Abstract
The World Health Organization–European Organization for Research and Treatment of Cancer has individualized three main categories among the primary cutaneous B cell lymphoma (PCBCL): leg-type primary cutaneous large B cell lymphoma (PCLBCL leg type), primary cutaneous follicle center lymphoma (PCFCL), and primary cutaneous marginal zone lymphoma (PCMZL). The genetic features of 21 PCBCL cases (six PCLBCL leg type four PCFCL large cells, seven PCFCL small cells, and four PCMZL) were investigated by comparative genomic hybridization (CGH array). Fluorescent in situ hybridization (FISH) analysis was performed to confirm CGH array data and to detect lymphoma-associated gene rearrangements. p14 ARF/p16 INK4a CDKN2A gene quantification, methylation analysis, and immunohistochemical detection were also performed. CGH array showed a higher number of recurrent genetic imbalances in PCLBCL leg type (mean 62) than in PCFCL large cells (mean 34). PCFCL small cells and PCMZL exhibited fewer chromosomal alterations (mean 24 and 9). FISH analysis provided concordant results with CGH array data in 97% (98 of 101) assays and demonstrated a t(8;14)(q24;q32) in two of six PCLBCL leg type. Recurrent deletions in 9p21 (p14 ARF/p16 INK4a CDKN2A) were a constant finding in PCLBCL leg type (six of six). Conversely, PCFCL large cells exhibited recurrent 1p36 deletions (four of four) without deletion in 9p21 (zero of four). The diagnostic and prognostic impact of the p16 INK4a CDKN2A gene status in PCBCL should therefore be confirmed on a larger series.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Primary cutaneous B cell lymphomas (PCBCL) are classified by the European Organization for Research and Treatment of Cancer (EORTC) into three main histopathological groups with distinct clinical and evolutive features [34]. While primary cutaneous follicle center cell lymphoma (PCFCL) and marginal zone B cell lymphoma (PCMZL) exhibit 97% and 100% 5-year survival, respectively, primary cutaneous large B cell lymphoma leg type (PCLBCL leg type) is considered an intermediately aggressive disease with 58% 5-year survival. Their relative frequency among cutaneous lymphomas is estimated at 13%, 2%, and 3%, respectively. However, the histological features of PCBCL largely differ from those of their nodal counterparts [15]. For example, BCL2 expression is generally absent from tumor cells of PCFCL that do not carry the t(14;18)(q32;q21) translocation, a cytogenetic hallmark of nodal follicular lymphomas [13, 30]. A great step toward uniform patient diagnosis and management was taken by the World Health Organization (WHO)–EORTC classification confirming that the overall majority of PCBCL fall into three diagnostic categories: PCMZL, PCFCL, and PCLBCL leg type [32, 33]. This classification takes into account recent advances on debated entities such as PCLBCL leg type that may occur outside the leg but typically exhibit a round cell morphology and a BCL2+ phenotype, unlike PCFCL [8, 11, 28].
However, clinicopathological variants include BCL2-negative PCLBCL leg type according to the site of involvement (7% at leg site vs 23% at nonleg site) [28], BCL2-positive large PCFCL (23%) [12] more frequently observed at the leg site (38%) that may represent a subgroup of PCFCL with worse prognosis [8, 28]. The lack of gold standard phenotypic criteria when using BCL-2, MUM-1, and FOXP1 markers may account for interobserver variation in large cell PCBCL diagnosis [11, 28]. Such cases may also be included in the other categories of PCBCL by some authors [20, 31]. However, other PCLBCL might correspond more to very rare diseases such as intravascular large B cell lymphoma, T cell/histiocyte-rich B cell lymphoma, or skin presentation of plasmablastic lymphoma rather than to a wait basket category [28, 32].
Therefore, additional analyses have been used for the subclassification of PCBCL, with a relative reproducibility between series that, in turn, may be due to differences in inclusion criteria. For example, the mutational pattern of the immunoglobulin gene of both PCFCL and PCLBCL leg type was primarily considered as germinal center derived [9]. In contrast, oligonucleotide microarray analysis identified a germinal center B cell-like signature in PCFCL and an activated B cell-like signature in PCLBCL leg type that was confirmed by immunohistochemistry [16, 17]. No IGH,myc, or bcl6 loci breakpoints were detected by interphase fluorescence in situ hybridization (FISH) in PCMZL or in PCFCL, whereas 11 out of 14 PCLBCL leg type harbored at least one of them [14]. In another report, PCLBCL leg type and nonleg type displayed the same genetic aberrations irrespective of classification, anatomic site, cell morphology, or BCL2 expression [31]. Using comparative genomic hybridization (CGH) or CGH array, the number of chromosomal alterations was generally found to be higher in PCLBCL leg type than in PCFCL, but the same recurrent chromosome abnormalities are rarely identified between series [10, 14, 24]. The use of a more sensitive and automatized CGH array procedure, restricted to academic institutions, showed high-level deoxyribonucleic acid (DNA) amplifications at 2p16.1 (63%) and deletion of chromosome 14q32.33 (68%) as the most recurrent alterations in PCFCL [6]. In the 12 PCLBCL leg-type cases, amplifications at 18q21.31-q21.33 were observed in 67% of cases. A small region at 9p21.3 involving the CDKN2A and CDKN2B loci was also observed, and the loss of CDKN2A by either deletion and/or promoter hypermethylation was associated with a poor outcome [6].
To extend these findings using commercially and worldwide available tools, we decided to select material from 21 PCBCL cases representative of the three main diagnostic categories of the WHO–EORTC classification. After a careful review of clinical and histopathological data, we performed CGH array. Recurrent imbalances were further investigated by FISH, and specific chromosomal losses such as 9p21 in PCLBCL leg type were confirmed. We also set up quantitative real-time polymerase chain reaction (PCR) assays to quantify the allelic loss of the p16 INK4a /CDKN2A and p14 ARF genes and to determine their methylation status.
Material and methods
Patient selection
We retrieved 21 patients with PCBCL displaying both clinical and immunohistochemical characteristic features (Table 1). Inclusion was based on the review of each case by a consensus panel according to the WHO–EORTC criteria and on the presence of at least 40% of tumor cells on frozen sections. This percentage was arbitrarily fixed to ensure a minimal amount of DNA from tumor cells for the CGH array analysis. Some cases were excluded when they did not exhibit such features or when the section of the frozen tissue revealed little tumoral infiltration. In accordance with morphological criteria used in a recent review of 300 PCBCL [28], six cases with confluent sheets of cells with a centroblast or immunoblast-like morphology corresponded to PCLBCL leg type. Four cases with confluent sheets of large cleaved cells (large centrocytes) corresponded to PCFCL large cells. Seven cases had a small cleaved cell morphology and were considered as PCFCL small cell. Four cases presented with PCMZL.
Immunohistochemistry
Immunohistochemistry was performed after a heat-induced antigen retrieval technique on paraffin-embedded sections using the biotin–streptavidin peroxidase LSAB kit (Dako, Trappes, France) on an automated Chemate (Dako). The primary antibodies were directed against CD20 (Dako), CD3 (Dako), BCL-2 (Dako), BCL-6 (Dako), and CD10 (Novocastra, Leica, Rueil-Malmaison, France), follicular dendritic cell antigens CNA42 (Dako), and CD21 (Novocastra), and MUM1 (Dako). For p16INK4a/CDKN2A immunohistochemical detection, two different antibodies and procedures were used on paraffin-embedded sections. The CINtec™ p16INK4ahistology kit including the primary antibody (clone E6H4), used at a 1:20 dilution for 1 h, was obtained from Dako together with the negative control antibody and visualization reagents. On parallel sections the monoclonal Pharmingen G175-405 monoclonal antibody (BD Biosciences, Le Pont-de-Claix, France) was used at a 1:10 dilution after a retrieval procedure at pH 8 in ethylenediamine tetraacetic acid (EDTA) buffer. Detection was further performed using the LSAB kit. On cervical intraepithelial lesions, both protocols provided a predominant nuclear staining of chromosome instability cells and a negative staining on adjacent normal epithelium. Occasional positive endothelial or fibroblast-like cells were found positive in the lamina propria. Positive controls also included tonsils with positive nuclear staining of basal epithelial cells and nodal large B cell lymphomas (n = 2) with more than 50% positive cells.
CGH array
For each sample, genomic DNA was isolated from ten fresh frozen tissue sections (20 μm) according to the manufacturer’s procedure after Proteinase K lysis, RNAse treatment, and isopropanol precipitation (Abbott-Vysis, Rungis, France). A qualitative control of DNA integrity was performed after electrophoresis on a 0.8% agarose gel. The DNA copy number changes in tumoral specimens were assessed by CGH array analysis using the Array 300 (Abbott-Vysis). The slides contained 287 clones spotted in triplicate including a set of subtelomeric sequences for each chromosome arm and 98 clones representing regions that have been reported to be amplified in human tumors. Genomic areas involved in homozygous or hemizygous deletion were also covered by 40 probes. DNA labeling and CGH array analysis were performed according to the manufacturer’s recommendations. Briefly, the DNAs were labeled by random priming using the Vysis kit. Then, cyanin 5 (Cy5)-isosex control and cyanin 3 (Cy3) test DNA were checked by gel analysis. After denaturation at 80°C, the DNAs were dropped on the chips, and hybridization was performed at 37°C for 60–72 h. Following hybridization and removal of the unhybridized probe, target spots were counterstained with diamidinophenylindole (DAPI). The analysis was performed using the GenoSensor Reader (Abbott-Vysis) which is a large-field multicolor fluorescence imaging system capturing an image of the hybridized chip in three color planes: Cy 3, Cy5, and DAPI blue. The included software automatically identifies each spot and, by analysis of the set of Cy3/Cy5 ratios on all targets, calculates the ratio most representative of the modal DNA copy number of the sample DNA. For each target, the normalized ratio of a target relative to the sample’s modal DNA copy number is provided, indicating the degree of gain or loss of copy number. The results were displayed as target reports, image, and coded image panes. The detection of copy ratio changes is highly dependent on the relative abundance of tumor cell DNA in the tissue specimen. As immunohistochemistry showed a high content of reactive inflammatory cells in some samples, we decided to set fluorescence ratios values at less than 0.9 for a genomic loss and at greater than 1.1 for a copy number gain to increase the sensitivity threshold.
FISH analysis
Interphase FISH assay was performed on touch imprints of frozen tissues of all cases, as previously described [1]. After fixation (methanol, formaldehyde) and enzymatic digestion (pepsin), specimens and probes were codenaturated and hybridized overnight according to the manufacturer’s instruction. After nuclei counterstaining by DAPI (Vector, Burlingame, USA), the cells were viewed using a fluorescent Axioplan II microscope (Zeiss, Le Pecq, France). Both dual-color dual-fusion (DCDF) and split probes (SP) were assayed. First, we sought systemic B cell lymphoma translocations t(11;14)(q13;q32), t(14;18)(q32;q21) and t(8;14)(q24;q32) by using DCDF probes LSI IGH-CCND1, IGH-BCL-2, and IGH-MYC (Abbott-Vysis). Breakpoints of IGH and MALT1 gene loci were also investigated with the corresponding Split Signal DNA Probes (SP, Dako). The FISH pattern of normal nuclei was two red (R) two green (G) spots (DCDF) or two fusion (F) signals using the SP probes, whereas positive nuclei typically exhibited a 1R1G2F pattern (DCDF) or a 1R1G1F pattern (SP). All variant patterns (1R2G1F, 2R1G1F, etc.) were taken into account for the exact determination of the percentage of normal and positive nuclei.
A second set of probes was employed on cases showing imbalances by CGH array for specific chromosomal regions. The following locus-specific probes were used: LSI AML1/ETO (21q22.3/8q22), MYC and MALT1, TOP2A/HER-2/CEP17 (17q21-q22/17q21.1/Centomere 17), p14 ARF/p16 INK4a /CDKN2A (9p21), 1p36/1q25, 7p tel, 19q tel. FISH patterns were determined by analyzing 200 nonoverlapped nuclei. The positive threshold to assess translocation or a gene breakpoint was calculated for each probe set as the mean + 3 SD of the percentage of nuclei with a positive FISH pattern in control samples. The calculated values for the IgH-BCL-2, IgH-CCND1, IgH-MYC MALT1, and IgH probe sets were 5%, 5%, 5%, 3%, and 10%, respectively. For the other probes, the cutoff values were 5% or less. This value (5%) was finally selected for all locus-specific probes with the exception of 10% for the IGH probes.
p14 ARF and p16 INK4a /CDKN2A allelic assay by quantitative real-time PCR
The abovementioned genes were assayed by quantitative PCR (Q-PCR) using a primer pair for the E2 exon shared by the p14 ARF and the p16 INK4a /CDKN2A genes and their corresponding probes [21]. Primers specific for the p16 INK4a /CDKN2A E1 exon and their specific probes did not provide reproducible Q-PCR results in our hands. The final primer and fluorescent probe concentration was 200 and 50 nM, respectively. Fifty nanograms genomic DNA was used as a template. The albumin gene served as control according to Hostein et al. [18]. Specificity was checked by melting curve analysis in preliminary experiments with SYBR green and efficiency by dilution curves. Healthy control DNA was used for normalization and showed a constant ratio between the target genes and the albumin control gene. The LN401 glioblastoma cell line (gift of Dr A. Merlo, University Hospital, Basel, Switzerland) exhibits a homozygous deletion at the INK4A locus but a normal Ct value for the control gene [21]. The Ct value for the albumin control gene was used to normalize DNA input around 10 ng (7.5 ± 5 ng). All samples exhibited the same Ct value for albumin (less than 1 Ct variation between samples and triplicates). The relative copy number of p16 INK4a /CDKN2A/p14ARF was expressed as the ratio of target gene Ct above the Ct values [18].
p14 ARF and p16 INK4a /CDKN2A methylation status
DNA methylation patterns in the CpG islands were determined by methylation-specific PCR (MSP) after bisulfite treatment of DNA to convert all unmethylated cytosines to uracils, according to Lehmann et al. with some modifications [22]. Two hundred nanograms of DNA were incubated in 200 mM NAOH at 42°C for 30 min in a total volume of 50 μL. After the addition of 275 μL of 3.6 M bisulfite containing 1 mM freshly prepared hydroquinone, samples were incubated for 5 h at 55°C in the dark. Then, 375 μL of 6 M sodium iodine (Sigma) containing 1 mM β-mercaptoethanol and 5 μL glass milk suspension (QIAEX II suspension, Qiagen, Hilden, Germany) were added and mixed thoroughly. After 10 min at room temperature and centrifugation (8,000 rpm, 3 min), the pellet was washed three times with 70% ethanol, once with 20 mM NAOH/90% ethanol 5 min at room temperature and twice with 90% ethanol. DNA was eluted from the dried pellet with 25 μL Tris–EDTA buffer for 15 min at 55°C. From this solution, 1 μL (8 ng) was used for MSP. After an initial denaturation step at 95°C for 10 min, MSP consisted in 40 cycles with 30 s at 95°C, 60 s at 60°C, and 30 s at 72°C.
For the p16 INK4a /CDKN2A promoter region, MSP was performed with E5/E6 primers amplifying methylated or unmethylated DNA together with specific probes for methylated or unmethylated DNA [22]. The primers and probe concentration were 200 and 100 nM, respectively. Per reaction, the Brilliant QPCR Core reagent kit was used with 2.5 mM MgCl2, 1.25 U SureStart Taq DNA polymerase, 0.8 mM deoxynucleotide triphosphates, and 30 nM reference dye in a total volume of 25 μL (Stratagene, Amsterdam, The Netherlands).
For p14 ARF MSP, the primer pairs amplified a region spanning six CpGs within the 5′ region of the gene [7]. Per reaction, the primer concentration was 200 nM in Brilliant QPCR Master Mix 30 nM reference dye in a total volume of 25 μL (Stratagene).
For all primers, melting curve analysis was performed, and the absence of amplification of native DNA samples prior to bisulfite treatment was checked. The American Type Culture Collection (ATCC) human genomic DNA (RPMI 8226; ATCC, Manassas, VA, USA) was used as the control for p16 INK4a /CDKN2A biallelic methylation. The HDLM2 cell line (gift from Dr J.F. Garcia, CNIO Madrid, Spain) was used as the methylated positive control for p14 ARF and p16 INK4a /CDKN2A [26]. A healthy control DNA showing no methylation for all loci was also included. The efficiency of Q-PCR for the amplification of methylated sequences was determined by serial dilutions of the methylated cell lines in the control unmethylated DNA. This allowed the detection of a 5% dilution of fully methylated DNA within the linear range.
Results
Clinical and histopathological data
Twenty-one PCBCL were included, and their characteristic features are summarized in Table 1.
Ten cases were characterized by a majority of large cells:
-
Six cases (Fig. 1d) were characterized by a round cell morphology and were classified as PCLBCL leg type. Five out of six were located on the leg, and one was located on the arm. BCL2 was highly expressed in all cases, BCL6 and CD10 were negative, and MUM1 was expressed in five out of six cases. Three of six patients presented with extracutaneous spread, and all had cutaneous relapse. The mean overall survival was 29 ± 26 months.
-
Four cases were classified as PCFCL large cells (Fig. 1c). They displayed a large cleaved cell morphology (large centrocytes) and were BCL2 negative. Two were BCL6 positive. The lesions were located on the face/scalp (two of four) or on the trunk (two of four). All patients are alive without disease. The mean follow-up was 77 31 months.
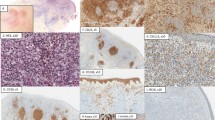
Histological and FISH features of PCBCL. a PCMZL: neoplastic proliferation with marginal zone features (×200). Insert: centrocytic-like cells and plasmacytoid differentiation (×1,000). b PCFCL small cells characterized by a follicular pattern (×400). Insert: predominance of small cleaved B-cells (×1,000). c PCFCL large cells with more than 80% of large cleaved B cells (×400). Insert (×1,000). d PCLBCL leg type characterized by a predominance of round large cells with prominent nucleoli (×400). Insert (×1,000). e FISH analysis using the LSI IGH-MYC DCDF probes (IGH: green spots; MYC: red spots). Nuclei with a 1R1G2F hybridization pattern (arrows) demonstrated a t(8;14)(q24;q32) in two of six PCLBCL leg-type cases (×630). f FISH analysis using the LSI p16 (red spots)/CEP 9 (green spots) probes. Nuclei with a 1R2G hybridization pattern indicated a hemizygous 9p21.3 deletion (p14 ARF/p16 INK4a gene) in five of six PCLBCL leg-type cases (×630)
Eleven cases were found to contain a majority of small tumoral cells:
-
Seven cases (Fig. 1b) were classified as PCFCL small cells because of a follicular architecture and a predominance of small cleaved cells (centrocytes) with a few large centroblastic cells. Their usual phenotype was BCL2+/−, BCL6+, and MUM1− (Table 1). The lesions were located predominantly on the trunk (four out of seven). All patients are alive. The mean follow-up was 85 23 months.
-
Four cases (Fig. 1a) were classified as PCMZL and were composed of a majority of plasma cells in one case or in three cases by a mixture of plasma cell, centrocytic-like cells with some residual reactive germinal centers, and a hyperplastic follicular dendritic network. Tumor cells had a BCL2-, BCL6-, and MUM1-negative phenotype. Three patients are alive without disease, and the last one has died of another cause. The mean follow-up was 54 18 months.
CGH array analysis
Both genomic losses (ratio less than 0.9) and copy number gains (ratio greater than 1.1) were detected by CGH array analysis. In the total group of PCBCL, the mean number of chromosomal aberrations was 34 (range 3 to 131 aberrations per case). Mean genomic gains (n = 22) were significantly more frequent than losses (n = 12, p < 0.05). The mean number of chromosomal imbalances (CI) was higher in large cell lymphomas (PCLBCL leg type and PCFCL large cells, n = 50) than in small cell lymphomas (PCFCL small cells and PCMZL, n = 18, p < 0.05). Some gains (Table 2) and losses (Table 3) were recurrently found in the total group of PCBCL. The most frequently observed anomalies were genomic gains in 7ptel (67%), 17q11.2 (48%), 3p21.3 (43%), 2p22.1-2p22.3 (38%), 11q13 (38%), 7q21.1 (29%), 12q12-q13 (29%), and losses in 9p11.2 (33%), 1p36 (29%), 1p36.31 (29%), 6q22-q23 (29%), 10q tel (29%), 1p36.32 (24%), 9p21 (24%), 9p21.3 (24%), 14q tel (24%) and Xp22.3 (24%).
The number of genomic alterations (mean ± SD) and the recurrence of these changes were also assessed for each PCBCL subtype. These different groups showed distinct patterns of hybridization (Fig. 2a–c). PCLBCL leg type exhibited more imbalances (62 ± 39) than PCFCL large cells (34 ± 5), PCFCL small cells (24 ± 19), and PCMZL (9 ± 2). PCLBCL leg type exhibited more genomic gains (37 ± 18) and losses (25 ± 21) than PCFCL large cells (24 ± 6, 10 ± 2), PCFCL small cells (17 ± 14, 7 ± 5), and PCMZL (7 ± 1, 2 ± 2). Within the PCFCL small cell group, patients with a unique skin lesion of the head or trunk showed more genomic changes (33 ± 21) than those with multiple lesions (13 ± 6).
CGH array analysis of PCBCL DNA was extracted from 21 tumoral skin biopsies and CGH analysis was performed using the Array 300 (Abbott-Vysis). The genomic alterations are indicated in a the group of PCLBCL leg type (N = 6), b the group of PCFCL large cells (N = 4), and c the group of PCFCL small cells (N = 7). Positive values represent gains, and negative values represent losses
In PCLBCL leg type, the most frequent findings (>50% of cases) were genomic gains in 7ptel (five of six), 17q11.2 (five of six), 1p36.33 (four of six), 3p21.3 (four of six), 7p21 (four of six), 7q11.23 (four of six), 7q21.1 (four of six), 11q13 (four of six), 12q12-q13 (four of six), 17q21-q22 (four of six), 19q tel (four of six), and losses in 9p21 (five of six), 9p21.3 (five of six), 6q22-q23 (four of six), and 17p11.2-p12 (four of six).
In the group of PCFCL large cells, only gains in 17q11.2 were found in more than 50% of the cases (three of four). Conversely, several recurrent losses were observed in 1p36 (four of four), 1p36.31 (four of four), 1p36.32 (three of four), 1p36.33 (three of four), and 1p tel (three of four). PCFCL small cells and PCMZL showed fewer genomic alterations than the other PCBCL groups. Gains in more than 50% of cases were observed only for PCFCL small cells: 7ptel (five of seven) and 2p22.1–2p22.3 (four of seven). No genomic loss was found in PCMZL or in the majority of PCFCL small cells.
FISH analysis
Analysis of all the PCBCL showed a t(8;14)(q24;q32) in 33% (two of six) PCLBCL leg type (Fig. 1e). Neither t(11;14)(q13;q32), t(14;18)(q32;q21), IGH, or MALT1 gene rearrangement was detected in the other PCBCL. FISH analysis was also performed to confirm the results of the CGH array analysis.
Both PCLBCL leg type (N = 6) and PCFCL large cells (N = 4) were assayed using commercially available locus-specific probes allowing the exploration of 11 loci for each of these ten cases: 1p36 (TP73), 7p tel, 8q22 (ETO), 8q24 (MYC), 9p21 (p14 ARF/p16 INK4a /CDKN2A), 11q13 (CCND1), 17q21 (TOP2A), 17q21 (HER-2), 18q21 (BCL2), 19q tel, and 21q22 (AML1). CGH array analysis found that these loci exhibited either a DNA loss (N = 7), DNA amplification (N = 35), or a normal (N = 68) pattern.
Hybridization of 101 of 110 loci was successfully performed by FISH assay. Concordant results between the two techniques were obtained in 97% (98 of 101) of the cases. In each case, abnormal FISH hybridization patterns were observed in a fraction of the nuclei (9–75%, mean 29%). These patterns always associated DNA losses with hemizygous deletions and DNA amplification with a low number gain (three copies) of the locus. The three discordant results corresponded to DNA amplification (1p36.33 in two PCLBCL leg type and 21q22 in one case of PCFCL large cells) found by CGH array but not by FISH analysis (normal hybridization pattern for all three cases).
Analysis of the chromosomal aberrations led to the identification of several oncogenes and/or tumor suppressor genes present in the regions of interest.
Deletion of chromosome 1p36
CGH array analysis showed a loss of the 1p36.22–1p tel chromosomal region (seven clones) in 50% (two of four) of the PCFCL large-cell cases. Such DNA loss was not found in the other PCBCL groups. The tumor suppressor genes PRKCZ, TP73, and CDC2L1 (p58) were covered by the deleted clones. According to CGH array analysis, a hemizygous deletion of TP73 was found by FISH (LSI 1p36/1q25 probe set) in 50% (two of four) of PCFCL large-cell cases but not in the PCLBCL leg-type group. The CDC2L1 (p58) gene was found deleted by CGH array in all the PCFCL large-cell cases (four of four).
Gain in chromosome 7
A DNA amplification of 5 of 12 clones hybridizing on the two arms of chromosome 7 was found by CGH array analysis in 50% (three of six) PCLBCL leg type. The oncogenes EGFR and MDR1 are located in the hybridized areas. A low copy number gain restricted to the chromosome 7p tel was also observed in 83% (five of six) of PCLBCL leg type, 50% (two of four) of PCFCL large cells, 71% (five of seven) of PCFCL small cells, and in 50% (two of four) of PCMZL. Five of these cases were assayed by FISH (LSI 7p tel probe), which demonstrated a trisomy of this locus in each case.
Gain in chromosome 11q13
CGH array analysis showed a gain in the chromosomal region 11q13 in 67% (four of six) of PCLBCL leg type, 50% (two of four) of PCFCL large cells, and in 29% (two of seven) of PCFCL small cells. FISH confirmed this finding and showed a trisomy of this locus covering the CCND1 gene.
Gain in chromosome 12q12-q13
A gain of 11 of 13 clones covering chromosome 12 was detected by CGH array analysis in 33% (two of six) of PCLBCL leg type. A common minimal region of gain (one clone) of the chromosomal region 12q12-q13 was found in 67% (four of six) PCLBCL leg type and in 50% (two of four) PCFCL large cells. This area covered the gene WNT1, which triggers oncogenic conversion. Another gain in 12q13-q14 encompassing the CDK2, CDK4, MDM2, and GLI oncogenes was found in 29–43% (two to three of seven) PCFCL small cells.
Gain in chromosome 17q11.2-q22
A DNA amplification of the chromosomal region 17q11.2-q22 (five clones) was found by CGH array analysis in 33% (two of six) of PCLBCL leg type. Two clones in this region more frequently showed a chromosomal alteration. One encompassing the THRA gene was associated with DNA amplification in 83% (five of six) of PCLBCL leg type, 75% (three of four) of PCFCL large cells, and in 29% (two of seven) of PCFCL small cells. The other clone covered the TOP2A gene and showed a gain in 67% (four of six) of PCLBCL leg type and in 50% (two of four) of PCFCL large cells. FISH experiments (LSI TOP2A/HER-2/CEP17) demonstrated a trisomy of the loci 17q11-q12 (TOP2A) and 17q11.2-17q12 (HER-2). FISH and CGH array results were strictly correlated.
Gain in chromosome 19q tel
A gain in the 19q tel chromosomal region was found by CGH array exclusively in 67% (four of six) of the PCLBCL leg type. FISH analysis (LSI 19q tel) confirmed this observation and showed a trisomy of this locus.
Deletion of chromosome 9p21.3 involving p14 ARF and p16 INK4a /CDKN2A
A loss of the 9p21.3 chromosomal region was observed in 83% (five of six) of PCLBCL leg type, in accordance with their CGH array analysis. FISH experiments using the LSI p14/p16 probe set identified a common altered region with a hemizygous 9p21.3 deletion encompassing the CDKN2A (p14 ARF/p16 INK4A), CDKN2B (p15 INK4B), and MTAP genes (Fig. 1f). The nondeleted PCLBCL leg-type case (no. 6) showed clinical lesions involving the arm and not the leg, as in the five other PCLBCL leg-type cases. None of the PCFCL large-cell cases (n = 4) exhibited 9p21.3 deletion, in accordance CGH array data (Table 4).
p14 ARF/p16 INK4a CDKN2A gene quantification
The genetic analysis of a common E2 exon of p14 ARF and p16 INK4a /CDKN2A genes was performed on PCLBCL leg type and PCFCL large cells. The relative copy number of p14 ARF/p16 INK4a /CDKN2A was expressed as the ratio of target exon Ct over the control gene Ct values [18]. Healthy control DNA as well as nondeleted samples were characterized by a Ct ratio = 1.1 ± 0.1. Cases with an exon E2-deleted ratio displayed a Ct ratio less than 0.5, and all corresponded to PCLBCL leg type, whereas PCFCL large cells displayed a normal range ratio. Due to a variable proportion between tumor and reactive cells, such global technique could not determine whether the p16 deletions were mono- or biallelic.
Finally, real-time Q-PCR correlated with both FISH and CGH array data in nine out of ten large PCBCL cases (Table 4). Case 6 displayed a deleted ratio for p14 ARF/p16 INK4a /CDKN2A exon2, which may correspond to cryptic deletion not detected by FISH or CGH array techniques using probes covering a much larger region.
p14 ARF and p16 INK4a /CDKN2A methylation analysis
After bisulfite modification of DNA, analysis was performed in parallel with different primer pairs specific for either the unmethylated DNA or methylated DNA. Despite the use of a very sensitive real-time PCR assay with specific probe detection for p16 INK4a /CDKN2A methylated DNA as shown by dilution experiments, no amplification for methylated sequences was achieved from PCLBCL leg type or PCFCL large cell samples.
p16 INK4a /CDKN2A immunohistochemical detection
Using two monoclonal antibodies with a different visualization system in parallel with several positive controls as described above, no labeling of tumor cell nuclei was observed in PCLBCL leg type or PCFCL large cell samples, except in case 8 with a minority of positive tumor cells (10%, data not shown). When no reactive or endothelial positive cell could be detected on consecutive sections, samples were interpreted as noninformative that could result from a sample over fixation or degradation.
Discussion
PCBCL is a heterogeneous group of B cell lymphomas falling into three main diagnostic categories: PCMZL, PCFCL, and PCLBCL leg type [32]. Our goal was therefore to analyze their genetic profiles on typical cases using commercially available tools that in turn could be used as ancillary diagnostic markers for borderline cases, especially between PCFCL and PCLBCL leg type. Therefore, we also separated large-cell PCFCL from small-cell PCFCL. The simple CGH array with a restricted set of genetic loci used in this study showed a higher number of CI (for gains or losses) in PCLBCL leg type (62 ± 39) than in PCFCL large cells (34 ± 5) or PCFCL small cells (24 ± 19). PCMZL exhibited few or no CI (9 ± 2). Moreover, FISH confirmed the CGH array data with a very high concordance (97%) when exploring the genetic status of 11 loci in ten cases of PCLBCL leg type (n = 6) and PCFCL large cells (n = 4). Our data are in broad accordance with previous studies using classical CGH showing a higher number of CI in PCLBCL leg type than in PCFCL [13, 14]. A combined CGH and CGH array study also found CI in 50% of diffuse large cell lymphoma, 33% of PCMZL, and in only one out of six PCFCL (8%) [23].
In our study, PCLBCL leg type was characterized in the majority of cases by a predominance of genomic gains at 1p36.33, 3p21.3, 7p21, 7ptel 7q11.23, 7q21.1, 11q13, 12q12-q13, 17q11.2, 17q21-q22, and 19q tel. Conversely, the most frequent losses were observed at 1p36, 1p36.31, 1p36.32, 1p36.33, 1p tel, 6q22-q23, 9p21, 9p21.3, and 17p11.2-p12. Our findings share some similarities with other series. Gains at 3p, 7p, 7q, and 12q and losses at 6q and 9p21 have also been reported in PCLBCL leg-type cases by other studies [6, 10, 14, 23].
In our study, PCFCL large cells displayed fewer genetic imbalances than PCLBCL leg type with rare recurrent gains at 2p, 12q12-q13, and 17q11.2 in accordance with other reports [14]. Specific gains at 2p13-p12 may indeed involve specific REL or BCL11A gene amplification as shown by FISH experiments [6, 14]. Strikingly, no other study has reported hemizygous losses at 1p36 observed by both CGH array and FISH as in our PCFCL large-cell cases.
PCFCL small cells and PCMZL showed even less frequent genomic alterations. Gains in the majority of cases were only observed for PCFCL small cells at 7ptel and 2p22.1-2p22.3: Differential diagnosis between PCMZL and PCFCL small cells may be further supported by their different genetic profiles. Some alterations such as 2p, 3q, and 11q13 gains were shared, although at different rates by PCLBCL leg type, PCFCL large cells, and PCFCL small cells, but were not seen in PCMZL cases. Despite selection of cases with marked dermal infiltration, the relative abundance of reactive cells may account for the negative CGH array results in PCMZL.
In contrast with the abovementioned reports, we did not detect any 18q21 gain in PCLBCL leg-type cases, and our CGH array data were confirmed by FISH experiments with both MALT1 and BCL2 probes. Indeed, 6q loss and 18q gain or amplification were primarily found in the majority of PCLBCL leg-type but not in PCFCL cases [10, 14, 24]. These imbalances were further detected in, respectively, 32% and 11% of PCFCL cases [6]. Differences in methodology (classical CGH vs. CGH array) and inclusion criteria prior to the WHO/EORTC classification make it difficult to correlate data obtained on different patients at heterogeneous clinical stages. BCL2 overexpression was detected by immunohistochemistry in six cases of PCLBCL leg type but was not correlated with a gain in 18q21. This observation is in accordance with a previous CGH report, which did not find any 18q gain in 9 of 16 diffuse large B cell lymphomas overexpressing BCL2 [2]. Conversely, chromosomal 18q amplifications were associated with the BCL2 overexpression in these lymphomas [25].
Interphase FISH experiments have suggested that t(14;18)(q32;q21) involving IGH–MALT1 could be observed in a subset of PCMZL with monocytoid morphology [27]. No MALT1 rearrangement was observed in our limited series of four MALT cases (three with monocytoid morphology), in accordance with larger reports showing the lack of MALT1 or IGH rearrangement in PCMZL [5, 13, 35]. Strikingly, Streubel et al. found by FISH the t(14;18)(q32;q21) involving IGH–BCL2 in a significant proportion (41%) of PCFCL, although consensus PCR with BIOMED-2 primers failed to amplify BCL2-JH rearrangement [29]. Such cases, which could represent borderline cases with systemic follicle center lymphoma and with unique skin presentation, were not observed in either our present or previous reports, in accordance with many other observers [3, 14, 19, 23, 30]. We also detected IGH breakpoints in two out of our six PCLBCL leg-type cases that were associated with MYC breakpoints without amplification in a reciprocal t(8;14)(q24;q32), as reported by others [13]. Conversely, no IGH breakpoint was detected in other PCBCL subtypes, as in our cases [13].
Interphase FISH was found to correlate with the CGH array technique for the detection of p16 INK4a /CDKN2A hemizygous deletion in five of six PCLBCL leg-type cases, whereas the remaining case was found to display a cryptic p16 INK4a /CDKN2A deletion by semiquantitative real-time PCR. Therefore, 9p21 loss not detected in other histological subtypes appears as the most recurrent genetic imbalance of PCLBCL leg type. Such loss was not found in conventional CGH studies, which may only detect larger deletions [10, 14]. Indeed, loss of heterozygosity at 9p21 was previously described in 2 out of 36 PCBCL cases [4]. In a recent array-based study, homozygous deletion of 9p21.3 was found in only 5 of 12 patients with PCLBCL leg type and was not observed in 19 patients with PCFCL [6]. In addition, complete methylation of the promoter region of p16 INK4a /CDKN2A but not of p15 INK4b /CDKN2B was also detected in two other PCLBCL leg-type cases, including one with hemizygous deletion. Such hypermethylation was not detected in our case despite the use of a sensitive real-time PCR technique. Our data and those of Dijkman et al. for p15 INK4b /CDKN2B [6] also contrast with the report of both p16 INK4a /CDKN2A and p15 INK4b /CDKN2B promoter hypermethylation in 43% and 23% of PCBCL cases of different subtypes [4]. In contrast with the latter report, we were unable to analyze p16 INK4a /CDKN2A immunohistochemical loss confidently according to the criteria used to calibrate our immunohistochemical procedure. More interestingly, all seven patients with PCLBCL leg type and p16 INK4a /CDKN2A inactivation of Dijkman et al. died like our patients, which would suggest that such genetic alterations are only seen in cases with adverse prognosis.
Finally, the genetic study of a restricted set of typical PCBCL strengthens their clinicopathological subclassification into three main categories. As the most aggressive disease, PCLBCL leg type requires therapeutic intensification, and its diagnosis may require further genetic studies, especially for borderline cases with PCFCL. Therefore, interphase FISH for the detection of IGH, MYC, or BCL6 breakpoints and especially p16 INK4a /CDKN2A deletion may be a valuable diagnostic tool. It would also be relevant to determine in a larger series whether p16 INK4a /CDKN2A deletion among PCBCL is truly specific of PCLBCL leg type.
References
Belaud-Rotureau MA, Parrens M, Dubus P, Garroste JC, de Mascarel A, Merlio JP (2002) A comparative analysis of FISH, RT-PCR, PCR, and immunohistochemistry for the diagnosis of mantle cell lymphomas. Mod Pathol 15:517–525
Berglund M, Enblad G, Flordal E, Lui WO, Backlin C, Thunberg U, Sundstrom C, Roos G, Allander SV, Erlanson M, Rosenquist R, Larsson C, Lagercrantz S (2002) Chromosomal imbalances in diffuse large B-cell lymphoma detected by comparative genomic hybridization. Mod Pathol 15:807–816
Child FJ, Russell-Jones R, Woolford AJ, Calonje E, Photiou A, Orchard G, Whittaker SJ (2001) Absence of the t(14;18) chromosomal translocation in primary cutaneous B-cell lymphoma. Br J Dermatol 144:735–744
Child FJ, Scarisbrick JJ, Calonje E, Orchard G, Russell-Jones R, Whittaker SJ (2002) Inactivation of tumor suppressor genes p15(INK4b) and p16(INK4a) in primary cutaneous B cell lymphoma. J Invest Dermatol 118:941–948
de la Fouchardiere A, Gazzo S, Balme B, Chouvet B, Felman P, Coiffier B, Salles G, Callet-Bauchu E, Berger F (2006) Cytogenetic and molecular analysis of 12 cases of primary cutaneous marginal zone lymphomas. Am J Dermatopathol 28:287–292
Dijkman R, Tensen CP, Jordanova ES, Knijnenburg J, Hoefnagel JJ, Mulder AA, Rosenberg C, Raap AK, Willemze R, Szuhai K, Vermeer MH (2006) Array-based comparative genomic hybridization analysis reveals recurrent chromosomal alterations and prognostic parameters in primary cutaneous large B-cell lymphoma. J Clin Oncol 24:296–305
Esteller M, Tortola S, Toyota M, Capella G, Peinado MA, Baylin SB, Herman JG (2000) Hypermethylation-associated inactivation of p14(ARF) is independent of p16(INK4a) methylation and p53 mutational status. Cancer Res 60:129–133
Geelen FA, Vermeer MH, Meijer CJ, Van der Putte SC, Kerkhof E, Kluin PM, Willemze R (1998) bcl-2 protein expression in primary cutaneous large B-cell lymphoma is site-related. J Clin Oncol 16:2080–2085
Gellrich S, Rutz S, Golembowski S, Jacobs C, von Zimmermann M, Lorenz P, Audring H, Muche M, Sterry W, Jahn S (2001) Primary cutaneous follicle center cell lymphomas and large B cell lymphomas of the leg descend from germinal center cells. A single cell polymerase chain reaction analysis. J Invest Dermatol 117:1512–1520
Gimenez S, Costa C, Espinet B, Sole F, Pujol RM, Puigdecanet E, Garcia-Moreno P, Sanchez J, Gallardo F, Estrach T, Garcia-Muret P, Romagosa V, Serrano S, Servitje O (2005) Comparative genomic hybridization analysis of cutaneous large B-cell lymphomas. Exp Dermatol 14:883–890
Grange F, Bekkenk MW, Wechsler J, Meijer CJ, Cerroni L, Bernengo M, Bosq J, Hedelin G, Fink Puches R, van Vloten WA, Joly P, Bagot M, Willemze R (2001) Prognostic factors in primary cutaneous large B-cell lymphomas: a European multicenter study. J Clin Oncol 19:3602–3610
Grange F, Petrella T, Beylot-Barry M, Joly P, D’Incan M, Delaunay M, Machet L, Avril MF, Dalac S, Bernard P, Carlotti A, Esteve E, Vergier B, Dechelotte P, Cassagnau E, Courville P, Saiag P, Laroche L, Bagot M, Wechsler J (2004) Bcl-2 protein expression is the strongest independent prognostic factor of survival in primary cutaneous large B-cell lymphomas. Blood 103:3662–3668
Hallermann C, Kaune KM, Gesk S, Martin-Subero JI, Gunawan B, Griesinger F, Vermeer MH, Santucci M, Pimpinelli N, Willemze R, Siebert R, Neumann C (2004) Molecular cytogenetic analysis of chromosomal breakpoints in the IGH, MYC, BCL6, and MALT1 gene loci in primary cutaneous B-cell lymphomas. J Invest Dermatol 123:213–219
Hallermann C, Kaune KM, Siebert R, Vermeer MH, Tensen CP, Willemze R, Gunawan B, Bertsch HP, Neumann C (2004) Chromosomal aberration patterns differ in subtypes of primary cutaneous B cell lymphomas. J Invest Dermatol 122:1495–1502
Harris NL, Jaffe ES, Diebold J, Flandrin G, Muller-Hermelink HK, Vardiman J, Lister TA, Bloomfield CD (1999) World Health Organization classification of neoplastic diseases of the hematopoietic and lymphoid tissues: report of the Clinical Advisory Committee meeting—Airlie House, Virginia, November 1997. J Clin Oncol 17:3835–3849
Hoefnagel JJ, Dijkman R, Basso K, Jansen PM, Hallermann C, Willemze R, Tensen CP, Vermeer MH (2005) Distinct types of primary cutaneous large B-cell lymphoma identified by gene expression profiling. Blood 105:3671–3678
Hoefnagel JJ, Mulder MM, Dreef E, Jansen PM, Pals ST, Meijer CJ, Willemze R, Vermeer MH (2006) Expression of B-cell transcription factors in primary cutaneous B-cell lymphoma. Mod Pathol 19:1270–1276
Hostein I, Pelmus M, Aurias A, Pedeutour F, Mathoulin-Pelissier S, Coindre JM (2004) Evaluation of MDM2 and CDK4 amplification by real-time PCR on paraffin wax-embedded material: a potential tool for the diagnosis of atypical lipomatous tumours/well-differentiated liposarcomas. J Pathol 202:95–102
Hsi ED, Mirza I, Gascoyne RD (2002) Absence of t(14,18) chromosomal translocation in primary cutaneous B- cell lymphoma. Br J Dermatol 146:1110–1111 discussion 1111–1112
Kodama K, Massone C, Chott A, Metze D, Kerl H, Cerroni L (2005) Primary cutaneous large B-cell lymphomas: clinicopathologic features, classification, and prognostic factors in a large series of patients. Blood 106:2491–2497
Labuhn M, Jones G, Speel EJ, Maier D, Zweifel C, Gratzl O, Van Meir EG, Hegi ME, Merlo A (2001) Quantitative real-time PCR does not show selective targeting of p14(ARF) but concomitant inactivation of both p16(INK4A) and p14(ARF) in 105 human primary gliomas. Oncogene 20:1103–1109
Lehmann U, Hasemeier B, Lilischkis R, Kreipe H (2001) Quantitative analysis of promoter hypermethylation in laser-microdissected archival specimens. Lab Invest 81:635–638
Mao X, Lillington D, Child F, Russell-Jones R, Young B, Whittaker S (2002) Comparative genomic hybridization analysis of primary cutaneous B-cell lymphomas: identification of common genomic alterations in disease pathogenesis. Genes Chromosomes Cancer 35:144–155
Mao X, Orchard G, Lillington DM, Child FJ, Vonderheid EC, Nowell PC, Bagot M, Bensussan A, Russell-Jones R, Young BD, Whittaker SJ (2004) BCL2 and JUNB abnormalities in primary cutaneous lymphomas. Br J Dermatol 151:546–556
Monni O, Joensuu H, Franssila K, Klefstrom J, Alitalo K, Knuutila S (1997) BCL2 overexpression associated with chromosomal amplification in diffuse large B-cell lymphoma. Blood 90:1168–1174
Sanchez-Aguilera A, Delgado J, Camacho FI, Sanchez-Beato M, Sanchez L, Montalban C, Fresno MF, Martin C, Piris MA, Garcia JF (2004) Silencing of the p18INK4c gene by promoter hypermethylation in Reed-Sternberg cells in Hodgkin lymphomas. Blood 103:2351–2357
Schreuder MI, Hoefnagel JJ, Jansen PM, van Krieken JH, Willemze R, Hebeda KM (2005) FISH analysis of MALT lymphoma-specific translocations and aneuploidy in primary cutaneous marginal zone lymphoma. J Pathol 205:302–310
Senff NJ, Hoefnagel JJ, Jansen PM, Vermeer MH, van Baarlen J, Blokx WA, Canninga-van Dijk MR, Geerts ML, Hebeda KM, Kluin PM, Lam KH, Meijer CJ, Willemze R (2007) Reclassification of 300 primary cutaneous B-cell lymphomas according to the new WHO–EORTC classification for cutaneous lymphomas: comparison with previous classifications and identification of prognostic markers. J Clin Oncol 25:1581–1587
Streubel B, Scheucher B, Valencak J, Huber D, Petzelbauer P, Trautinger F, Weihsengruber F, Mannhalter C, Cerroni L, Chott A (2006) Molecular cytogenetic evidence of t(14;18)(IGH;BCL2) in a substantial proportion of primary cutaneous follicle center lymphomas. Am J Surg Pathol 30:529–536
Vergier B, Belaud-Rotureau MA, Benassy MN, Beylot-Barry M, Dubus P, Delaunay M, Garroste JC, Taine L, Merlio JP (2004) Neoplastic cells do not carry bcl2-JH rearrangements detected in a subset of primary cutaneous follicle center B-cell lymphomas. Am J Surg Pathol 28:748–755
Wiesner T, Streubel B, Huber D, Kerl H, Chott A, Cerroni L (2005) Genetic aberrations in primary cutaneous large B-cell lymphoma: a fluorescence in situ hybridization study of 25 cases. Am J Surg Pathol 29:666–673
Willemze R (2006) Primary cutaneous B-cell lymphoma: classification and treatment. Curr Opin Oncol 18:425–431
Willemze R, Jaffe ES, Burg G, Cerroni L, Berti E, Swerdlow SH, Ralfkiaer E, Chimenti S, Diaz-Perez JL, Duncan LM, Grange F, Harris NL, Kempf W, Kerl H, Kurrer M, Knobler R, Pimpinelli N, Sander C, Santucci M, Sterry W, Vermeer MH, Wechsler J, Whittaker S, Meijer CJ (2005) WHO–EORTC classification for cutaneous lymphomas. Blood 105:3768–3785
Willemze R, Kerl H, Sterry W, Berti E, Cerroni L, Chimenti S, Diaz-Perez JL, Geerts ML, Goos M, Knobler R, Ralfkiaer E, Santucci M, Smith N, Wechsler J, van Vloten WA, Meijer CJ (1997) EORTC classification for primary cutaneous lymphomas: a proposal from the Cutaneous Lymphoma Study Group of the European Organization for Research and Treatment of Cancer. Blood 90:354–371
Wongchaowart NT, Kim B, Hsi ED, Swerdlow SH, Tubbs RR, Cook JR (2006) t(14;18)(q32;q21) involving IGH and MALT1 is uncommon in cutaneous MALT lymphomas and primary cutaneous diffuse large B-cell lymphomas. J Cutan Pathol 33:286–292
Acknowledgments
This work was supported by a grant from the Programme Hospitalier de Recherche Clinique (CHU Bordeaux) and from the Ligue Nationale contre le Cancer (comité Aquitaine Charentes). We thank Dr. Ray Cooke for his linguistic advice.
Conflict of interest statement
We declare that we have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Belaud-Rotureau, MA., Marietta, V., Vergier, B. et al. Inactivation of p16 INK4a /CDKN2A gene may be a diagnostic feature of large B cell lymphoma leg type among cutaneous B cell lymphomas. Virchows Arch 452, 607–620 (2008). https://doi.org/10.1007/s00428-008-0593-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00428-008-0593-x