Abstract
The bone morphogenetic proteins (BMPs) are a family of the transforming growth factor-β (TGF-β) superfamily that perform multiple roles during vertebrate and invertebrate development. Here, we report the molecular cloning of a novel BMP from regenerating arms of the ophiuroid Amphiura filiformis. The theoretically translated amino acid sequence of this novel BMP has high similarity to that of the sea urchin BMP univin. This novel BMP has been named afuni. Whole-mount in situ hybridisation implicates afuni in arm regeneration. Expression occurs in distinct proximal and distal regions of late regenerates (3- and 5-week postablation). These sites are at different stages of regeneration, suggesting multiple roles for this gene in adult arm development. Cellular expression of this gene occurs in migratory cells within the radial water canal (RWC) of regenerating and nonregenerating arms. These migrating coelomocytes suggest a key role for the coelomic RWC as a source of the cellular material for use in arm regeneration by A. filiformis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The bone morphogenetic proteins (BMPs) are one of the largest families of signalling molecules yet described (Ueno and Moos 1997). They make up almost one third of the known members of the transforming growth factor-β (TGF-β) superfamily, with over 30 BMPs having been identified to date (Ducy and Karsenty 2000). The term bone morphogenetic protein was first used by Urist et al. (1973), who used bone extracts to induce bone formation when implanted in ectopic sites in rats. Cloning of BMP genes, however, was not achieved until Wozney et al. (1988) isolated three human cDNA clones corresponding to BMPs and found them independently capable of inducing the formation of cartilage in vivo. BMPs are now known to perform diverse biological functions in a range of processes including wound healing (see review by O’Kane and Ferguson 1997) and both vertebrate and invertebrate embryonic development (see review by Graff 1997). They do this by acting as morphogens that specify cell fates in a concentration-dependent manner.
During early vertebrate embryonic development, BMP4 signalling induces epidermal cell fate in the ectoderm. Suppression of BMP4 signalling by dorsally expressed antagonists (including chordin, noggin, and follistatin) leads to ectodermal cells adopting a neural fate (Graff 1997; Chang and Hemmati-Brivanlou 1998). Later in vertebrate development, BMPs are involved in patterning of the neural tube.
TGF-βs have been isolated from numerous phyla throughout the animal kingdom from cnidarians (e.g., Reinhardt et al. 2004) to vertebrates (e.g., Wozney et al. 1988). The first TGF-β cloned from an echinoderm was univin, a BMP family member isolated from embryos of the sea urchin Strongylocentrotus purpuratus (Stenzel et al. 1994). High levels of univin expression were seen in the egg and prehatched blastula with expression becoming more restricted later in development. At gastrulation, univin expression is restricted to the foregut and ciliated band. In later functional studies, microinjection of synthetic univin mRNA into Paracentrotus lividus embryos rescued defects in skeleton elongation caused by interfering with cell–extracellular matrix (ECM) interactions engineered by blocking nectin with a monoclonal antibody (Zito et al. 2003). More recently, members of the BMP2/4 (Angerer et al. 2000) and BMP5/8 (Ponce et al. 1999) subfamilies have been cloned from sea urchin. Functional analysis of BMP2/4 in sea urchin embryos revealed a role for this molecule in patterning the animal-vegetal axis. Here, BMP2/4 and its antagonist noggin play critical roles in positioning of the ectoderm–endoderm boundary and in specifying epidermal versus nonepidermal cell fate in the ectoderm.
Other echinoderm BMPs have now been characterised from starfish (Shih et al. 2002), sea cucumber (Harada et al. 2002), and feather star (Patruno et al. 2003). As invertebrate deuterostomes, echinoderms occupy an important phylogenetic position with respect to chordates. Moreover, many echinoderms exhibit maximal indirect development. Maximal indirect development is defined as being where the construction of embryos and adults are, mechanistically, separated in time. The adults arise from a subgroup of pluripotential cells in the embryo that are set aside during embryogenesis and used later to generate the adult structures. Patterning of adult structures occurs only after embryonic development is complete (Davidson et al. 1995; Peterson et al. 1997). It should be noted that maximal indirect development is distinct from the indirect development seen in some vertebrates (such as various species of amphibian) and terrestrial insects, in which the ‘secondary larvae’ possess fundamental features of the adult body plan. In these indirect developers with secondary larvae and in direct developers, the major features of body plans are already patterned during embryogenesis (for example, the head and A–P axis). It is therefore postulated that these indirect developers with secondary larvae are essentially direct developers (Davidson et al. 1995).
In either case (‘direct’ or maximal indirect), the period of development that has received least attention is that of postembryonic and postmetamorphic development in the adult. Part of the reason is that the majority of the commonly studied “models” (Caenorhabditis elegans, arthropods, vertebrates) are essentially direct developers. Furthermore, in those animals that undergo maximal indirect development, attention has focused on the embryo because for diverse technical reasons, adult morphogenesis is generally more inaccessible to experimental manipulation than embryogenesis. For example, the highly calcified nature of adult echinoderm tissue in relation to embryos increases the difficulty of achieving successful nucleic acid probe penetration into tissues.
The phenomenon of adult regeneration in echinoderms therefore provides a unique opportunity to explore development in the adult cellular and tissue environment. The expression of a BMP2/4 in echinoderm regeneration was investigated for the first time in arms of the feather star Antedon bifida (Patruno et al. 2003). AnBMP2/4 is expressed in both nonregenerating and regenerating arms with upregulation detected during the wound repair (24- to 48-h postamputation) and early regenerative phases (48- to 72-h postamputation). In the present report, we describe the identification and sequence analysis of afuni, a TGF-β isolated from regenerating arm tissue of the brittle star Amphiura filiformis. This is the first known reported TGF-β from a brittle star and we demonstrate by in situ hybridisation analysis that this gene is upregulated in specific cell types in arm regeneration.
Materials and methods
Animal collection and induction of regeneration
Adult A. filiformis were collected at Kristineberg Marine Research Station, Sweden. Brittle stars were obtained by sediment grab and maintained in marine sediment in a natural circulating seawater system at 12°C. Arm autotomy was induced by gently grasping the midproximal region of the arm using forceps. Breakage typically occurred along a natural autotomy plane two to ten segments proximal to the site of manipulation. To reduce the likelihood of loss of material due to later manipulative damage, two arms per individual underwent induced autotomy. Regenerating animals were held in sediment in seawater at 12°C supplied with aeration. Regenerated arm tissue was removed along with a few segments of the nonregenerating arm using a scalpel.
RNA isolation, cDNA synthesis, reverse transcriptase-polymerase chain reaction (RT-PCR), and cloning
Total RNA was isolated from regenerating arms of A. filiformis using tri-reagent (Sigma) and reverse-transcribed into single-stranded cDNA using Superscript II (Invitrogen). Degenerate primers were designed based on a conserved region of TGF-β genes for use in PCR (forward, 5′-ACTCTAGATGGATYRTNGCNCC-3′; reverse, 5′-ACGAATTCTTANCGRCANCCNCA-3′; modified from Stenzel et al. 1994). The PCR reaction mix consisted of 1 μl DNA, 13.2 μl water, 2.5 μl 10× PCR buffer (DNAmp), 1.5 μl 25 mM Mg(OAc)2, 0.5 μl 10 mM dNTP mix, 1 μl of each 10 μmol primer solution, and 0.3 μl accurase DNA polymerase (DNAmp). Cycling conditions were 94°C for 1 min, followed by 45 cycles of 94°C for 1 min, 55°C for 2 min, and 72°C for 3 min. A final extension step of 72°C for 10 min was used. A 229-base PCR product was purified from an agarose gel using the QIAquick gel extraction kit (QIAgen) and cloned into the pCR 2.1-TOPO vector (Invitrogen). Plasmids were transformed into TOP10 chemically competent Escherichia coli cells (Invitrogen). Plasmid DNA from selected colonies was purified using the QIAprep spin miniprep kit (QIAgen) and sequenced at MWG-Biotech AG Laboratories.
PCR screen of cDNA library
A gene-specific primer (5′-CCGGCACCCGCATGCTTCTACAACCATG-3′) was used in conjunction with the M13 forward primer (5′-GTAAAACGACGGCCAG-3′) to PCR probe an A. filiformis regenerating arm Unizap cDNA library (Stratagene) to attain the full-coding sequence (930 bases) and 5′ untranslated region (156 bases) of afuni. The titer of the amplified library was 2.7×1011 pfu/ml. The PCR reaction mix consisted of 0.5 μl library DNA, 16.04 μl water, 2 μl 10× PCR buffer, 1.2 μl 25 mM Mg(OAc)2, 0.16 μl 10 mM dNTP mix, 0.2 μl of each 10 μmol primer solution, and 0.2 μl accurase DNA polymerase (DNAmp). Cycling conditions were 94°C for 2 min, then 35 cycles of 94°C for 1 min, 60.8°C for 1 min, and 72°C for 1 min. A final extension step of 72°C for 10 min was used. PCR product was purified from an agarose gel using the QIAquick gel extraction kit (QIAgen), cloned into the pGEM-T easy vector system (Promega) and transformed into XL-1 Blue supercompetent E. coli cells (Stratagene). Plasmid DNA from selected colonies was purified using the QIAprep spin miniprep kit (QIAgen) and sequenced at MWG-Biotech AG Laboratories.
Computer analyses of amino acid sequence
Searches were conducted for genes with high-sequence similarity to the obtained clones using DNA>protein-translated basic local alignment search tool (BLAST) located on the National Centre for Biotechnology Information web site (http://www.ncbi.nlm.nih.gov/BLAST). Theoretically translated amino acid sequences were aligned using NPS@: network protein sequence analysis ClustalW (Combet et al. 2000). Phylogenetic analyses and tree construction were performed using the neighbour-joining method (Saitou and Nei 1987) on PAUP v4.0.
Whole-mount in situ hybridisation
The whole-mount in situ hybridisation protocol used here was adapted from the method described by Nieto et al. (1996). A 795-base fragment of afuni-translated sequence was used as a template to synthesise antisense and sense RNA probes. The identified region was isolated from insert-containing plasmid by PCR using gene specific primers (5′-CAACGTATCAACGATCCCTCG-3′, a sense primer designed against nucleotides 129–149 of the coding region, and 5′-CCCGCATGCTTCTACAACCAT-3′, an antisense primer designed to hybridise to nucleotides 904–924). Reaction mix and cycling conditions were as described for use in the PCR screen of the cDNA library above. The PCR product was purified from an agarose gel using the QIAquick gel extraction kit (QIAgen), ligated into the pGEM-T easy vector system (Promega) and transformed into XL-1 Blue supercompetent E. coli cells (Stratagene). The sequence and orientation of DNA inserts was checked by sequencing (MWG-Biotech AG Laboratories). Plasmid DNA was linearised by restriction digestion with either NcoI (antisense) or SpeI (sense) to produce DNA templates for RNA probe transcription. RNA probes were synthesised by transcription using Sp6 (antisense) and T7 (sense) RNA polymerases on phenol/chloroform/isoamyl alcohol (25:24:1)-purified plasmid DNA in a reaction containing a digoxigenin-labelled nucleotide mix (Roche). Probes were treated with DNase I and purified using 4 M LiCl2 prior to use.
Tissue samples from nonregenerating arms and regenerating arms 2-day, 1-week, 2-week, 3-week, and 5-week postamputation were examined. Numerous replicate arms per sample (at least 5) were included in each sample. Fixation of A. filiformis arms and hybridisation of antisense and sense (control) probes were performed essentially as described by Holland et al. (1996). In addition to this protocol, tissue was treated with 6% hydrogen peroxide in phosphate-buffered Tween 20 (PBT) for 20 min and PBT washed prior to RNA hybridisation. Transcripts were identified by colour development of an anti DIG-AP antibody (Dako). Whole-arm tissue examination was conducted using a Zeiss Axioskop 2 compound optical microscope connected to a camera (Olympus). Tissue was embedded in 20% gelatin/PBS and 30-μm-thick sections were cut using a Vibratome Classic 220 for optical microscope examination using the Zeiss Axioskop 2 compound optical microscope. Material was also embedded in medium hardness Agar 100 resin, ca. 2-μm-thick sections cut using a Reichert OMU3 Ultramicrotome and examined using a Leica DMR optical microscope.
Results
Isolation of afuni from regenerating A. filiformis arms
A 229-base fragment (excluding sequence derived from degenerate primers) of a novel TGF-β gene was isolated from regenerating arm tissue from A. filiformis by degenerate primer PCR. Further, the 5′ untranslated region, full-coding region and 3′ stop codon was obtained by a PCR screen of a cDNA library. This sequence has been submitted to Genbank (accession number AY954372). The theoretically translated amino acid sequence contains a carboxy-terminal region consisting of seven cysteines and a putative arg–X–X–arg site (where X is any amino acid) where proteolytic cleavage occurs. These features are characteristic of the BMPs (Ponce et al. 1999). Theoretically translated amino acid sequence of the conserved carboxy-terminal region shows high-sequence similarity to numerous BMPs, the highest of which is to sea urchin univin (85%, Fig. 1). Identities were lower with members of the BMP2/4 and BMP5/7 subgroups (67% and 60% with sea urchin BMP2/4 and BMP5/7, respectively). This novel gene was named afuni. It should also be noted that afuni has 66% amino acid sequence identity with another BMP known to be involved in echinoderm regeneration, anBMP2/4 (Patruno et al. 2003), across comparable regions. However, anBMP2/4 has been excluded from further phylogenetic analyses as sequence of only part of the C-terminal domain of this gene is known.
Alignment of the carboxy-terminal region of afuni with corresponding sequences from other members of the TGF-β superfamily. Differences between afuni and compared sequences are indicated in the body of the table. Percentage sequence identities with afuni are shown on the right of the table. Asterisk indicates conservation of amino acids between all sequences. Seven conserved cysteines, characteristic of BMPs, are highlighted. Sequences shown (and Genbank accession numbers) are (1) afuni, (2) Strongylocentratus purpuratus univin (U10533), (3) Homo sapiens BMP2 (M22489), (4) H. sapiens BMP4 (M22490), (5) S. purpuratus BMP2/4 (AF119713), (6) H. sapiens BMP5 (M60314), (7) H. sapiens BMP7 (X51801), (8) Xenopus laevis GDF6 (U08338), (9) S. purpuratus BMP5-7 (Z48313), (10) X. laevis Vg1 (M18055), (11) Drosophila melanogaster decapentaplegic (M30116), (12) D. melanogaster 60A (M77012), (13) H. sapiens inhibinαb (M13436), and (14) H. sapiens TGF-β1 (X02812)
Phylogenetic analyses using the neighbour-joining method confirmed the position of afuni within the BMP family of the TGF-β superfamily (Fig. 2). Bootstrap analyses of clades in Fig. 2 indicate that afuni is phylogenetically closest to sea urchin univin. The tree shown places afuni and univin close to the clade containing members of the BMP2/4-DPP subgroup and phylogenetically more distant from the BMP5/8-60A subgroup.
Phylogenetic tree showing the relationships between afuni and other vertebrate and invertebrate BMPs. The tree was constructed using neighbour joining to compare distances between the theoretically translated conserved carboxy-terminal region (110 amino acids) of each BMP beginning at the first conserved cysteine. Numbers associated with branches indicate the percentage of 1,000 bootstrap replicates supporting the topology shown. hsTGF-β1 was defined as an outgroup for tree construction. Sequences used (and accession numbers not previously stated) are afuni; D. melanogaster (dm) DPP and 60A; Halocynthia roretzi (hr) BMPa (D83183) and BMPb (D85464); H. sapiens (hs) BMP2, BMP4, BMP5, BMP7, and TGF-β1; Mus musculus (mm) GDF1 (M51639) and GDF3 (L06443); S. purpuratus (sp) BMP2/4, BMP5/7, and univin; X. laevis (xl) Vg1. See Fig. 1 legend for Genbank accession numbers not given here
A. filiformis arm regeneration
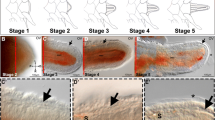
Figure 3 illustrates a simplified profile of the development of the regenerating A. filiformis arm at 12°C. Following autotomy, wound healing occurs and a blastema can be seen at the site of amputation at 2-day postablation (Fig. 3a). This blastema increases in length over the ensuing days (Fig. 3b). This increase in length continues, and morphological features, such as recognisable segments and associated spines and podia, appear around 2- to 3-week postablation and can be clearly seen after 1 month (Fig. 3c). It should be noted that arm regeneration occurs by ‘terminal growth’, with cell proliferation and differentiation occurring at the distal tip of the regenerate. Ophiuroid arm regeneration is characterised by an initial increase in arm length followed by later increase in width of the then-proximal regenerated arm (Stancyk et al. 1994).
Arm regeneration in A. filiformis. Arm autotomy was induced and regeneration occurred as described in “Materials and Methods.” a Following ablation, wound healing occurs, and by 2 days, a blastema has formed (arrow). b After 1 week, the regenerate has increased in length. Arrow indicates the transition between the previously established arm and the regenerate. c At 1 month, the arm has increased in length, and morphological features including clearly defined segments with associated spines can be seen, particularly in the wider proximal-most region of the regenerate (arrow)
Expression of afuni during arm regeneration by A. filiformis
In situ hybridisation was performed to elucidate the expression of afuni both in normal adult arms and during arm regeneration. The time points used in this study include early regeneration following wound healing when the blastema is formed (2 days and 1 week), and later stages of regeneration when arm structures, such as spines and podia, are forming (2 and 3 weeks) and differentiating into the fully mature structures (3 and 5 weeks). Application of the afuni antisense RNA probe to nonregenerating arms revealed an expression pattern running in a proximal-distal direction on the oral side of the arm (Fig. 4a). This pattern was not seen in arms treated with the afuni sense probe (Fig. 4b). Longitudinal sections (30 μm) taken from colour-developed nonregenerating arms indicate that this afuni antisense probe staining is localised in the radial water canal (RWC, Fig. 4c).
Afuni expression in A. filiformis arms. a Whole mount in situ hybridisation shows expression of afuni running in a proximal–distal direction along the oral side of the arm (×10 magnification). Arrowheads indicate nonspecific staining at the bases of podia, attributable to colour trapping by valves linking the RWC to the podia at these locations. b Expression is not seen in nonregenerating arms treated with the afuni sense probe (×10 magnification). Arrowheads indicate colour-trapping at the bases of podia as also seen in the antisense afuni probe development. c Longitudinal 30-μm section taken from the oral side of nonregenerating arm shows that the observed staining (arrow) is localised in a region containing the RWC (×10 magnification). Arrowheads indicate colour trapping at the bases of podia, which can be seen in this section as circular structures. d Afuni expression in 3-week regenerates (×5 magnification). Two unique sites of expression can be seen at the proximal region (black arrow) and toward the distal tip (red arrow). Arrowhead indicates the point of transition between the original arm and the regenerate. An oral view of this arm is shown; however, it should be noted that the tip of the regenerate is contorted so that the lateral side of this region is visible. e The proximal region of expression in the regenerate shows a clear segmental distribution (arrows) (×10 magnification, oral view). f The expression observed near the distal tip of the regenerate (black arrow) occurs in a clearly defined region at a site where clear segmental definition is first observed distally (red arrow) (×20 magnification, lateral view). Scale bars represent 200 μm, except F (100 μm). Proximal (p) and distal (d) ends of the arm are indicated
In regenerating arms, afuni expression was not detected by in situ hybridisation until 3 weeks after amputation. Here, expression was observed in two separate regions of the regenerate (Fig. 4d). In the proximal portion of the regenerate, afuni expression was seen occurring in a pattern following the newly formed segments of the arm (Fig. 4e). This expression coincides with the position of the newly formed segmental skeletal vertebrae. Toward the distal tip of the regenerate, afuni expression was also observed in a region where segmental patterning of the regenerate is first observed (Fig. 4f). There is a distinct region between the proximal and distal sites where no afuni expression is detected (Fig. 4d). No expression was observed in arms treated with the sense control probe (data not shown). This expression pattern was also observed in regenerating arms 5-week postamputation (data not shown).
Resin sections (2 μm) taken from 3-week regenerates confirm that both the proximal and distal staining is cellular (Fig. 5). Expression in the proximal region of the regenerate is observed in the mid-oral region of the arm when viewed in transverse section (Fig. 5a). When viewed at higher magnification, this proximal staining is localised to cells located in the area of the radial nerve cord (RNC) and in the RWC including branches of this structure (Fig. 5b). Distal staining occurs exclusively and extensively in the centre of the regenerating arm (Fig. 5c) in the newly developing RWC but not the RNC (Fig. 5d). At both proximal and distal locations, the subset of cells expressing afuni all appear elongated in comparison to the other more rounded cells seen in the sections (Fig. 5b and d). These elongate cells also appear to possess processes (Fig. 5d), which we use as indicative evidence to suggest that these cells are plausibly migratory in nature. However, unequivocal identification of these cells as migratory would require use of an appropriate marker. At present, no such marker is available.
Transverse sections (2 μm) showing afuni expression in regenerating arms 3-week postablation. a Transverse section of staining in the proximal region of regenerate (×40 magnification). Cells expressing afuni (dark colour) are localised in the region identified by the box at the centre of the oral side of the arm. b ×60 magnification of area within black box in a. Black arrow indicates dark elongate cells in the RNC/RWC region of the arm showing expression of afuni. White arrow indicates a lateral branch of the RWC containing dark elongate cells. c Transverse section of staining in the distal region of regenerate (×40 magnification). Cells expressing afuni (dark colour, representative cells indicated by arrows) are localised in a region containing the RWC (boxed). d ×80 magnification of area within black box in c. Arrows indicate dark elongate cells showing expression of afuni. Left arrow indicates an acute surface angle of the cell; this is a putative cell ‘process’, suggestive that the stained cells are migratory. O Oral side of arm, A aboral side of arm. Scale bars are 20 μm
Discussion
Afuni is a novel TGF-β with high-sequence identity with sea urchin univin
The molecular approach utilised in this study employed RT-PCR and PCR screening of a cDNA library to isolate afuni, a novel TGF-β gene from regenerating arms of the brittle star A. filiformis. This gene has highest sequence similarity to sea urchin univin, the percentage identity of the theoretically translated conserved carboxy-terminal regions being 85%, with the overall sequence identity being 35%. To date, the full-coding sequence for a univin gene has only been isolated from S. purpuratus. A gene fragment of around 250 bases, with high-sequence similarity to part of the C-terminal region of univin, has been isolated from another sea urchin, P. lividis (Zito et al. 2003). This gene has been excluded from figures and tables, as only part of the C-terminal sequence is known. However, sequence similarity of comparable regions of this molecule and afuni is high (88%). Phylogenetic analyses using the neighbour-joining method (Saitou and Nei 1987) confirmed that afuni belongs to the BMP family of the TGF-βs and forms a strongly supported clade with univin. The topology of the constructed tree is supported by sequence alignment, which reveals that afuni has higher similarity to univin (85%) than to other BMPs including members of the BMP2/4-DPP (e.g., 69% with human BMP2) and BMP5/8-60A (e.g., 66% with human BMP5) subgroups.
The neighbour-joining analyses conducted here place afuni and univin phylogenetically close to the BMP2/4-DPP subgroup. Previous phylogenetic analyses have indicated that univin is phylogenetically close to or within proposed BMP2/4-DPP subgroups (Shih et al. 2002; Patruno et al. 2003). However, these analyses were not extensive and did not include members of other BMP subgroups including the BMP5/8-60As. Another analysis placed univin in a clade containing Vg1 (Harada et al. 2002). A detailed phylogenetic analysis of BMPs conducted by Ponce et al. (1999) places univin in a unique clade separate from both the Vg1 and BMP2/4-DPP clades (Ponce et al. 1999). A precise, unified, phylogenetic position for univin and afuni is therefore unclear. However, our current analyses suggest that these unique echinoderm univin genes may be highly divergent members of the BMP2/4-DPP subgroup.
The lack of information about univin genes from other species makes it difficult to make detailed inferences on the evolution and radiation of this gene. It should be noted that although there is high-sequence similarity in the theoretically translated C-terminal regions, there is considerably less similarity of sequence outside this C-terminal domain. Not surprisingly, aligned DNA sequences show even less similarity across the entire known sequences (data not shown); indeed, the translated region of afuni (930 bases) is somewhat shorter than that of univin (1149 bases). The exceedingly small number of known univin genes means that very little is known of sequence divergence of this gene between different echinoderm classes or indeed species.
Afuni is expressed during arm regeneration
Our major interest in afuni is the role of this TGF-β superfamily member in regeneration. To date, no studies on the function of BMPs in echinoderm regeneration have been published. It is therefore difficult to predict the precise function of afuni in brittle star regeneration. The lack of expression observed in regenerates earlier than 3-week postablation indicates that this gene is probably not involved in the early processes of regeneration such as the differentiation of the newly formed blastema. However, BMP family members influence neural/epidermal cell fate during embryonic development in vertebrates and invertebrates (De Robertis and Sasai 1996; Hogan 1996; Graff 1997; Chang and Hemmati-Brivanlou 1998; Chitnis 1999). In crinoid arm regeneration, anBMP2/4 was expressed in the coelomic epithelium near to the site of amputation 24 h after arm ablation (Patruno et al. 2003). Expression remained high until 1-week postablation when the pattern changed slightly to a localised expression in the proximal region of the regenerate. At 2-week postablation, the mRNA signal was no longer detectable. AnBMP2/4 was thus implicated in performing a role in early patterning of the regenerating arm. A similar function for afuni during regeneration cannot be implied from the results presented here. However, the spatiotemporal expression of afuni at two sites in the regenerate (3- and 5-week postablation) suggests that it may be influencing more than one process in later regenerative development. It should be noted that the two distinct regions of expression are separated temporally as well as spatially. The distal site, where morphological and segmental definition is not clear, is at an earlier stage of regeneration than the proximal site, where segments are well defined and possess developed spines and podia.
The expression of afuni toward the distal tip of the regenerate occurred at the point where the distal-most segmental definition is observed. The location of this expression suggests that this gene may be playing a role in arm segmentation. Expression was detected in putative migrating cells at this site, and it is hypothesised that these cells have migrated to this distal site from the proximal and/or nonregenerating region of the RWC. Afuni is only expressed at or near the distal site of action of these cells where differentiation is thought to occur. This hypothesis ties in with the theory that afuni is involved in the segmentation of the newly regenerated distal arm and also accounts in part for the region between proximal and distal expression where no expression is detected. Afuni expression at this distal site was restricted to cells in the RWC. The proximal region of the regenerate cells expressing afuni was detected in the RWC and RNC. This is suggestive of different roles for afuni at the proximal and distal sites.
The afuni expression in the proximal regenerate may imply a putative role in the increase in arm width following the initial increase in length. However, the localisation of cells in the RNC of the proximal regenerate also suggests a possible role for afuni in late regenerative neurogenesis. It is unclear whether these cells in the RNC have migrated from the RWC or if the expression observed in this structure is a local event. If afuni expression in the proximal regenerated radial nerve cord is a unique event then this gene may be playing a role in late neural regeneration, perhaps in late neural cellular differentiation. Such a role may be analogous to BMP functions in late neural development of the vertebrate embryo, which influence the dorsal–ventral patterning and differentiation of cells in the vertebrate neural tube (Gilbert 2000).
Afuni expression was also detected in adult nonregenerating arms. It therefore seems likely that afuni plays a role in the normal somatic growth of the arm. The microinjection studies conducted by Zito et al. (2003) implicated univin as a key factor in the skeletogenesis of P. lividus embryos. It must be considered that afuni may be playing a similar role in ophiuroid arm regeneration. This appears particularly plausible at the proximal location of regenerating arms 3- and 5-week postablation, where afuni expression appears to coincide with the position of skeletal vertebrae.
The region of the arm between the proximal and distal sites of expression where no mRNA signal is detected can be accounted for by the biphasic nature of afuni expression and is consistent with afuni playing distinct roles at the proximal and distal sites. The initial role for afuni at the distal site is in the region where segmentation is being established. Hence, no afuni expression is observed prior to 3 weeks due to the absence of segmentation in the regenerating arm buds. The region of the arm immediately adjacent to the distal site of expression represents a site where the segmentation pattern is established. However, the cells in this region are not yet involved in the processes of arm thickening or the later stages of neural regeneration, and hence no afuni expression is observed. The temporally older cells in the proximal region are actively engaged in these processes, and hence a second phase of afuni expression is observed.
The afuni expression observed in the RWC implies that this structure may be a major source of cellular material utilised in the regeneration process. In regenerates, afuni expression was observed in elongate cells possessing morphological distinct processes located in the RWC. We suggest that these cells are migratory coelomocytes, originating from the coelomic epithelium of the RWC. The nerve cord and coelomic epithelial sites are known sources of cells migrating to the blastema/regenerate during echinoderm regeneration (Candia Carnevali et al. 1995). It should be noted that despite several attempts, it was not possible to section nonregenerating arms due to their highly calcified nature; thus, the nature of the cells expressing afuni in the nonregenerating RWC remains unresolved. Previous observations have noted that in ophiuroids, coelomic cells migrate to distal parts of the regenerate where proliferation and differentiation may occur (Thorndyke et al. 2001). The expression of anBMP2/4 in the coelomic canal of the crinoid A. bifida (Patruno et al. 2003) similarly indicates that the role of the coelom in regeneration may be not restricted to the ophiuroids and may be a common factor throughout echinoderm regeneration. Indeed, the organogenetic potential of the crinoid coelom was recognised in a review by Candia Carnevali and Bonasoro (2001).
Interestingly, studies conducted on the sea urchin, S. purpuratus have shown that the cellular material used in the formation of the echinoderm adult body form derives, at least in part, from larval coelomic cavities. These pluripotent coelomic cells have been termed set-aside cells (Davidson et al. 1995; Peterson et al. 1997). The hypothesis that the cells used in ophiuroid regeneration are derived from the coelomic RWC provides an interesting parallel to the larval coelomic set-aside cell theory in echinoderms and perhaps suggests functional lineage continuity between embryonic, larval and adult coelomic cells. This serves to highlight the importance of the coelom in echinoderm development in terms of both adult body construction through an embryonic and larval stage, and adult body reconstruction via tissue regeneration. It is clear that the evolution of coelomic structures facilitated the modes of development observed in modern echinoderms. The coelomic network of adult ophiuroids (and likely other echinoderm classes including crinoids) appears to be a key structure in regeneration and is likely to be at least partly responsible for this group’s impressive regenerative capacity. It also seems that the plasticity of the coelom may have been a vital evolutionary adaptation facilitating the emergence of this unique pentamerously symmetrical phylum.
References
Angerer LM, Oleksyn DW, Logan CY, McClay DR, Dale L, Angerer RC (2000) A BMP pathway regulates cell fate allocation along the sea urchin animal-vegetal embryonic axis. Development 127:1105–1114
Candia Carnevali MD, Bonasoro F (2001) Microscopic overview of crinoid regeneration. Microsc Res Tech 55:403–426
Candia Carnevali MD, Bonasoro F, Lucca E, Thorndyke MC (1995) Pattern of cell proliferation in the early stages of arm regeneration in the feather star Antedon mediterranea. J Exp Zoolog 272:464–474
Chang C, Hemmati-Brivanlou A (1998) Cell fate determination in embryonic ectoderm. J Neurobiol 36:128–151
Chitnis AB (1999) Control of neurogenesis—lessons from frogs, fish and flies. Curr Opin Neurobiol 9:18–25
Combet C, Blanchet C, Geourjon C, Deleage G (2000) NPS@: network protein sequence analysis. Trends Biochem Sci 25:147–150
Davidson EH, Peterson KJ, Cameron RA (1995) Origin of bilaterian body plans: evolution of developmental regulatory mechanisms. Science 270:1319–1325
De Robertis EM, Sasai Y (1996) A common plan for dorsoventral patterning in bilateria. Nature 380:37–40
Ducy P, Karsenty Y (2000) The family of bone morphogenetic proteins. Kidney Int 57:2207–2214
Gilbert SF (2000) Developmental biology, 6th edn. Sinauer Associates, Sunderland, USA
Graff JM (1997) Embryonic patterning: to BMP or not to BMP, that is the question. Cell 89:171–174
Harada Y, Shoguchi E, Taguchi S, Okai N, Humphreys T, Tagawa K, Satoh N (2002) Conserved expression pattern of BMP-2/4 in hemichordate acorn worm and echinoderm sea-cucumber embryos. Zool Sci 19:1113–1121
Hogan BLM (1996) Bone morphogenetic proteins: multifunctional regulators of vertebrate development. Genes Dev 10:1580–1594
Holland LZ, Holland PWH, Holland ND (1996) Protocols: whole-mount in situ hybridisation applicable to amphioxus and other small larvae. In: Ferraris JD, Palumbi SR (eds) Molecular zoology. Wiley-Liss, New York, pp 476–483
Nieto MA, Patel K, Wilkinson DG (1996) In situ hybridization analysis of chick embryos in whole mount and tissue sections. Methods Cell Biol 51:219–235
O’Kane S, Ferguson MJW (1997) Transforming growth factor βs and wound healing. Int J Biochem Cell Biol 29:63–78
Patruno M, McGonnell IM, Graham A, Beesley PW, Candia Carnevali MD, Thorndyke MC (2003) Anbmp2/4 is a new member of the transforming growth factor-β superfamily isolated from a crinoid and involved in regeneration. Proc R Soc Lond B 270:1341–1347
Peterson KJ, Cameron RA, Davidson EH (1997) Set-aside cells in maximal indirect development: evolutionary and developmental significance. Bioessays 19:623–631
Ponce MR, Micol JL, Peterson KJ, Davidson EH (1999) Molecular characterization and phylogenetic analysis of SpBMP5–7, a new member of the TGF-β superfamily expressed in sea urchin embryos. Mol Biol Evol 16:634–645
Reinhardt B, Broun M, Blitz IL, Bode HR (2004) HyBMP5–8b, a BMP5–8 orthologue, acts during axial patterning and tentacle formation in hydra. Dev Biol 267:43–59
Saitou N, Nei M (1987) The neighbor-joining met hod: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Shih LJ, Chen CA, Chen CP, Hwang SP (2002) Identification and characterization of bone morphogenetic protein 2/4 gene from the starfish Archaster typicus. Comp Biochem Physiol B Biochem Mol Biol 131:143–151
Stancyk SE, Golde HM, Papelindstrom PA, Dobson WE (1994) Born to lose. 1. Measures of tissue loss and regeneration by the brittlestar Microphiopholis gracillima (Echinodermata, Ophiuroidea). Mar Biol 118:451–462
Stenzel P, Angerer LM, Smith BJ, Angerer RC, Vale WW (1994) The univin gene encodes a member of the transforming growth factor-β superfamily with restricted expression in the sea urchin embryo. Dev Biol 166:149–158
Thorndyke MC, Patruno M, Beesley PW, Mallefet J (2001) Cellular and molecular bases of arm regeneration in brittlestars. In: Barker M (ed) Echinoderms 2000. Swets and Zeitlinger, Lisse, pp. 323–326
Ueno N, Moos M (1997) Bone morphogenetc proteins. Genes Funct 1:287–288
Urist MR, Iwata H, Ceccotti PL, Dorfman RL, Boyd SD, McDowell RM, Chien C (1973) Bone morphogenesis in implants of insoluble bone gelatin. Proc Natl Acad Sci U S A 70:3511–3515
Wozney JM, Rosen V, Celeste AJ, Mitsock LM, Whitters MJ, Kriz RW, Hewick RM, Wang EA (1988) Novel regulators of bone formation: molecular clones and activites. Science 242:1528–1534
Zito F, Costa C, Sciarrino S, Poma V, Russo R, Angerer LM, Matranga V (2003) Expression of univin, a TGF-β growth factor, requires ectoderm-ECM interaction and promotes skeletal growth in the sea urchin embryo. Dev Biol 264:217–227
Acknowledgements
We thank the Kristineberg Marine Research Station’s staff for support during this work. R.B. and M.C.T. also acknowledge the support of the EU Transnational Access to Infrastructures Programme and V.R. (Sweden) for work conducted at Kristineberg. We would also like to thank Patricia Goggin at the Electron Microscopy Unit, Royal Holloway, University of London, for assistance with preparation of resin sections. R.B. was funded by a NERC studentship.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by N. Satoh
Rights and permissions
About this article
Cite this article
Bannister, R., McGonnell, I.M., Graham, A. et al. Afuni, a novel transforming growth factor-β gene is involved in arm regeneration by the brittle star Amphiura filiformis. Dev Genes Evol 215, 393–401 (2005). https://doi.org/10.1007/s00427-005-0487-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00427-005-0487-8