Abstract
TMPRSS2–ERG, the most common gene fusion in prostate cancer, is associated with expression of a truncated protein product of the oncogene ERG. A novel anti-ERG monoclonal antibody has been recently characterized. We investigated the correlation between ERG rearrangement assessed by fluorescence in situ hybridization (FISH) and ERG expression detected by immunohistochemistry in a large cohort of patients treated with radical prostatectomy for clinically localized prostate cancer. Thirteen tissue microarrays comprising 305 tumors and a subset of 112 samples of nonneoplastic prostatic tissue were assessed for ERG rearrangement status by FISH and for ERG expression by immunohistochemistry. Accuracy of ERG detection by immunohistochemistry in predicting ERG status as assessed by FISH (criterion standard) was calculated in terms of sensitivity, specificity, positive and negative predictive values. Of 305 tumor foci, 103 (34%) showed ERG rearrangement by FISH. ERG was detected by immunohistochemistry in 100 (33%) cases, 99 of which were FISH positive. ERG detection by immunohistochemistry demonstrated a sensitivity and specificity of 96% and 99%, respectively, with positive and negative predictive values of 99% and 98%, respectively. None of the 112 samples of nonneoplastic prostatic tissue was rearranged by FISH or showed any ERG expression. In conclusion, ERG detection by immunohistochemistry in prostate cancer was highly predictive of ERG rearrangement as assessed by FISH in a large cohort of prostatectomy patients. Given the high yield and the easier task of performing immunohistochemistry vs. FISH, ERG assessment by immunohistochemistry may be useful for characterizing ERG status in prostate cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Tumor-associated molecular alterations occurring in leukemias, lymphomas, sarcomas and some epithelial tumors have been useful in defining distinct pathologic entities and may also serve as specific diagnostic and prognostic markers [1]. Several highly sensitive and specific techniques have been used for the detection of such molecular alterations. Among these, polymerase chain reaction (PCR) and fluorescence in situ hybridization (FISH) are the most commonly applied. However, because of their relatively high costs and need of qualified personnel, they cannot always be easily implemented as routine tests in clinical laboratories.
Some genetic alterations result in the expression of novel, disease-specific protein products, which may represent tumor-specific targets. Antibodies directed against proteins encoded by genetic alterations are useful tools in the characterization of diagnostic subtypes of hematologic malignancies, such as acute myeloid leukemia and many lymphomas [2].
Immunohistochemistry is a routine laboratory procedure and a relatively simple, low-cost method compared to molecular techniques. However, to be effective in replacing or complementing the molecular characterization of genetic alterations, the sensitivity and specificity of the immunohistochemical test need to be assessed against the criterion standard for the detection of such alterations.
Recurrent chromosomal rearrangements leading to the fusion of the androgen receptor-regulated gene TMPRSS2 and ETS transcription factors are present in about half of prostate cancers from radical prostatectomy series [3–5]. v-ets erythroblastosis virus E26 oncogene homolog (avian) (ERG) is the most common fusion partner and its rearrangements have been extensively investigated by FISH [3, 4], which shows deletion or translocation of its 5′end in fusion-positive cases.
TMPRSS2–ERG fusion by FISH is highly specific for prostate cancer. FISH signal patterns of ERG rearrangement have never been detected in nonneoplastic prostate tissue samples, including benign prostatic hyperplasia and several forms of prostate atrophy [4, 6], although detection of fusion transcripts by PCR has occasionally been reported in nonneoplastic tissue [7, 8]. Moreover, in a recent study, Scheble et al. [9] screened 54 different tumor types other than prostate cancer and found FISH signal patterns specific of ERG rearrangements in none of them.
The genetic alteration leads to the juxtaposition of androgen-responsive regulatory elements of TMPRSS2 to ERG sequence, with consequent overexpression of the rearranged ERG gene [5, 10]. In the most commonly detected fusion transcripts of TMPRSS2–ERG positive cases, TMPRSS2 contributes only untranslated sequences, thus resulting in the overproduction of a truncated ERG protein product, rather than a chimeric protein [5, 11]. Studies exploring the potential role of a truncated ERG protein, analogous to ERG rearrangement product, in prostate tumorigenesis reported controversial results, with overexpression of ERG being found to induce a neoplastic phenotype of prostate cells (mouse prostatic intraepithelial neoplasia) in some [11, 12], but not in others [13].
The truncated protein product of ERG rearrangement may be targeted as immunohistochemical surrogate for the gene fusion and may help elucidate its role in tumor development and progression, as previously hypothesized [11].
Two independent studies recently addressed the immunohistochemical detection of the fusion product by testing two different monoclonal antibodies [14, 15]. Furusato et al. [14] developed a mouse anti-ERG monoclonal antibody and found it to be highly specific in distinguishing prostate cancer foci from benign glands on whole mount sections of prostate. They also noticed a strong concordance between mRNA levels of fusion transcripts detected by branched-chain DNA signal amplification and ERG immunohistochemistry, as well as between ERG rearrangement detected by FISH and ERG protein expression in a smaller sample of prostate specimens (10). Park et al. [15] characterized a novel rabbit anti-ERG monoclonal antibody directed against the C-terminus of the most common gene fusion product. They found that ERG protein expression determined by immunohistochemistry with this antibody highly correlated with the ERG rearrangement status determined by FISH on prostate cancer tissue microarrays from two independent patient cohorts.
In this study, we aimed to provide independent confirmation of the accuracy of ERG expression detected by the rabbit anti-ERG monoclonal antibody used by Park et al. [15] in predicting ERG rearrangement as assessed by FISH (criterion standard) in a large cohort of patients treated with radical prostatectomy for clinically localized prostate cancer.
Materials and methods
Study population and tissue microarray construction
Two hundred and seventy-one patients treated with radical prostatectomy for clinically localized prostate cancer were included in the study. Cases were selected in part based on previous assessment of ERG rearrangement status by FISH, as previously reported [16, 17]. Thirteen tissue microarrays, composed of up to three representative 1.5 mm tissue cores for each tumor and one 1.5 mm core for paired nonneoplastic prostate tissue in a subset of 112 cases, were constructed from formalin-fixed paraffin-embedded radical prostatectomy blocks. Two of 13 tissue microarrays (46 patients) were from an outside institution (Nagoya Daini Red Cross Hospital, Nagoya, Japan). Concomitant separate tumors were sampled when identified, as previously described [16]. Patients' clinical data (including age and pre-operative PSA) and radical prostatectomy pathologic parameters (including Gleason score and pathologic stage), when available, were collected in an institutional board approved database.
ERG gene rearrangement assessment by FISH
A dual-color interphase break-apart FISH assay was performed on the 13 formalin-fixed paraffin-embedded tissue microarrays to assess the ERG gene rearrangement status, as previously described [4, 5, 16]. Briefly, two differentially labeled Bacterial artificial chromosome (BAC) FISH probes were used: the Biotin-14-dCTP labeled BAC clone RP11-24A11 (eventually conjugated to produce a red signal) for the neighboring 3′ centromeric region of the ERG locus and the Digoxigenin-dUTP labeled BAC clone RP11-372O17 (eventually conjugated to produce a green signal), for the neighboring 5′ telomeric region. According to this break-apart probe system, a nucleus without ERG rearrangement shows 2 pairs of juxtaposed red and green signals, forming yellow signals (class N, or negative for fusion); a nucleus with ERG rearrangement through translocation (class Esplit) shows one pair of still combined red and green signals (a yellow signal), while the other breaks into a single red and a single green signal (split signal); a nucleus with ERG rearrangement through deletion (class Edel) shows one yellow signal and a single red signal, while the green signal (5′ region) is lost. Any case with ERG signal abnormalities in >10% of the neoplastic cells was scored as positive and classified accordingly, as previously described [16]. In addition, the presence of multiple copies of the ERG rearrangement sequence, with or without retention of the green signal (class 2 + Esplit and 2 + Edel, respectively) was assessed in all evaluated nuclei. Tissue hybridization, washing, and fluorescence detection were performed as previously described [18].
The samples were analyzed under a 100× oil immersion objective using a Leica DM-RB fluorescence microscope equipped with appropriate filters. The corresponding hematoxylin and eosin section was available for side-by-side comparison with the FISH section. Evaluation of the slides was performed by a pathologist (SMF) and a medical technologist (KS) with experience in analyzing FISH. For each case at least 50 morphologically intact, nonoverlapping nuclei were scored. Technical difficulties included insufficient (<50 cells) or absent diagnostic material to evaluate and very weak or absent probe signals.
Evaluation of ERG protein expression by immunohistochemistry
Immunohistochemistry was performed on 4 μm sections of the 13 formalin-fixed paraffin-embedded tissue microarrays using a Discovery XT automated stainer (Ventana Medical Systems, Tucson, AZ). Antigen retrieval was performed in Tris/Borate/EDTA buffer, pH 8.0–8.5, for 8 min at 95°C, 12 min at 100°C, and then 8 min at room temperature. The slides were then incubated with an anti-ERG rabbit monoclonal antibody (1:100 dilution, clone ID: EPR3864, Catalog Number 2805-1, Epitomics, San Diego, CA) for 32 min at room temperature. A secondary antibody (OmniMap anti-Rabbit HRP; Tuscon, AZ) was applied for 16 min at room temperature. The chromogenic substrate (ChromoMap DAB, Tucson, AZ) was applied for 8 min at room temperature. Slides were counterstained with hematoxylin II. Using the vascular endothelial cells as internal positive control, the expression of ERG protein was evaluated as negative or positive in prostate glands, regardless of the intensity of staining. A tumor was judged positive if any of the evaluable cores showed a positive staining.
Statistical analysis
To compare the predicted accuracy of immunohistochemistry ERG protein detection in determining the ERG rearrangement status as assessed by FISH (criterion standard), sensitivity, specificity, positive and negative predictive values and 95% Confidence Intervals (CI) were calculated using the NCSS 2007 software (Kaysville, UT). Associations for ERG expression status and clinicopathologic features were also explored using the SPSS software (Chicago, IL). Equality of continuous variables was tested using the Student t test and that of categorical variables was tested using chi-square test.
Results
Study population
Patients' mean age at time of diagnosis was 61 years (range: 40–81). Mean pre-operative PSA was 8.3 ng/mL (range: 0.9–66.8). Prostatectomy Gleason score was 6 in 76 (28%), 7 in 161 (59%), ≥8 in 33 (12%) and indeterminate in 1 (1%) patient who received neoadjuvant androgen deprivation therapy. Pathological stage was available in 264 (97%) cases, and was pT2 in 179 (68%), pT3a in 74 (28%) and pT3b in the remaining 11 (4%) cases.
Comparison of ERG expression detected by immunohistochemistry and ERG status assessed by FISH
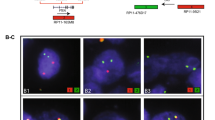
Three hundred and five tumors were evaluable for both immunohistochemistry and FISH, one for each of the 271 patients and a concomitant separate tumor focus in 34 (12%) cases. ERG rearrangement by FISH was found in 103 (34%) tumors. Of the FISH positive tumors, 49 (47%) were classified as Edel, 47 (46%) as Esplit, and 7 (7%) as combined Edel + Esplit. Duplication of the rearranged sequence with deletion of the 5′ sequence was observed in 4 cases (2 + Edel) (Fig. 1).
ERG rearrangement by break-apart FISH. For the non-rearranged ERG locus, the set of probes appear either as juxtaposed red and green signals or a yellow spot due to the overlap between the red or green probes (arrows). The red and green signals are separated when an ERG gene rearrangement occurs. Loss of the green signal complementary to the 5′ telomeric region of ERG represents a deletion in this chromosomal region and indirectly indicates a fusion of the TMPRSS2 and ERG loci (Edel). Duplication of the red signal complementary to the 3′ ERG region may also occur (2 + Edel) a. Separate and distinct red and green signals indicate ERG rearrangement by translocation (Esplit) b. (a and b, ×100)
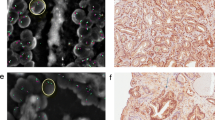
ERG expression by immunohistochemistry was detected in 100 (33%) cases, 99 of which were FISH positive (Fig. 2). We identified one (<1%) tumor demonstrating ERG protein expression in one of the 2 informative cores without any detectable ERG rearrangement as assessed by FISH. Conversely, four (1%) cases with ERG rearrangement by FISH, two Edel and two Esplit, had no detectable ERG protein expression in any of the informative cores. The endothelial cells (positive control) were strongly positive for ERG immunohistochemistry in these cores.
Immunohistochemical detection of ERG protein expression. ERG protein expression by immunohistochemistry is detected in nuclei of prostate cancer cells (a). Vascular endothelial cells (arrows) and infiltrating stromal lymphocytes (arrowheads) show positive ERG expression in both ERG-positive (a) and ERG-negative (b) tumor samples (×20)
The overall sensitivity and specificity of ERG protein expression by immunohistochemistry in predicting ERG rearrangement status were 96% (95% CI 90% to 99%) and 99% (95% CI 97% to 100%), respectively, with a positive predictive value of 99% (95% CI 94% to 100%) and a negative predictive value of 98% (95% CI 95% to 99%).
Of the 34 patients with two separate concomitant tumor foci, 24 (71%) were homogeneous for ERG rearrangement status (present or absent in both tumor foci), whereas 10 (29%) were heterogeneous, with one tumor focus harboring the ERG rearrangement and not the other. ERG protein was concordantly expressed in the ERG rearranged focus, whereas no expression was found in the ERG fusion-negative focus.
Except for 6 (2%) cases, all TMA cores of a given tumor revealed homogeneity for rearrangement status and corresponding ERG expression. In the 6 cases, 1 of the informative tumor cores showed rearrangement by FISH (5 Edel, 1 Esplit) as well as protein expression by immunohistochemistry, and the remaining cores showed no rearrangement or protein expression.
None of the 112 tissue samples of benign prostatic tissue was rearranged by FISH or showed ERG protein expression by immunohistochemistry.
No significant association was found between ERG expression and clinicopathological parameters, such as age, pre-operative PSA, Gleason score, and pathologic stage.
Discussion
Chromosomal translocations often result in the expression of either hybrid gene products with new properties or inappropriately expressed normal proteins. Monoclonal antibodies to these proteins are useful tools in the characterization of diseases, such as hematologic and mesenchymal malignancies [2, 19].
Recurrent chromosomal aberrations resulting in overexpression of ETS gene family members, primarily ERG, have been identified and characterized in prostate cancer [11]. ERG rearrangements mostly lead to the production of a truncated protein that can be detected by immunohistochemistry.
Two groups recently conducted a comparative evaluation of TMPRSS2–ERG gene fusion status and ERG oncoprotein expression by immunohistochemistry using two different anti-ERG monoclonal antibodies. Park et al. [15] compared ERG gene rearrangement analysis by FISH and ERG detection by immunohistochemistry using a novel rabbit anti-ERG monoclonal antibody, in a combined multi-institutional cohort consisting of 207 patient tumors. Furusato et al. [14] evaluated the expression of ERG in 261 tumors using a mouse anti-ERG monoclonal antibody: in a subset of 35 specimens the authors made the comparative analysis with TMPRSS2–ERG gene fusion status assessed by branched-chain DNA assay, and in a subset of 10 with ERG gene rearrangement assessed by FISH. Both groups found a high concordance between fusion status and ERG protein expression. They also showed that both antibodies recognize all the protein products resulting from genomic fusions of ERG protein-coding sequences and regulatory sequences of any of the known 5′ fusion partners (TMPRSS2, SLC45A3 and NDRG1), as well as the wild type ERG [14, 15].
Similar to Park et al. [15], we found that immunohistochemical detection of ERG protein in prostate cancer using the same rabbit anti-ERG monoclonal antibody is highly predictive of ERG rearrangement status assessed by FISH. In our study group of 305 tumors, we found a sensitivity of 96%, which is comparable to the 95.7% reported by Park et al., whereas the specificity we found was slightly higher than the one reported by the same authors (99% vs. 96.5%). The specificity of a test depends on the number of “false positive” that the test picks up compared against the criterion standard. We found only one (1/305 ≤ 1%) case demonstrating ERG protein expression without ERG rearrangement by FISH (false positive), whereas Park et al. found 4 (4/207 = 2%) of such discordant cases. Technical reasons may in part explain this finding. Alternatively, ERG overexpression in prostate cancer may occur through mechanisms other than gene rearrangements detectable by the FISH break-apart system, as previously hypothesized [15]. Together with chromosomal translocations, amplification, point mutations, and activation by insertions of new regulatory sequences may be responsible for oncogenic activation of cellular genes, including the ETS family (reviewed by Seth et al. [20]). Although consistent amplification of ERG or ETV1 in prostate cancer samples with respective transcript overexpression has not been found [5], further studies may help characterize alternative mechanisms of ERG overexpression.
We also observed 4 cases with ERG rearrangement as assessed by FISH and no expression of ERG protein (false negative). Likewise, in the study by Park et al. [15], 4 false negative cases from one of the two evaluated cohorts were reported. Besides technical issues, the inefficient transcription and/or translation of the gene fusion product may be responsible for cases with gene rearrangement by FISH which do not express detectable levels of the protein. The transcription of TMPRSS2–ERG fusion gene is driven by androgen signaling pathway and it is conceivable that, if the androgen pathway is defective, prostate cancer cells with TMPRSS2–ERG gene fusion may not express ERG protein at a level high enough to be detected by immunohistochemistry. Although Park et al. showed no difference in protein expression between ERG rearranged tumors harboring SLC45A3–ERG or NDRG1–ERG gene fusions and those harboring TMPRSS2–ERG gene fusion [15], still unknown 5′ partners might result in gene fusions without ERG protein overexpression, thus explaining some of the “false negative” cases [21, 22]. In the present study, we did not evaluate the 5′ partners of ERG rearranged cases.
Importantly, we also found high positive and negative predictive values (99% and 98%, respectively) of ERG protein detection by immunohistochemistry, which are comparable to those reported by Park et al. [15]. The high positive predictive value denotes the high probability that a positive ERG immuno-expression reflects the underlying presence of ERG rearrangement in the tumor tested; on the other hand a high negative predictive value implies a high likelihood that if prostate cancer cells do not express ERG protein as detected by immunohistochemistry, the tumor does not harbor the gene rearrangement. Those values do however depend on the overall prevalence of ERG rearrangement in the population under study. In our study population, 34% (103/305) of tumors showed ERG rearrangement by FISH. This frequency is similar to the value of 30% (134/445) found by Attard et al. [23], although lower than the value of 47% (60/128) and 40% (32/79) reported by Park et al. in their cohorts [15], or the commonly referred value of 48.5% (115/237) found by Perner et al. [4], who also assessed ERG gene alterations using a FISH break-apart assay. Since the incidence of ERG rearrangements may vary in relation to different clinicopathological characteristics, such as clinical stage [23], patient race [16], Gleason score [23] and prostate cancer zone of origin [4, 16], the distinct compositions of the prostate tumor sets surveyed in each study may in part explain these differences.
In line with prior studies [24–26], we observed interfocal heterogeneity for ERG rearrangement between distinct tumors within the same prostate gland. The overall incidence of 29% reported herein was slightly lower than those reported by others (41% to 51%) [24–26]. However, the cases evaluated in this study with more than one tumor per prostate gland included patients with a predominant transition zone tumor and a secondary peripheral zone tumor [16]. Transition zone prostate cancer harbor TMPRSS2–ERG fusion in lower percentages of cases, as previously reported [16], which may artificially increase the number of concordantly negative tumor foci in each patient, thus resulting in a lower rate of intertumoral heterogeneity detected.
All but 6 of our cases demonstrated intrafocal homogeneity for rearrangement status and concordant ERG protein expression. Intrafocal heterogeneity for ERG rearrangement has been reported in literature, ranging from 1 to 3% [4, 17, 26].
Similar to Park et al., we found no significant association between ERG expression and clinicopathological parameters, such as age, pre-operative PSA, Gleason score, and pathologic stage. Furusato et al. found that although ERG expression did not correlate with most clinicopathological features, when individual tumors were considered, higher Gleason score showed significant correlation with ERG-positive immunostaining [14]. Clinicopathological variables have been previously reported to be inconsistently correlated with fusion status [3, 4, 16, 17, 23, 25]. The different compositions of patient populations may explain the controversial results in literature.
ERG protein expression in vascular endothelial cells was herein used as positive internal control for the immunostaining, as endothelial cells lining blood vessels and capillaries show positive reactivity to anti-ERG antibodies in tumors as well as in nonneoplastic tissue samples [15, 27]. Infiltrating stromal lymphocytes also expressed ERG protein, as reported previously [15]. Since ERG rearrangement by FISH has never been found in lymphocytes or endothelial cells of either benign or neoplastic tissue specimens, the observed positive staining is consistent with antibody detection of a wild type ERG protein, as previously hypothesized [15]. ERG gene has in fact been shown to be constitutively expressed in vascular endothelial cells and related to endothelial homeostasis and angiogenesis; in addition ERG seems to play a role in lymphocyte development and differentiation [28, 29].
A potential limitation to this study is that cases were selected in part based on previous assessment of ERG rearrangement by FISH [16, 17] thus not representing temporally consecutive, unselected radical prostatectomy series.
Our findings independently confirmed the high concordance of ERG protein expression by immunohistochemistry with ERG rearrangement by FISH in a larger cohort of cases of clinically localized prostate cancer from radical prostatectomy specimens. Although the cases were not from a single institution, both FISH and immunohistochemistry results and interpretation were reviewed by a single pathologist.
Given the specificity of ERG rearrangement FISH patterns for neoplastic prostate, immunostaining with the anti-ERG antibody can be utilized as a surrogate for the genetic alteration. ERG immunohistochemistry has the advantage of being easily implemented as a routine laboratory technique; it has a high yield and can be rapidly performed and simply interpreted on tissue sections by light microscopy. The internal control represented by endothelial cells and infiltrating stromal lymphocytes contributes to the straightforward interpretation of immunostaining results.
The evaluation of ERG protein expression pattern in prostate cancer by immunohistochemistry may improve the understanding of its role in prostate tumorigenesis. Additional efforts are needed to further characterize the rare prostate cancer cases with discordant results between FISH rearrangement and ERG protein expression.
References
Rabbitts TH, Stocks MR (2003) Chromosomal translocation products engender new intracellular therapeutic technologies. Nat Med 9(4):383–386
Falini B, Martelli MP, Tiacci E, Ascani S, Thiede C, Pileri SA (2010) Immunohistochemical surrogates for genetic alterations of CCDN1, PML, ALK, and NPM1 genes in lymphomas and acute myeloid leukemia. Best Pract Res Clin Haematol 23(3):417–431
Mehra R, Tomlins SA, Shen R, Nadeem O, Wang L, Wei JT, Pienta KJ, Ghosh D, Rubin MA, Chinnaiyan AM, Shah RB (2007) Comprehensive assessment of TMPRSS2 and ETS family gene aberrations in clinically localized prostate cancer. Mod Pathol 20(5):538–544
Perner S, Mosquera JM, Demichelis F, Hofer MD, Paris PL, Simko J, Collins C, Bismar TA, Chinnaiyan AM, De Marzo AM, Rubin MA (2007) TMPRSS2–ERG fusion prostate cancer: an early molecular event associated with invasion. Am J Surg Pathol 31(6):882–888
Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, Lee C, Montie JE, Shah RB, Pienta KJ, Rubin MA, Chinnaiyan AM (2005) Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 310(5748):644–648
Mosquera JM, Mehra R, Regan MM, Perner S, Genega EM, Bueti G, Shah RB, Gaston S, Tomlins SA, Wei JT, Kearney MC, Johnson LA, Tang JM, Chinnaiyan AM, Rubin MA, Sanda MG (2009) Prevalence of TMPRSS2–ERG fusion prostate cancer among men undergoing prostate biopsy in the United States. Clin Cancer Res 15(14):4706–4711
Clark J, Merson S, Jhavar S, Flohr P, Edwards S, Foster CS, Eeles R, Martin FL, Phillips DH, Crundwell M, Christmas T, Thompson A, Fisher C, Kovacs G, Cooper CS (2007) Diversity of TMPRSS2–ERG fusion transcripts in the human prostate. Oncogene 26(18):2667–2673
Furusato B, Gao CL, Ravindranath L, Chen Y, Cullen J, McLeod DG, Dobi A, Srivastava S, Petrovics G, Sesterhenn IA (2008) Mapping of TMPRSS2–ERG fusions in the context of multi-focal prostate cancer. Mod Pathol 21(2):67–75
Scheble VJ, Braun M, Beroukhim R, Mermel CH, Ruiz C, Wilbertz T, Stiedl AC, Petersen K, Reischl M, Kuefer R, Schilling D, Fend F, Kristiansen G, Meyerson M, Rubin MA, Bubendorf L, Perner S (2010) ERG rearrangement is specific to prostate cancer and does not occur in any other common tumor. Mod Pathol 23(8):1061–1067
Petrovics G, Liu A, Shaheduzzaman S, Furusato B, Sun C, Chen Y, Nau M, Ravindranath L, Dobi A, Srikantan V, Sesterhenn IA, McLeod DG, Vahey M, Moul JW, Srivastava S (2005) Frequent overexpression of ETS-related gene-1 (ERG1) in prostate cancer transcriptome. Oncogene 24(23):3847–3852
Tomlins SA, Laxman B, Varambally S, Cao X, Yu J, Helgeson BE, Cao Q, Prensner JR, Rubin MA, Shah RB, Mehra R, Chinnaiyan AM (2008) Role of the TMPRSS2–ERG gene fusion in prostate cancer. Neoplasia 10(2):177–188
Klezovitch O, Risk M, Coleman I, Lucas JM, Null M, True LD, Nelson PS, Vasioukhin V (2008) A causal role for ERG in neoplastic transformation of prostate epithelium. Proc Natl Acad Sci USA 105(6):2105–2110
Carver BS, Tran J, Gopalan A, Chen Z, Shaikh S, Carracedo A, Alimonti A, Nardella C, Varmeh S, Scardino PT, Cordon-Cardo C, Gerald W, Pandolfi PP (2009) Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet 41(5):619–624
Furusato B, Tan SH, Young D, Dobi A, Sun C, Mohamed AA, Thangapazham R, Chen Y, McMaster G, Sreenath T, Petrovics G, McLeod DG, Srivastava S, Sesterhenn IA (2010) ERG oncoprotein expression in prostate cancer: clonal progression of ERG-positive tumor cells and potential for ERG-based stratification. Prostate Cancer Prostatic Dis 13(3):228–237
Park K, Tomlins SA, Mudaliar KM, Chiu YL, Esgueva R, Mehra R, Suleman K, Varambally S, Brenner JC, MacDonald T, Srivastava A, Tewari AK, Sathyanarayana U, Nagy D, Pestano G, Kunju LP, Demichelis F, Chinnaiyan AM, Rubin MA (2010) Antibody-based detection of ERG rearrangement-positive prostate cancer. Neoplasia 12(7):590–598
Falzarano SM, Navas M, Simmerman K, Klein EA, Rubin MA, Zhou M, Magi-Galluzzi C (2010) ERG rearrangement is present in a subset of transition zone prostatic tumors. Mod Pathol 23(11):1499–1506
Falzarano SM, Zhou M, Hernandez AV, Klein EA, Rubin MA, Magi-Galluzzi C (2011) Single focus prostate cancer: pathological features and ERG fusion status. J Urol 185(2):489–494
Rubin MA, Varambally S, Beroukhim R, Tomlins SA, Rhodes DR, Paris PL, Hofer MD, Storz-Schweizer M, Kuefer R, Fletcher JA, Hsi BL, Byrne JA, Pienta KJ, Collins C, Sellers WR, Chinnaiyan AM (2004) Overexpression, amplification, and androgen regulation of TPD52 in prostate cancer. Cancer Res 64(11):3814–3822
Espinosa I, Lee CH, Kim MK, Rouse BT, Subramanian S, Montgomery K, Varma S, Corless CL, Heinrich MC, Smith KS, Wang Z, Rubin B, Nielsen TO, Seitz RS, Ross DT, West RB, Cleary ML, van de Rijn M (2008) A novel monoclonal antibody against DOG1 is a sensitive and specific marker for gastrointestinal stromal tumors. Am J Surg Pathol 32(2):210–218
Seth A, Watson DK (2005) ETS transcription factors and their emerging roles in human cancer. Eur J Cancer 41(16):2462–2478
Esgueva R, Perner S, LaFargue CJ, Scheble V, Stephan C, Lein M, Fritzsche FR, Dietel M, Kristiansen G, Rubin MA (2010) Prevalence of TMPRSS2–ERG and SLC45A3-ERG gene fusions in a large prostatectomy cohort. Mod Pathol 23(4):539–546
Pflueger D, Rickman DS, Sboner A, Perner S, LaFargue CJ, Svensson MA, Moss BJ, Kitabayashi N, Pan Y, de la Taille A, Kuefer R, Tewari AK, Demichelis F, Chee MS, Gerstein MB, Rubin MA (2009) N-myc downstream regulated gene 1 (NDRG1) is fused to ERG in prostate cancer. Neoplasia 11(8):804–811
Attard G, Clark J, Ambroisine L, Fisher G, Kovacs G, Flohr P, Berney D, Foster CS, Fletcher A, Gerald WL, Moller H, Reuter V, De Bono JS, Scardino P, Cuzick J, Cooper CS (2008) Duplication of the fusion of TMPRSS2 to ERG sequences identifies fatal human prostate cancer. Oncogene 27(3):253–263
Mehra R, Han B, Tomlins SA, Wang L, Menon A, Wasco MJ, Shen R, Montie JE, Chinnaiyan AM, Shah RB (2007) Heterogeneity of TMPRSS2 gene rearrangements in multifocal prostate adenocarcinoma: molecular evidence for an independent group of diseases. Cancer Res 67(17):7991–7995
Perner S, Demichelis F, Beroukhim R, Schmidt FH, Mosquera JM, Setlur S, Tchinda J, Tomlins SA, Hofer MD, Pienta KG, Kuefer R, Vessella R, Sun XW, Meyerson M, Lee C, Sellers WR, Chinnaiyan AM, Rubin MA (2006) TMPRSS2:ERG fusion-associated deletions provide insight into the heterogeneity of prostate cancer. Cancer Res 66(17):8337–8341
Barry M, Perner S, Demichelis F, Rubin MA (2007) TMPRSS2–ERG fusion heterogeneity in multifocal prostate cancer: clinical and biologic implications. Urology 70(4):630–633
Mohamed AA, Tan SH, Mikhalkevich N, Ponniah S, Vasioukhin V, Bieberich CJ, Sesterhenn IA, Dobi A, Srivastava S, Sreenath TL (2010) Ets family protein, erg expression in developing and adult mouse tissues by a highly specific monoclonal antibody. J Cancer 1:197–208
Birdsey GM, Dryden NH, Amsellem V, Gebhardt F, Sahnan K, Haskard DO, Dejana E, Mason JC, Randi AM (2008) Transcription factor Erg regulates angiogenesis and endothelial apoptosis through VE-cadherin. Blood 111(7):3498–3506
Anderson MK, Hernandez-Hoyos G, Diamond RA, Rothenberg EV (1999) Precise developmental regulation of Ets family transcription factors during specification and commitment to the T cell lineage. Development 126(14):3131–3148
Disclosure/Conflict of Interest
The authors declare that they have no conflict of interests.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Falzarano, S.M., Zhou, M., Carver, P. et al. ERG gene rearrangement status in prostate cancer detected by immunohistochemistry. Virchows Arch 459, 441–447 (2011). https://doi.org/10.1007/s00428-011-1128-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00428-011-1128-4