brain areas that mediate specific actions of the hormone. GHSR is highly expressed in the nucleus of the solitary tract (NTS), which is located in the medulla oblongata and controls essential functions, including orofacial, autonomic, neuroendocrine and behavioral responses. Here, we used a mouse model, in which the expression of enhanced green fluorescent protein (eGFP) is controlled by the promoter of GHSR (GHSR-eGFP mice), to gain neuroanatomical and functional insights of the GHSR-expressing neurons of the NTS. We found that GHSR-expressing neurons of the NTS are segregated in clusters that were symmetrically distributed to the midline: (1) a pair of rostral clusters, and (2) a caudal and medially located cluster. We also identified that a subset of GHSR neurons of the caudal NTS are GABAergic. Finally, we found that rostral NTS GHSR neurons increase the levels of the marker of neuronal activation c-Fos in mice exposed to fasting/refeeding or high-fat diet bingeing protocols, while caudal NTS GHSR neurons increase the levels of c-Fos in mice exposed to gastric distension or LiCl-induced malaise protocols. Thus, current data provide evidence that ghrelin receptor signaling seems to target segregated clusters of neurons within the NTS that, in turn, may be activated by different stimuli.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ghrelin is an octanoylated peptide hormone predominantly produced in the stomach (Kojima et al. 1999). Ghrelin acts in the central nervous system, where it displays a variety of effects including the modulation of food intake, blood glucose homeostasis and gastrointestinal tract motility, among others (Kojima and Kangawa 2005). Actions of ghrelin are mediated by a G-protein-coupled receptor named growth hormone secretagogue receptor (GHSR) (Howard et al. 1996). The neuroanatomical distribution of GHSR in the mouse brain is well established, and some neuronal targets of ghrelin have been linked to specific actions of the hormone (Guan et al. 1997; Zigman et al. 2006; Perello et al. 2012; Cabral et al. 2013; Mani et al. 2014). For instance, some GHSR-expressing neurons (hereafter referred as GHSR neurons) of the hypothalamic arcuate nucleus (ARC) are agouti-related protein (AgRP)-producing neurons that mediate some of the orexigenic and glucoregulatory actions of ghrelin while GHSR neurons of the area postrema (AP) are GABA neurons involved in the modulation of gastric emptying (Wang et al. 2014; Cabral et al. 2017a). Catecholaminergic GHSR neurons mediate some of the orexigenic, anti-depressant, and food reward-related effects of ghrelin (Chuang et al. 2011). The specific functions of several other populations of GHSR neurons are not as well characterized. Interestingly, GHSR neurons are also present in the nucleus of the solitary tract (NTS), which is a major integrative center located in the medulla oblongata within the caudal brainstem. The NTS, which together with the AP and the dorsal motor nucleus of the vagus (DMV) constitute the dorsal vagal complex, plays a key role controlling many essential functions, including orofacial, autonomic, neuroendocrine and behavioral responses (Barraco 1994; King 2007). Thus, NTS GHSR neurons are likely a critical target of ghrelin signaling to mediate some of its effects on such physiological functions.
The distribution, chemical phenotype and function of NTS GHSR neurons have been only rudimentarily described or theorized in the past. Thus, the current study was performed to gain some neuroanatomical and functional insights of NTS GHSR neurons. First, we mapped the distribution of GHSR neurons within the NTS using several strategies, including a transgenic reporter mouse, in which the enhanced green fluorescent protein (eGFP) is under the control of the GHSR promoter (GHSR-eGFP mice). Using these reporter mice, we also investigated the phenotype of NTS eGFP-positive (eGFP+) neurons using red fluorescent labeling for markers of specific neuronal types known to be present in this area. Since a clustering analysis of the 3D distribution of the NTS eGFP+ neurons of the GHSR-eGFP mice suggested the presence of a spatial grouping pattern of these neurons, we hypothesized that NTS GHSR neurons may be arranged in segregated clusters that respond to different stimuli. To test this possibility, we exposed GHSR-eGFP mice to a variety of experimental conditions known to involve activation of NTS neurons as well as to be modulated by ghrelin signaling, and we then looked for the induction of the marker of neuronal activation c-Fos in eGFP+ cells.
Materials and methods
Mice
This study was performed using 3–5-month-old female and male mice, which provided similar results. Experimental mice were generated in the animal facility of the IMBICE and included: (1) wild-type mice on a pure C57BL/6 background, (2) GHSR-eGFP mice, in which eGFP is under the control of the GHSR promoter, and (3) GHSR-eGFP/Gad2-tdTomato mice, in which tdTomato is expressed in glutamic acid decarboxylase 2 (Gad2)-expressing GABA cells and eGFP is expressed in GHSR-expressing cells. GHSR-eGFP mice [Mutant Mouse Resource & Research Center, Tg(Ghsr)-EGFP KZ65Gsat; Stock #030942] were derived from crosses between hemizygous mice (Mani et al. 2014). GHSR-eGFP/Gad2-tdTomato mice were generated by crossing Gad2-tdTomato mice and GHSR-eGFP/Ai14 mice. Gad2-tdTomato mice were generated by crossing Gad2-CreER mice and Ai14 mice, and GHSR-eGFP/Ai14 mice were generated by crossing GHSR-eGFP mice and Ai14 mice. Gad2-CreER mice [Jackson Laboratory, Gad2tm1(cre/ERT2)Zjh/J; Stock #010702] express a tamoxifen-inducible Cre recombinase under the endogenous promoter elements of the Gad2 gene (Taniguchi et al. 2011). Ai14 mice [Allen Institute, 129S6-Gt(ROSA)26Sortm14(CAGtdTomato)Hze/J; Stock #007908] harbor a targeted mutation of the Gt(ROSA)26Sor locus with a loxP-flanked STOP cassette preventing the transcription of the tdTomato, which is expressed only following Cre-mediated recombination (Madisen et al. 2010). Two-month-old GHSR-eGFP/Gad2-tdTomato mice received a daily dose of tamoxifen (70 mg/kg body weight (BW), IP), for four consecutive days to induce Cre recombination. After 3 weeks, the pattern of red fluorescent cells displayed the distribution of Gad2 expression, as previously shown (Taniguchi et al. 2011). All genetically-modified mice were backcrossed for more than ten generations onto a C57BL/6 genetic background. Mice were housed in a 12-h light/dark cycle in standard environmentally controlled conditions with regular chow and water available ad libitum, except when indicated. This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Research Council, USA, and all efforts were made to minimize mice suffering. All experimentation received approval from the institutional Animal Care and Use Committee of the IMBICE (approval ID 16–0112).
Neuroanatomical distribution of GHSR neurons within the NTS
The distribution of GHSR cells of the NTS was analyzed by comparing its expression as detected using the following methods: (1) review of GHSR mRNA expression reported in the in situ hybridization histochemistry (ISHH) coronal dataset of wild-type mice according to the Allen Mouse Brain Atlas, (2) endogenous fluorescence in brain samples of GHSR-eGFP mice (without any staining), (3) immunohistochemistry for eGFP in brain samples of GHSR-eGFP mice, and (4) binding studies with fluorescein-labeled ghrelin in brain samples of wild-type mice. For the analysis, all images were loaded into the software Fiji using the TrakEM2 plugin, and the number of positive cells was quantified (Cardona et al. 2012; Schindelin et al. 2012). Areas corresponding to the NTS were delineated according to a mouse brain atlas (Paxinos and Franklin 2001) and to a detailed neuroanatomical description of the mouse NTS (Ganchrow et al. 2014). Data were corrected for double counting, according to the method of Abercrombie (Abercrombie 1946), where the ratio of the actual number of neurons to the observed number is represented by T/(T + h) where T = section thickness, and h = the mean diameter of the neuron. For this, eGFP+ cell diameter of at least 30 cells was quantified using Fiji. The number of positive cells was expressed per µm along the rostro-caudal axis.
The distribution of neurons expressing GHSR mRNA in the NTS was assessed by the analysis of the coronal ISHH dataset available at http://mouse.brain-map.org/experiment/show/74511739 from the Allen Institute collection (Lein et al. 2007). In this case, 19 full sized images from the slice subset encompassing the NTS region were used. Images corresponded to 25-µm coronal brain sections that were 100 µm apart from each other.
The distribution of eGFP+ neurons was also evaluated in the NTS of GHSR-eGFP mice (n = 5) either by direct examination of green fluorescent cells or by chromogenic immunohistochemistry against eGFP. Here, GHSR-eGFP mice were anesthetized and perfused as previously described (Cabral et al. 2012). Then, brains were removed, post-fixed in formalin for 2 h at 4 °C and immersed in 20% sucrose in phosphate-buffered saline, pH 7.4 at 4 °C overnight. Next day, brains were frozen on dry ice and serially cut on a cryostat into three interleaving equivalent series of 35-µm coronal sections from bregma − 5.34 to bregma − 8.24 mm, to encompass the entire NTS. During the sectioning, each series of brain sections, which were 105 µm apart from each other, was harvested in phosphate-buffered saline at room temperature and then placed in an antifreeze solution at − 20 °C until further histological processing. One series of sections containing the NTS was mounted on glass slides and microphotographed with a laser scanning confocal microscope (Olympus FV1000, Japan) with emission filters of 490–540 nm (laser 473 nm). Other series of sections containing the NTS was processed for immunohistochemistry. Briefly, sections were pretreated with 0.5% H2O2, treated with blocking solution (3% normal donkey serum and 0.25% Triton X-100 in phosphate-buffered saline) and incubated with a rabbit anti-eGFP antibody (Molecular Probes, cat# A-6455; 1:20,000) for 48 h at 4 °C. Then, sections were incubated with a biotinylated anti-rabbit antibody (Vector Laboratories, cat# BA-1000; 1:2000) for 1 h and with Vectastain Elite ABC kit (Vector Laboratories, cat# PK6200) for 1 h, according to manufacturer’s protocols. Then, visible signal was developed with 3–3′-diaminobenzidine (DAB, Sigma Aldrich, cat# D8001), giving a brown precipitate that was only present in eGFP+ cells. Afterwards, sections were mounted and microphotographed with a bright-field microscope.
The distribution of ghrelin-binding cells of the NTS was mapped using the central administration of a fluorescein-labeled ghrelin analog (F-ghrelin), as previously described (McGirr et al. 2011; Cabral et al. 2013, 2014). F-ghrelin was provided by Dr. Luyt from the University of Western Ontario (Canada). Briefly, anesthetized wild-type mice (n = 6) were stereotaxically implanted with a single indwelling guide cannula into the lateral ventricle and intracerebroventricularly (ICV) injected with 2 µl of F-ghrelin (60 pmol/mouse). The placement coordinates for the lateral ventricle were: antero-posterior: − 0.34 mm; lateral: + 1 mm and ventral: − 2.3 mm. Mice were perfused with formalin 30 min after treatment and their brains processed as described above to generate three equivalent series of 35-µm sections. Correct location of ICV cannula injectors was verified at the end of the experiment by histological analysis. One series of sections containing the NTS was used for chromogenic immunohistochemistry, as described above. Here, the primary antibody was a goat anti-fluorescein antibody (Invitrogen, cat# A-11,095, 1:1500, 48 h at room temperature) and the secondary antibody was a biotinylated anti-goat antibody (Vector Laboratories, cat# BA-5000, 1:1000). Visible signal was developed with DAB/nickel solution, which gives a purple precipitate. Finally, sections were mounted and microphotographed with a bright-field microscope.
Detailed neuroanatomical analysis of eGFP+ neurons of NTS of GHSR-eGFP mice
Here, formalin-fixed brain of GHSR-eGFP mice (n = 2) were cut into six interleaving series of sequential 35-µm coronal sections and integrally processed for immunohistochemistry against eGFP, as described above. Each series was separately mounted on glass slides, and microphotographed with a bright-field microscope. Imaging was performed at low magnification, to capture the gross anatomy of the sections, and at intermediate magnification to resolve cellular distribution in the different regions of the NTS. Using Fiji, images were flat-field corrected and assembled into low and intermediate magnification mosaics. For analysis, low-magnification mosaics were sequentially loaded into trakEM2, following their original rostro-caudal order (Cardona et al. 2012; Schindelin et al. 2012). Then, intermediate magnification mosaics were registered within each section, ultimately producing a complete 3D volume of a medulla segment containing a detailed coverage of the whole NTS. Afterwards, areas corresponding to the NTS were delineated using the above-described reference atlases. Finally, the position of each eGFP+ neuron within the NTS volume was manually annotated. To estimate the total number of eGFP+ cells, all cells containing cytoplasmic brown staining were quantified; counting was corrected by Abercrombie as described above. The resulting spatial distribution of eGFP+ neurons within the NTS was inspected for 3D clustering patterns using the DBSCAN algorithm (Ester et al. 1996) implemented in Fiji as a macro. Algorithm parameters used for the clustering were minPts = 4 and Eps = 60 µm, based on the analysis of the distribution of n-distances and preliminary scans. All delineated structures and cells were exported as 3D models and imported into Blender (http://www.blender.org/) for volumetric rendering.
Phenotypical characterization of eGFP+ neurons of the NTS of GHSR-eGFP mice
To study the phenotype of eGFP+ neurons of the NTS, three independent sets of mice were used: (a) a set of naïve GHSR-eGFP mice (n = 6); (b) a set of GHSR-eGFP mice (n = 6) that had been previously (48 h) ICV injected with colchicine (16 µg in 4 µL per mouse); and (c) GHSR-eGFP/Gad2-tdTomato mice (n = 5). All mice were euthanized and perfused. Brains were dissected and processed as described above to generate three equivalent series of coronal 35-µm brain sections.
Brain sections of naïve GHSR-eGFP mice were used for immunohistochemistry using either rabbit anti-tyrosine hydroxylase (anti-TH, Santa Cruz, H-196, cat# sc-14,007, 1:5,000) or goat anti-choline acetyltransferase (anti-ChAT, Chemicon, cat# AB144P, 1:20,000) antibodies. Brain sections of colchicine-treated GHSR-eGFP mice were used for immunohistochemistry using either rabbit anti-neuropeptide Y (anti-NPY, Abcam, cat# ab30914, 1:7000), rabbit anti-prothyrotropin-releasing hormone (anti-proTRH, made against KQSPQVEPWDKEPLEE sequence of the TRH precursor and previously characterized in our laboratory (Perello et al. 2008), 1:3000) or rabbit anti-Met-enkephalin (anti-M-ENK, Millipore, cat# AB502, 1:10,000) antibodies. Specifically, sections of naïve and colchicine-treated GHSR-eGFP mice were treated with blocking solution for 1 h at room temperature and then incubated with each of the above-listed primary antibodies for 48 h at 4 °C. Then, sections were incubated for 2 h with either an anti-rabbit (Molecular Probes, cat# A21207, 1:1000) or an anti-goat (Molecular Probes, A11048, 1:1000) antibody conjugated to red fluorescent Alexa Fluor 594, depending on the primary antibody used in each case. Sections of GHSR-eGFP/Gad2-tdTomato mice were analyzed without any further staining. All brain sections containing the NTS of each series were sequentially mounted on glass slides and cover-slipped with mounting media containing Hoechst. Fluorescent images were acquired with a laser scanning Olympus FV1000 confocal microscope with emission filters of: 490–540 nm for eGFP detection (laser 473 nm), 575–675 nm for Alexa Fluor 594 or TdTomato detection (laser 559 nm) and 430–455 nm for Hoechst detection (laser 405 nm). For each mouse, the total number of green and/or red fluorescent cells in the NTS was bilaterally quantified in 9–11 brain interleaving sections between bregma − 6.24 mm and bregma − 7.92 mm. Results were expressed as a percentage, which represents cells positive for both green and red signals compared to the total number of cells labeled with either red or green signals alone.
Experimental manipulations of GHSR-eGFP mice
Here, different cohorts of 2–3-month-old GHSR-eGFP mice weighting 20–25 g were single housed and exposed to a series of experimental manipulations described below. All experiments were performed in the morning, starting at 9:30 −10:00 am, using naïve mice, which were perfused at the end of the trial. Importantly, at least two different trials were performed for each of the following experimental paradigms.
-
1.
ICV administration of ghrelin Mice were first stereotaxically implanted with an ICV guide cannula in the lateral ventricle, as described above, and allowed to recover for 7 days. On the experimental day, mice (n = 6 per group) were ICV injected with 2 µl of vehicle alone or containing 300 pmol of ghrelin (Global Peptide, cat. #C-et-004). This dose corresponds to the smaller dose of ghrelin that induces the maximal food intake response in our experimental conditions (Cabral et al. 2016). After 2 h, when food intake was verified to increase, mice were euthanized and perfused. As described above, correct location of ICV cannula injectors was also verified at the end of the experiment by histological analysis.
-
2.
Fasting/refeeding Mice were fasted by removing the food from their cages at 10:00 am and maintaining ad libitum access to water. Forty-eight hours later, mice were either perfused (n = 5) or re-fed with previously weighed pellet of standard chow (n = 6). After 2 h of refeeding, mice were euthanized and perfused, and food intake was quantified by weighing the remaining food. The length of food deprivation used in this experiment was based on our previous studies showing that 48-h fasting triggers more robust and consistent hyperphagic events and that GHSR mRNA levels as well as the central sensitivity to ghrelin are higher in mice fasted for longer periods of time; importantly, 48-h fasting is a manipulation fully tolerated by wild-type mice that display a normal overall health status during fasting (Fernandez et al. 2017).
-
3.
High-fat diet (HFD) bingeing Ad libitum-fed mice were daily and time-limited exposed to HFD for 2 h during four consecutive days (n = 6). HFD (Gepsa, Grupo Pilar, http://www.gepsa.com) provided 3.9 kcal/g energy and its percent weight composition was as follows: carbohydrate 22.5, proteins 22.8, fat 21.1, fibers 23.0, minerals 5.6 and water content 5.0 (For details see Valdivia et al. 2014). HFD intake was recorded and verified to increase over successive days, as previously shown (Valdivia et al. 2015). Control mice remained with access to regular chow (n = 6). Regular chow intake and body weight were daily recorded for both experimental groups. All mice were perfused the fourth experimental day 2 h after HFD intake.
-
4.
Gastric distension Mice were given one intra-gastric infusion of 500 µl vaseline via gavage using a stomach tube (n = 5). Control mice were exposed to the same procedure without having any volume infused in the stomach (n = 2). Oral gavage procedure was performed by trained personnel as previously done in the past (Valdivia et al. 2014). Briefly, the mouse’s head was extended backwards to create a straight line through the neck and esophagus. The gavage tube was placed in the diastema of the mouth and then gently advanced along the upper palate until the end of the esophagus was reached. Mice were made accustomed to handling and the procedure daily for at least 4 days prior to experimentation to reduce stress. Mice were perfused 2 h after treatment.
-
5.
LiCl-induced malaise Ad libitum-fed mice were injected with LiCl (12 ml/kg, 150 mM, IP, n = 6) or saline (n = 3). We have already shown that this dose of LiCl induces malaise in mice as indicated by the fact that they develop conditioned taste aversion when this dose of LiCl is paired to a novel appetitive stimulus (De Francesco et al. 2015).
-
6.
Insulin-induced hypoglycemia Mice were injected with insulin (1.0 IU/Kg BW, IP, Lilly, cat# U-100, n = 6) or saline (n = 4), as we reported previously (Enriori et al. 2007), and perfused 2 h after treatment. Blood glucose was measured from cheek blood samples using a glucometer (Accu-chek, Roche Diagnostic Corporation) before and 30 min after injection.
-
7.
Glucose-induced hyperglycemia Mice were fasted for 24 h by removing the food from their cages at 10:00 am. On the morning of the experimental day, mice were injected with glucose (2 g/kg BW, IP, 50% w/v, Biopack, n = 7) or saline (n = 4 ), as we reported previously (Enriori et al. 2007), and perfused 2 h after treatment. Here, the length of food deprivation used was based on previous studies showing that 24-h fasted mice display a smaller variability in blood glucose levels during this test (Ayala et al. 2010). Blood glucose was measured from cheek blood samples using a glucometer (Accu-chek, Roche Diagnostic Corporation) before and 30 min after injection.
Assessment of c-Fos in eGFP+ neurons
Brains were cut into three equal series of 35-µm coronal sections, as described above. Double c-Fos/eGFP immunostaining was performed in one complete series containing the NTS, as described (Cabral et al. 2017a). Briefly, sections were pretreated with 0.5% H2O2, treated with blocking solution and incubated with a rabbit anti-c-Fos antibody (Santa Cruz Biotechnology, cat# SC-7202, 1:4,000) for 48 h at 4 °C. Then, sections were incubated with a biotinylated anti-rabbit antibody and the Vectastain Elite ABC kit, as described above. Purple visible signal was developed with DAB/nickel solution. Then, c-Fos immunostained sections were incubated with a rabbit anti-eGFP antibody, and sequentially incubated with a secondary antibody and the Vectastain Elite ABC kit, as detailed above. Finally, cytoplasmic brown visible signal was developed with DAB solution. Sections were sequentially mounted on glass slides and cover-slipped with mounting media. Bright-field images were acquired with a DS-Ri1 Nikon digital camera. All images were taken in comparable areas and under the same optical and light conditions. Quantitative analyses were performed between bregma − 6.24 and bregma − 7.81 mm; thus, at least nine sections containing the NTS were analyzed per mouse. Based on the data described in the “Results” section, NTS was subdivided into rostral and caudal parts, which included coronal brain sections between bregma − 6.24 and bregma − 7.08 mm and between bregma − 7.08 and bregma − 7.92 mm, respectively. Total number of c-Fos-positive (c-Fos+) cells in the NTS as well as the fraction of eGFP+ cells of the NTS that were also positive for c-Fos (eGFP+/c-Fos+) were counted. Results were expressed as: (1) total c-Fos+ cells per coronal section, (2) the percentage of eGFP+/c-Fos+ compared to the total number eGFP+ cells in the rostral or caudal portion of the NTS, or (3) fold change vs. their respective control group.
Statistical analyses
Blind quantitative analysis was performed independently by, at least, two observers. Data were expressed as the mean±SEM. One-way ANOVA followed by the Tukey’s post-test was used to compare the quantitative analysis of neuroanatomical data from different groups. The ROUT (Robust regression and Outlier removal) method was used to identify and remove outliers, which ended up being two mice from control groups, one fasted mouse and one LiCl-treated mouse. Analyses were performed using GraphPad Prism 6.0. Significant differences were considered when P < 0.05.
Results
Neuroanatomical analysis of GHSR neurons of the NTS
First, we performed a neuroanatomical analysis of GHSR cells in the NTS as visualized by: (1) ISHH in wild-type mice, reviewing the dataset reported by the Allen Mouse Brain Atlas (Fig. 1a, b), (2) direct detection of eGFP fluorescence in GHSR-eGFP mice (Fig. 1c, d), (3) chromogenic immunostaining against eGFP in GHSR-eGFP mice (Fig. 1e, f) and (4) binding of F-ghrelin in wild-type mice (Fig. 1g, h). All four strategies revealed the presence of GHSR cells in the NTS. Analysis of the Allen Mouse Brain Atlas ISHH dataset indicated that ~ 2100 GHSR-expressing cells exist in the NTS and that ~ 87% of them are concentrated between bregma − 7.0 mm and bregma − 8.0 mm (Fig. 1i). Notably, the manual analysis of the GHSR dataset revealed that the automated detection method to quantify gene expression used by the Allen Mouse Brain Atlas underestimated the number of GHSR-expressing neurons in the NTS. Analysis of fluorescent cells in brain sections of GHSR-eGFP mice indicated that ~ 1400 eGFP+ cells exist in the NTS and that ~ 86% of them are enriched between bregma − 7.0 mm and bregma − 8.0 mm (Fig. 1j). Analysis of eGFP immunostaining in brain sections of GHSR-eGFP mice indicated that ~ 1800 eGFP+ cells exist in the NTS and that ~ 60% of them are concentrated between bregma − 7.0 mm and bregma − 8.0 mm but also present in the rostral NTS (Fig. 1k). Finally, the analysis of the distribution of F-ghrelin-binding cells in the NTS of wild-type mice indicated that ~ 3300 positive cells exist in the NTS and that ~ 80% of them are concentrated between bregma − 7.0 mm and bregma − 8.0 mm (Fig. 1l). Notably, F-ghrelin-binding cells in the rostral NTS displayed a fainter staining compared to positive cells of the caudal NTS. Importantly, all methods displayed a good level of agreement in terms of the coronal distribution of the putative GHSR neurons of the NTS and provide a more detailed accounting of the numbers of NTS GHSR neurons, compared to our previous reports (Zigman et al. 2006; Perello et al. 2012; Cabral et al. 2013; Mani et al. 2014).
GHSR neurons of the NTS. Panels a-h display representative photomicrographs of coronal mouse brain sections containing a rostral (bregma − 6.64/− 6.72 mm, upper row) and a caudal (bregma − 7.48 mm, lower row) section of the NTS showing the presence of GHSR-expressing neurons by ISHH (a, b, Allen Mouse Brain Atlas), eGFP fluorescence (c, d) and anti-eGFP immunostaining (e, f) in GHSR-eGFP mice and anti-fluorescein immunostaining in F-Ghr-injected mice (g, h). Inserts in each panel show high-magnification images of the areas marked in low-magnification photomicrographs. Delineation of the NTS is superimposed in each panel; T: solitary tract. Arrowheads point at positively identified neurons in each inset. Scale bars: panels, 200 µm; insets, 20 µm. Bar graphs i–l show the quantitative analysis of the number of GHSR-expressing cells per µm in the rostro-caudal axis evidenced by ISHH (i Allen Mouse Brain Atlas), eGFP fluorescence (j) and anti-eGFP immunostaining (k) in GHSR-eGFP mice and anti-fluorescein immunostaining in F-Ghr-injected mice (l)
Detailed neuroanatomical analysis of eGFP+ neurons of NTS of GHSR-eGFP mice
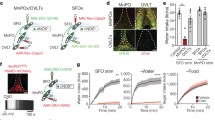
Then, we used the GHSR-eGFP mice to perform a detailed neuroanatomical analysis of the cytoarchitectonic features and distribution of eGFP+ neurons throughout the rostro-caudal axis of the nucleus. In the rostral NTS, eGFP+ neurons were centrally located and enriched in the rostral central (RC) subnucleus (Fig. 2a). In the caudal NTS, eGFP+ neurons were medially located and enriched in the central (Ce), medial (M) and parvicellular (PC) sub-nuclei (Fig. 2a). The number of eGFP+ neurons in each NTS subnucleus could not be estimated given the undefined boundaries that separate them. Most eGFP+ cells of the NTS displayed a similar shape and cytoarchitectonic features that included an ovoid soma (8.4 ± 0.2 µm of diameter) with two or three short sparsely branched primary dendrites, which extend between ~ 17 and ~ 45 µm in the coronal plane without extensive terminal arborizations or dendritic spines (Fig. 2b). The 3D reconstruction of coronal sections between bregma − 6.34 mm and bregma − 8.23 mm showed that eGFP + neurons display a V-shaped distribution in the horizontal plane, closely resembling the shape of the NTS, with their rostral and the caudal portions forming the apices and the base of the V, respectively (Fig. 3a, b). To test if NTS eGFP+ neurons were differentially clustered in the NTS, a clustering analysis on the cell’s coordinates was performed using the DBSCAN algorithm. This analysis detected two types of populated clusters that were symmetrically distributed to the midline: (1) a pair of laterally symmetric rostral clusters that ranged from bregma − 6.5 mm to bregma − 7.0 mm and contained a total of ~ 650 cells, and (2) a caudal cluster that was medially located from bregma − 7.1 mm to bregma − 7.7 mm and contained ~ 750 neurons. The rest of eGFP+ cells of the NTS were classified as either not clustered (~ 300) or forming small clusters of less than 20 cells (~ 130 in total).
Pattern distribution of eGFP+ neurons of the NTS of the GHSR-eGFP mice. a Depicts a set of representative coronal mouse brain sections subjected to chromogenic anti-eGFP immunostaining (brown) containing rostral to caudal levels of the NTS. For each level, a diagram of the NTS is overlayed following the nomenclature and delineation described in Ganchrow et al. (2014). High-magnification images were obtained from the areas marked in low-magnification images. Rostro-caudal coordinates are expressed in mm in reference to bregma. Scale bars: high magnification, 200 µm; low magnification, 500 µm. b Shows photomicrographs of representative “ovoid” eGFP+ neurons present in the rostral and caudal NTS. High-magnification images were obtained from the areas marked in low magnification images. Scale bars: low magnification, 200 µm; high magnification, 20 µm. Ce central subnucleus, CL caudal lateral subnucleus, COM commissural subnucleus, DL dorsal lateral subnucleus, M medial subnucleus, PC parvicellular subnucleus, RC rostral central subnucleus, RL rostral lateral subnucleus, RM rostral medial subnucleus, V ventral subnucleus, VL ventral lateral subnucleus, T solitary tract
Spatial reconstruction of the eGFP+ neuron ensemble of the NTS. a Shows a representative dorsal projection of all eGFP+ neurons (shown as circles) found in 55 serial sections of a GHSR-eGFP mouse, along with a projection of the NTS outline for reference. The cells are color coded according to their clustering: magenta, rostral cluster; cyan, caudal cluster; gray, other or no cluster. The bregma coordinates are indicated to the left in mm. b Shows different 3D-renderings of the cell ensemble and surrounding structures. From left to right: top row, lateral (left) and caudal orthogonal views; bottom row, inclined dorsal and ventral views. Cells are shown as spheres, color coded as in a. The limits of the NTS (light gray) and the area postrema (green) are shown in all views. The insets provide a wider view of each rendering, where the outline of the medulla (orange) is included, and the cell clusters are represented by their enclosing surfaces. A anterior, P posterior, D dorsal, V ventral, L left, R right
A subset of eGFP+ neurons of the NTS are GABAergic
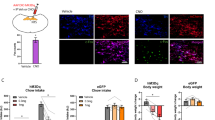
To test if eGFP+ cells of the NTS are GABAergic, we studied GHSR-eGFP/Gad2-tdTomato transgenic mice that express eGFP in GHSR cells and tdTomato in Gad2-expressing (GABAergic) cells. The distribution pattern of eGFP+/tdTomato+ cells in the GHSR-eGFP/Gad2-tdTomato reporter mice was similar as that seen in each independent mouse model (not shown). Notably, few tdTomato+ cells were present in the rostral NTS while most of them were located in the caudal NTS. In the caudal NTS, we found a significant amount of tdTomato+/eGFP+ cells that were enriched in the PC subnucleus (Fig. 4). The quantitative analysis of the degree of co-localization indicated that tdTomato+/eGFP+ cells represented 30.1 ± 8.6% of the caudal NTS eGFP+ cells and 4.8 ± 0.8% of the rostral NTS tdTomato+ cells. To further characterize other neurotransmitters produced by eGFP+ cells of the NTS, we performed red fluorescent immunostainings against a number of neuronal markers in brain sections of GHSR-eGFP mice. In particular, we tested antibodies against the neuropeptides M-ENK, NPY and TRH as well as antibodies against the enzymes TH and ChAT, which label catecholaminergic and cholinergic neurons, respectively. All neuronal markers labeled specific cell populations within the dorsal vagal complex; however, no eGFP+ neurons were double labeled within the NTS (not shown).
A subset of eGFP+ cells of the NTS of GHSR-eGFP mice are GABAergic. Panel shows representative confocal photomicrographs of a coronal section containing the NTS (bregma − 7.48 mm) of a GHSR-eGFP/Gad2-tdTomato mouse; the left photomicrograph shows eGFP+ cells (green), middle photomicrograph shows tdTomato+ cells (red) and cell nuclei (blue), and right photomicrograph shows the merge of the respective green, red and blue signals. Inserts in each panel shows in high magnification the areas marked in low-magnification photomicrographs. Arrowheads point at dual-labeled cells. Scale bars: panels, 100 µm; insets, 10 µm. T solitary tract
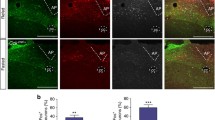
eGFP+ neurons of the rostral and caudal NTS of GHSR-eGFP mice increase c-Fos levels in response to different stimuli
To gain insights of the potential role of NTS GHSR neurons, we hypothesized that eGFP+ neurons of the NTS of the GHSR-eGFP mice would increase the expression levels of the marker of neuronal activation c-Fos when the neuronal circuit engaging them is recruited. To test if c-Fos is a good marker for this cell type, we first investigated if ICV ghrelin treatment increases the level of c-Fos in eGFP+ cells of the NTS of GHSR-eGFP mice. We found that the percentage of eGFP+/c-Fos+ cells significantly increased in both the rostral and caudal NTS of ghrelin-treated GHSR-eGFP mice, compared to vehicle-treated mice (Fig. 5a). In particular, eGFP+/c-Fos+ represented 1.0 ± 0.3 and 1.0 ± 0.4% of eGFP+ cells in the rostral and caudal NTS of vehicle-treated GHSR-eGFP mice, respectively, while they represented 4.9 ± 1.0 and 38.7 ± 8.1% of all eGFP+ cells in the same regions of the NTS of ghrelin-treated mice.
eGFP+ cells of the NTS increase c-Fos in response to different stimuli. a Shows representative low-magnification images of double chromogenic immunostaining for c-Fos (black) and eGFP (brown) in coronal brain slices containing the rostral (bregma − 6.64 mm, upper row) and caudal (bregma − 7.48 mm, lower row) NTS of GHSR-eGFP mice ICV-treated with vehicle or 300 pmol of ghrelin. For each level, a diagram of the NTS is overlayed following the delineation described in Paxinos and Franklin (2001). T solitary tract. Insets in each image show high-magnification photomicrographs of the areas marked in low-magnification images. Arrowheads point at dual-labeled cells. Scale bars: panels, 100 µm; insets, 10 µm. b and c Show the quantitative analysis of the percentage of eGFP+/c-Fos+ relative to the number of eGFP+ cells found in the rostral (b) and caudal (c) clusters of the NTS and normalized by the percentage found in control animals. *p < 0.05; **p < 0.01; ****p < 0.0001; One-way ANOVA, Tukey’s post-test
To test if GHSR neurons of the rostral and caudal NTS respond to different stimuli, we assessed the induction of c-Fos in eGFP+ neurons of GHSR-eGFP mice exposed to a variety of experimental paradigms. Since a subset of NTS neurons are recruited in re-fed mice after a fasting event (Wu et al. 2014), we studied c-Fos induction in eGFP+ neurons of the NTS of GHSR-eGFP mice exposed to a fasting–refeeding protocol. The body weight of fasted mice significantly decreased at the end of the fasting period, compared with fed mice (18.2 ± 1.3 g%). Compared to fed mice, the total number of c-Fos+ cells (data not shown) and the fraction of eGFP+/c-Fos+ cells were not affected in the rostral and caudal NTS of fasted GHSR-eGFP mice (Fig. 5b, c). Re-fed mice ate 915 ± 125 mg of regular chow 2 h after refeeding and the number of c-Fos+ cells significantly increased, compared to the numbers found in fasted mice, in the rostral (118 ± 20 vs. 16 ± 4 cells/section) and caudal (146 ± 27 vs. 49 ± 14 cells/section) NTS. The percentage of eGFP+/c-Fos + cells in the rostral NTS significantly increased in re-fed mice, compared to the values found in fasted GHSR-eGFP mice (Fig. 5b), while the percentage of eGFP+/c-Fos+ cells in the caudal NTS remained unaffected (Fig. 5c). An independent set of GHSR-eGFP mice was exposed to a HFD bingeing protocol, in which mice are daily exposed to HFD during 2 h for four consecutive days. Regular chow intake and body weight of mice exposed to the HFD bingeing protocol were not significantly changed throughout the experiment. In particular, mice ate 8.58 ± 0.27, 8.54 ± 0.49 and 8.93 ± 0.63 kcal/day of regular chow and weighed 24.0 ± 0.4, 24.2 ± 0.5 and 24.7 ± 0.4 g after the first, second and third day of the test, respectively. After the last HFD eating event, mice ate 625 ± 70 mg of HFD. The number of c-Fos + cells significantly increased in mice exposed to HFD, compared to control mice, in both the rostral (68 ± 10 vs. 30 ± 6 cells/section) and caudal (64 ± 13 vs. 27 ± 9 cells/section) NTS. The percentage of eGFP+/c-Fos+ cells in the rostral NTS significantly increased in GHSR-eGFP mice exposed to HFD, compared to the values found in control mice (Fig. 5b), while the percentage of eGFP+/c-Fos + cells in the caudal NTS also increased but such difference did not reach statistical significance (P = 0.1371, Fig. 5c).
Since a subset of NTS neurons are regulated by sensory information from the gastrointestinal tract, we studied GHSR-eGFP mice that underwent gastric distention. Compared to control mice, the number of c-Fos+ cells significantly increased in both the rostral (110 ± 22 vs. 29 ± 6 cells/section) and caudal (179 ± 26 vs. 30 ± 10 cells/section) NTS of mice that underwent gastric distention. The percentage of eGFP+/c-Fos+ cells in the caudal NTS significantly increased in GHSR-eGFP mice that underwent gastric distention, compared to the values found in control mice, while the percentage of eGFP+/c-Fos+ cells in the rostral NTS did not differ among groups (Fig. 5b-c). Since the feeling of gastrointestinal discomfort is also known to activate NTS neurons, we studied GHSR-eGFP mice that were injected with LiCl to induce a malaise (De Francesco et al. 2015). Compared to control mice, the number of c-Fos+ cells significantly increased only in the caudal NTS (96 ± 23 vs. 26 ± 8 cells/section) of LiCl-treated GHSR-eGFP mice, while the number of c-Fos+ cells was unaffected in the rostral NTS (32 ± 10 vs. 29 ± 6 cells/section). Similarly, the percentage of eGFP+/c-Fos+ cells significantly increased in the caudal NTS of LiCl-treated GHSR-eGFP mice, compared to control mice, while the percentage of eGFP+/c-Fos+ cells in the rostral NTS did not differ among groups (Fig. 5b, c).
Since a subset of NTS neurons is able to sense glucose levels, we studied c-Fos induction in eGFP+ neurons of GHSR-eGFP mice subjected to hypo- or hyperglycemia. Insulin-treated GHSR-eGFP mice displayed ~ 75% decrease of glucose levels 30 min after treatment and did not show significant differences in the number of c-Fos + cells in the rostral (25 ± 5 vs. 27 ± 5 cells/section) and caudal (48 ± 11 vs. 28 ± 10 cells/section) NTS, compared to vehicle-treated mice. Similarly, the percentage of eGFP+/c-Fos + cells in the rostral and caudal NTS did not differ in insulin-treated mice, compared to vehicle-treated mice (Fig. 5b, c). Another set of fasted GHSR-eGFP mice was injected with glucose to induce a ~ five-fold increase of glycaemia. The number of c-Fos in the rostral (34 ± 11 vs. 16 ± 3 cells/section) and caudal (61 ± 16 vs. 49 ± 14 cells/section) NTS as well as the percentage of eGFP+/c-Fos+ cells were unaffected by glucose treatment (Fig. 5b, c).
Discussion
NTS neurons are organized along the rostro-caudal axis in accordance with their function: rostral NTS neurons mainly receive gustatory and orotactile afferents, while caudal NTS neurons receive cardiovascular, visceral and respiratory afferents (King 2007). NTS neurons integrate these afferent pathways and then regulate different orofacial, autonomic, neuroendocrine and behavioral responses via projections to different brain targets (Rinaman 2010). It is well established that some NTS neurons express GHSR and that ghrelin action on this nucleus induces some effects. For instance, administration of ghrelin into the caudal brainstem, via injection into the fourth ventricle or directly into the dorsal vagal complex, induces gastric contractions via vagal cholinergic outputs and food intake (Faulconbridge et al. 2003, 2008; Swartz et al. 2014). Additionally, intra-NTS administration of ghrelin decreases both sympathetic activity and arterial blood pressure in rats (Lin et al. 2004). Notably, 3D modeling approaches have shown that NTS neurons display distinct neuronal activity patterns, depending on the stimulus, suggesting a functional segregation of neurons within the NTS (Schwarz et al. 2010). To the best of our knowledge, the neuroanatomical distribution of NTS GHSR neurons and their potential segregation in different clusters with specific roles have not been previously studied.
Reporter mice are a valuable tool to visualize neurons expressing receptors, such as the GHSR, which are hard to label by other methods (e.g. due to the lack of reliable antibodies). Previously, we used dual-label chromogenic immunohistochemistry against eGFP and ISHH for GHSR mRNA to validate GHSR-eGFP mice as an accurate model to visualize GHSR cells in the brain (Mani et al. 2014). We concluded that GHSR-eGFP mice are useful for studying GHSR function in some brain areas, where most eGFP+ cells displayed GHSR mRNA signal. In the NTS, however, only ~ 50% of all eGFP+ cells of the NTS displayed GHSR mRNA expression (Mani et al. 2014). Here, we performed a comprehensive analysis of the NTS GHSR cells using four different strategies. The quantification of a single available sample of the Allen Mouse Brain Atlas for GHSR ISHH data indicated the NTS contains ~ 2100 GHSR-expressing cells. In the NTS of GHSR-eGFP mice, quantitative analysis of fluorescent cells revealed the presence of ~ 1400 eGFP+ cells while eGFP immunostaining revealed the presence of ~ 1800 positive cells, which is close to the ISHH estimation. A higher number of cells detected after chromogenic immunostaining were expected since the latter strategy is more sensitive due to the enzymatic amplification steps of this method, while direct eGFP fluorescence displays a lower signal-to-noise ratio due to the presence of green autofluorescence of the tissue that could undermine the identification of faint eGFP+ cells. According to the current analysis, most eGFP+ cells of the NTS of the GHSR-eGFP mice should represent GHSR cells while a small fraction of NTS GHSR cells seem to lack eGFP signal. The reason why our previous study failed to detect GHSR mRNA in all eGFP+ cells of the NTS is uncertain; it can be hypothesized, however, that the correct identification of cells positive for GHSR mRNA may have been impaired by the precipitation of DAB on cell bodies in the last step of the chromogenic immunostaining or the diffused localization of the silver stain signal. The estimated number of ghrelin-binding cells in the NTS (~ 3300) was higher than the numbers of GHSR cells estimated by the other strategies. This overestimation could be attributed to a higher background staining of cells observed in the in vivo binding experiments, which was more prominent in regions nearer to the fourth ventricle due to the diffusion of the tracer. In addition, the existence of other ghrelin-binding proteins, different from GHSR, in the NTS cannot be completely ruled out (Callaghan et al. 2014). Notably, all strategies indicated that most GHSR cells are located in the caudal NTS and enriched around bregma − 7.5 mm. Importantly, GHSR-eGFP mice represent a valuable tool to study NTS GHSR cells because it not only provides more unambiguously labeled cells in a reproducible and simple manner but also allows the visualization of the cytoarchitectonic features of positive cells. In this regard, it is interesting to stress that the cytoarchitectonic features of eGFP+ cells in the NTS of GHSR-eGFP mice strongly suggest the neuronal nature of these cells; even though we cannot completely rule out that some GHSR-expressing cells of the NTS may be something other than neurons.
The distribution of eGFP+ cells in GHSR-eGFP mice suggests that NTS GHSR neurons may be arranged in segregated clusters that seem to respond to different stimuli. In particular, we found a rostrally-located laterally-symmetric pair of clusters of GHSR neurons that are responsive to rapid and spontaneous eating induced by either a previous fasting event or HFD bingeing. These GHSR neurons are located mainly in the RC subnucleus, which receives afferent gustatory inputs from cranial nerves that innervate the oral cavity (King 2007). We also found a caudally-located cluster of GHSR neurons that is activated by gastric distention, a mechanical stimulus that activates vagal mechanoreceptors, or the administration of the nauseogenic agent LiCl, which is a interoceptive stressor (Curtis et al. 1994; Traub et al. 1996). Notably, both maneuvers suppress food intake via activation of glucagon-like peptide-1 (GLP-1)-producing neurons of the NTS (Rinaman 1999; Vrang et al. 2003). Thus, GHSR neurons of the rostral NTS may be linked to feeding-related behaviors while GHSR neurons of the caudal NTS may be a part of a neuronal circuit that integrates gastrointestinal sensory inputs and modulates food intake.
To the best of our knowledge, the chemical phenotype of NTS GHSR neurons was uncertain. Previously, we had shown that GHSR neurons of the rat NTS do not produce cholecystokinin (Zigman et al. 2006). In addition, it was shown that ghrelin acts on NTS catecholaminergic neurons via an indirect pathway that involves an inhibition of the presynaptic glutamate release (Cui et al. 2011). Here, we not only confirmed that NTS GHSR neurons are not catecholaminergic but also found that eGFP+ neurons do not synthesize acetylcholine, M-ENK, NPY or TRH. Interestingly, we found that ~ 30% of NTS GHSR neurons are GABAergic and enriched in the PC subnucleus of the caudal NTS. The PC subnucleus belongs to a gastric distension-responsive region of the NTS, and a subset of PC neurons is known to innervate the lateral parabrachial nucleus, which potently regulates food intake (Whitehead 1990; Wu et al. 2012; Ganchrow et al. 2014). Cytoarchitectonic features of NTS eGFP+ cells resemble previously described “ovoid” neurons of the rostral NTS, which are the most abundant set of inhibitory GABA interneurons within this nucleus (King and Bradley 1994). NTS GABA interneurons play a key regulatory role inhibiting the excitatory transmission from solitary tract afferents to NTS neurons (Fawley et al. 2011; McDougall and Andresen 2012; Chen et al. 2016). In addition, some NTS GABA neurons innervate the DMV and influence vagal motor activity (Davis et al. 2004; Gao et al. 2009). Notably, GABA neurons are usually heterogeneous (Lovett-Barron and Losonczy 2014), and we found here that only a fraction of NTS GABA neurons express GHSR. Thus, it is possible to hypothesize that ghrelin signaling targets a subset of NTS GABA neurons that, in turn, regulates other neurons that control dorsal vagal complex-mediated functions. It is important to stress that the specificity and efficiency of Gad2-CreER mice to label GABA neurons is higher than 90% (Taniguchi et al. 2011). In addition, Gad2-tdTomato mice may underestimate the visualization of all GABA neurons since some GABA neurons express the Gad1 isoform of GAD (Feldblum et al. 1993). To visualize GABA cells, some studies have taken advantage of the fact that the vesicular GABA transporter (VGAT) is expressed in all GABA neurons (Wojcik et al. 2006); however, the use of VGAT promoter to target GABA cells is also associated with some caveats since VGAT is also expressed in glycinergic neurons (Dumoulin et al. 1999). Thus, the use of traditional dual-labeling studies in GHSR-eGFP mice using ISHH for different markers of GABA neurons may provide a better estimate of the actual number and distribution of ghrelin responsive GABA cells. Despite our efforts, the identity of some NTS GHSR neurons remains to be elucidated. Potential targets of ghrelin signaling in the NTS may include neurons producing glutamate, glycine, somatostatin, GLP-1, nesfatin-1, galanin, among others, which are present in the NTS and play important physiological roles (Coelho et al. 2004; Goebel-Stengel et al. 2011; Shi et al. 2017). Further studies using traditional strategies as well as state-of-the-art genetically modified mice are required to establish the chemical phenotypes and functions of all GHSR-expressing neurons of the NTS.
NTS contains glucose sensing neurons, which regulate feeding and glucose homeostasis, and a subset of those glucose-responsive cells is GABAergic (Ritter et al. 2000)(Boychuk et al. 2015). Here, we did not find induction of c-Fos in NTS eGFP+ neurons of GHSR-eGFP mice exposed to acute hyper- or hypoglycemia. These observations do not necessarily mean that NTS GHSR neurons do not respond to glycemia since glucose can regulate rapid neuronal events (e.g. electrical activity) independent of gene transcription (Boychuk et al. 2015). Previously, we reported that mice with GHSR expression limited to the hindbrain (within Phox2b-expressing neurons) normalize the relative hypoglycemia detected in GHSR-deficient mice upon fasting suggesting that GHSR signaling in the hindbrain regulates glucose homeostasis (Scott et al. 2012). GHSR neurons of the hindbrain are present in the NTS as well as in the DMV, AP, nucleus ambiguous and facial motor nucleus (Scott et al. 2012). Thus, the hindbrain neuronal circuits mediating the effects of GHSR signaling on glucose homeostasis require further studies.
Finally, it is interesting to stress that peripherally administered ghrelin does not reach the NTS, nor does it increase c-Fos expression in NTS GHSR neurons (Cabral et al. 2017a). In contrast, circulating ghrelin seems to indirectly act on NTS neurons via its action on AP neurons that, in turn, activate some NTS neurons lacking GHSR (Li et al. 2006; Cabral et al. 2014, 2017a). In addition, ghrelin is not synthetized in the brain (Cabral et al. 2017b). Thus, the physiological relevance of the presence of GHSR in NTS neurons is uncertain. Notably, GHSR displays a high constitutive activity, in the absence of ghrelin (Holst et al. 2003), and GHSR heterodimerizes with other G protein coupled receptors to modulate their signaling (Schellekens et al. 2015). Thus, constitutive GHSR activity and/or GHSR interactions with other receptors in the NTS may impact the physiological roles of this nucleus, independently of plasma ghrelin.
References
Abercrombie M (1946) Estimation of nuclear population from microtome sections. Anat Rec 94:239–247
Ayala JE, Samuel VT, Morton GJ et al (2010) Standard operating procedures for describing and performing metabolic tests of glucose homeostasis in mice. Dis Model Mech 3:525–534. https://doi.org/10.1242/dmm.006239
Barraco IRA (1994) Nucleus of the solitary tract. CRC Press, Boca Raton
Boychuk CR, Gyarmati P, Xu H, Smith BN (2015) Glucose sensing by GABAergic neurons in the mouse nucleus tractus solitarii. J Neurophysiol 114:999–1007. https://doi.org/10.1152/jn.00310.2015
Cabral A, Suescun O, Zigman JM, Perello M (2012) Ghrelin indirectly activates hypophysiotropic CRF neurons in rodents. PloS One 7:e31462. https://doi.org/10.1371/journal.pone.0031462
Cabral A, Fernandez G, Perello M (2013) Analysis of brain nuclei accessible to ghrelin present in the cerebrospinal fluid. Neuroscience 253:406–415. https://doi.org/10.1016/j.neuroscience.2013.09.008
Cabral A, Valdivia S, Fernandez G et al (2014) Divergent neuronal circuitries underlying acute orexigenic effects of peripheral or central ghrelin: critical role of brain accessibility. J Neuroendocrinol 26:542–554. https://doi.org/10.1111/jne.12168
Cabral A, Portiansky E, Sánchez-Jaramillo E et al (2016) Ghrelin activates hypophysiotropic corticotropin-releasing factor neurons independently of the arcuate nucleus. Psychoneuroendocrinology 67:27–39. https://doi.org/10.1016/j.psyneuen.2016.01.027
Cabral A, Cornejo MP, Fernandez G et al (2017a) Circulating ghrelin acts on GABA neurons of the area postrema and mediates gastric emptying in male mice. Endocrinology 158:1436–1449. https://doi.org/10.1210/en.2016-1815
Cabral A, López Soto EJ, Epelbaum J, Perelló M (2017b) Is ghrelin synthesized in the central nervous system? Int J Mol Sci 18:638. https://doi.org/10.3390/ijms18030638
Callaghan B, Kosari S, Pustovit RV et al (2014) Hypotensive effects of ghrelin receptor agonists mediated through a novel receptor. Br J Pharmacol 171:1275–1286. https://doi.org/10.1111/bph.12527
Cardona A, Saalfeld S, Schindelin J et al (2012) TrakEM2 software for neural circuit reconstruction. PloS One 7:e38011. https://doi.org/10.1371/journal.pone.0038011
Chen Z, Travers SP, Travers JB (2016) Inhibitory modulation of optogenetically identified neuron subtypes in the rostral solitary nucleus. J Neurophysiol 116:391–403. https://doi.org/10.1152/jn.00168.2016
Chuang J-C, Perello M, Sakata I et al (2011) Ghrelin mediates stress-induced food-reward behavior in mice. J Clin Invest 121:2684–2692. https://doi.org/10.1172/JCI57660
Coelho EF, Ferrari MFR, Maximino JR et al (2004) Decreases in the expression of CGRP and galanin mRNA in central and peripheral neurons related to the control of blood pressure following experimental hypertension in rats. Brain Res Bull 64:59–66. https://doi.org/10.1016/j.brainresbull.2004.05.003
Cui RJ, Li X, Appleyard SM (2011) Ghrelin inhibits visceral afferent activation of catecholamine neurons in the solitary tract nucleus. J Neurosci Off J Soc Neurosci 31:3484–3492. https://doi.org/10.1523/JNEUROSCI.3187-10.2011
Curtis KS, Sved AF, Verbalis JG, Stricker EM (1994) Lithium chloride-induced anorexia, but not conditioned taste aversions, in rats with area postrema lesions. Brain Res 663:30–37
Davis SF, Derbenev AV, Williams KW et al (2004) Excitatory and inhibitory local circuit input to the rat dorsal motor nucleus of the vagus originating from the nucleus tractus solitarius. Brain Res 1017:208–217. https://doi.org/10.1016/j.brainres.2004.05.049
De Francesco PN, Valdivia S, Cabral A et al (2015) Neuroanatomical and functional characterization of CRF neurons of the amygdala using a novel transgenic mouse model. Neuroscience 289:153–165. https://doi.org/10.1016/j.neuroscience.2015.01.006
Dumoulin A, Rostaing P, Bedet C et al (1999) Presence of the vesicular inhibitory amino acid transporter in GABAergic and glycinergic synaptic terminal boutons. J Cell Sci 112:811–823
Enriori PJ, Evans AE, Sinnayah P et al (2007) Diet-induced obesity causes severe but reversible leptin resistance in arcuate melanocortin neurons. Cell Metab 5:181–194. https://doi.org/10.1016/j.cmet.2007.02.004
Ester M, Kriegel H, Sander J, Xu X (1996) A density-based algorithm for discovering clusters in large spatial databases with noise. AAAI Press, Palo Alto, pp 226–231
Faulconbridge LF, Cummings DE, Kaplan JM, Grill HJ (2003) Hyperphagic effects of brainstem ghrelin administration. Diabetes 52:2260–2265
Faulconbridge LF, Grill HJ, Kaplan JM, Daniels D (2008) Caudal brainstem delivery of ghrelin induces fos expression in the nucleus of the solitary tract, but not in the arcuate or paraventricular nuclei of the hypothalamus. Brain Res 1218:151–157. https://doi.org/10.1016/j.brainres.2008.04.068
Fawley JA, Peters JH, Andresen MC (2011) GABAB-mediated inhibition of multiple modes of glutamate release in the nucleus of the solitary tract. J Neurophysiol 106:1833–1840. https://doi.org/10.1152/jn.00476.2011
Feldblum S, Erlander MG, Tobin AJ (1993) Different distributions of GAD65 and GAD67 mRNAS suggest that the two glutamate decarboxylases play distinctive functional roles. J Neurosci Res 34:689–706. https://doi.org/10.1002/jnr.490340612
Fernandez G, Cabral A, Andreoli MF et al (2017) Evidence supporting a role for constitutive ghrelin receptor signaling in fasting-induced hyperphagia in male mice. Endocrinology. https://doi.org/10.1210/en.2017-03101
Ganchrow D, Ganchrow JR, Cicchini V et al (2014) Nucleus of the solitary tract in the C57BL/6J mouse: Subnuclear parcellation, chorda tympani nerve projections, and brainstem connections. J Comp Neurol 522:1565–1596. https://doi.org/10.1002/cne.23484
Gao H, Glatzer NR, Williams KW et al (2009) Morphological and electrophysiological features of motor neurons and putative interneurons in the dorsal vagal complex of rats and mice. Brain Res 1291:40–52. https://doi.org/10.1016/j.brainres.2009.07.024
Goebel-Stengel M, Wang L, Stengel A, Taché Y (2011) Localization of nesfatin-1 neurons in the mouse brain and functional implication. Brain Res 1396:20–34. https://doi.org/10.1016/j.brainres.2011.04.031
Guan XM, Yu H, Palyha OC et al (1997) Distribution of mRNA encoding the growth hormone secretagogue receptor in brain and peripheral tissues. Brain Res Mol Brain Res 48:23–29
Holst B, Cygankiewicz A, Jensen TH et al (2003) High constitutive signaling of the ghrelin receptor—identification of a potent inverse agonist. Mol Endocrinol Baltim Md 17:2201–2210. https://doi.org/10.1210/me.2003-0069
Howard AD, Feighner SD, Cully DF et al (1996) A receptor in pituitary and hypothalamus that functions in growth hormone release. Science 273:974–977
King MS (2007) Anatomy of the rostral nucleus of the solitary tract. In: Bradley RM (ed) The role of the nucleus of the solitary tract in gustatory processing. CRC Press/Taylor & Francis, Boca Raton
King MS, Bradley RM (1994) Relationship between structure and function of neurons in the rat rostral nucleus tractus solitarii. J Comp Neurol 344:50–64. https://doi.org/10.1002/cne.903440105
Kojima M, Kangawa K (2005) Ghrelin: structure and function. Physiol Rev 85:495–522. https://doi.org/10.1152/physrev.00012.2004
Kojima M, Hosoda H, Date Y et al (1999) Ghrelin is a growth-hormone-releasing acylated peptide from stomach. Nature 402:656–660. https://doi.org/10.1038/45230
Lein ES, Hawrylycz MJ, Ao N et al (2007) Genome-wide atlas of gene expression in the adult mouse brain. Nature 445:168–176. https://doi.org/10.1038/nature05453
Li Y, Wu X, Zhao Y et al (2006) Ghrelin acts on the dorsal vagal complex to stimulate pancreatic protein secretion. Am J Physiol Gastrointest Liver Physiol 290:G1350-1358. https://doi.org/10.1152/ajpgi.00493.2005
Lin Y, Matsumura K, Fukuhara M et al (2004) Ghrelin acts at the nucleus of the solitary tract to decrease arterial pressure in rats. Hypertens 43:977–982. https://doi.org/10.1161/01.HYP.0000122803.91559.55
Lovett-Barron M, Losonczy A (2014) Behavioral consequences of GABAergic neuronal diversity. Curr Opin Neurobiol 26:27–33. https://doi.org/10.1016/j.conb.2013.11.002
Madisen L, Zwingman TA, Sunkin SM et al (2010) A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13:133–140. https://doi.org/10.1038/nn.2467
Mani BK, Walker AK, Lopez Soto EJ et al (2014) Neuroanatomical characterization of a growth hormone secretagogue receptor-green fluorescent protein reporter mouse. J Comp Neurol 522:3644–3666. https://doi.org/10.1002/cne.23627
McDougall SJ, Andresen MC (2012) Low-fidelity GABA transmission within a dense excitatory network of the solitary tract nucleus. J Physiol 590:5677–5689. https://doi.org/10.1113/jphysiol.2012.241976
McGirr R, McFarland MS, McTavish J et al (2011) Design and characterization of a fluorescent ghrelin analog for imaging the growth hormone secretagogue receptor 1a. Regul Pept 172:69–76. https://doi.org/10.1016/j.regpep.2011.08.011
Paxinos G, Franklin KBJ (2001) The mouse brain in stereotaxic coordinates, 2nd edn. Academic Press, San Diego
Perello M, Stuart R, Nillni EA (2008) Prothyrotropin-releasing hormone targets its processing products to different vesicles of the secretory pathway. J Biol Chem 283:19936–19947. https://doi.org/10.1074/jbc.M800732200
Perello M, Scott Michael M, Ichiro S et al (2012) Functional implications of limited leptin receptor and ghrelin receptor coexpression in the brain. J Comp Neurol 520:281–294. https://doi.org/10.1002/cne.22690
Rinaman L (1999) A functional role for central glucagon-like peptide-1 receptors in lithium chloride-induced anorexia. Am J Physiol 277:R1537-1540
Rinaman L (2010) Ascending projections from the caudal visceral nucleus of the solitary tract to brain regions involved in food intake and energy expenditure. Brain Res 1350:18–34. https://doi.org/10.1016/j.brainres.2010.03.059
Ritter S, Dinh TT, Zhang Y (2000) Localization of hindbrain glucoreceptive sites controlling food intake and blood glucose. Brain Res 856:37–47
Schellekens H, De Francesco PN, Kandil D et al (2015) Ghrelin’s orexigenic effect is modulated via a serotonin 2C receptor interaction. ACS Chem Neurosci 6:1186–1197. https://doi.org/10.1021/cn500318q
Schindelin J, Arganda-Carreras I, Frise E et al (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. https://doi.org/10.1038/nmeth.2019
Schwarz J, Burguet J, Rampin O et al (2010) Three-dimensional macronutrient-associated Fos expression patterns in the mouse brainstem. PloS One 5:e8974. https://doi.org/10.1371/journal.pone.0008974
Scott MM, Perello M, Chuang J-C et al (2012) Hindbrain ghrelin receptor signaling is sufficient to maintain fasting glucose. PloS One 7:e44089. https://doi.org/10.1371/journal.pone.0044089
Shi X, Chacko S, Li F et al (2017) Acute activation of GLP-1-expressing neurons promotes glucose homeostasis and insulin sensitivity. Mol Metab 6:1350–1359. https://doi.org/10.1016/j.molmet.2017.08.009
Swartz EM, Browning KN, Travagli RA, Holmes GM (2014) Ghrelin increases vagally mediated gastric activity by central sites of action. Neurogastroenterol Motil 26:272–282. https://doi.org/10.1111/nmo.12261
Taniguchi H, He M, Wu P et al (2011) A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron 71:995–1013. https://doi.org/10.1016/j.neuron.2011.07.026
Traub RJ, Sengupta JN, Gebhart GF (1996) Differential c-fos expression in the nucleus of the solitary tract and spinal cord following noxious gastric distention in the rat. Neuroscience 74:873–884
Valdivia S, Patrone A, Reynaldo M, Perello M (2014) Acute high fat diet consumption activates the mesolimbic circuit and requires orexin signaling in a mouse model. PloS One 9:e87478. https://doi.org/10.1371/journal.pone.0087478
Valdivia S, Cornejo MP, Reynaldo M et al (2015) Escalation in high fat intake in a binge eating model differentially engages dopamine neurons of the ventral tegmental area and requires ghrelin signaling. Psychoneuroendocrinology 60:206–216. https://doi.org/10.1016/j.psyneuen.2015.06.018
Vrang N, Phifer CB, Corkern MM, Berthoud H-R (2003) Gastric distension induces c-Fos in medullary GLP-1/2-containing neurons. Am J Physiol Regul Integr Comp Physiol 285:R470-478. https://doi.org/10.1152/ajpregu.00732.2002
Wang Q, Liu C, Uchida A et al (2014) Arcuate AgRP neurons mediate orexigenic and glucoregulatory actions of ghrelin. Mol Metab 3:64–72. https://doi.org/10.1016/j.molmet.2013.10.001
Whitehead MC (1990) Subdivisions and neuron types of the nucleus of the solitary tract that project to the parabrachial nucleus in the hamster. J Comp Neurol 301:554–574. https://doi.org/10.1002/cne.903010406
Wojcik SM, Katsurabayashi S, Guillemin I et al (2006) A shared vesicular carrier allows synaptic corelease of GABA and glycine. Neuron 50:575–587. https://doi.org/10.1016/j.neuron.2006.04.016
Wu Q, Clark MS, Palmiter RD (2012) Deciphering a neuronal circuit that mediates appetite. Nature 483:594–597. https://doi.org/10.1038/nature10899
Wu Q, Lemus MB, Stark R et al (2014) The temporal pattern of cfos activation in hypothalamic, cortical, and brainstem nuclei in response to fasting and refeeding in male mice. Endocrinology 155:840–853. https://doi.org/10.1210/en.2013-1831
Zigman JM, Jones JE, Lee CE et al (2006) Expression of ghrelin receptor mRNA in the rat and the mouse brain. J Comp Neurol 494:528–548. https://doi.org/10.1002/cne.20823
Acknowledgements
MPC was supported by CONICET.
Funding
This work was funded by the PICTO2013-0065 grant, which is co-financed by the National Agency of Scientific and Technological Promotion of Argentina and GlaxoSmithKline, to MP.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. All procedures performed in studies involving animals were in accordance with the ethical standards of the institution or practice at which the studies were conducted.
Rights and permissions
About this article
Cite this article
Cornejo, M.P., De Francesco, P.N., García Romero, G. et al. Ghrelin receptor signaling targets segregated clusters of neurons within the nucleus of the solitary tract. Brain Struct Funct 223, 3133–3147 (2018). https://doi.org/10.1007/s00429-018-1682-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00429-018-1682-5