Abstract
Genes in the odd-skipped (odd) family encode a discrete subset of C2H2 zinc finger proteins that are widely distributed among metazoan phyla. Although the initial member (odd) was identified as a Drosophila pair-rule gene, various homologs are expressed within each of the three germ layers in complex patterns that suggest roles in many pathways beyond segmentation. To further investigate the evolutionary history and extant functions of genes in this family, we have initiated a characterization of two homologs, odd-1 and odd-2, identified in the genome of the nematode, Caenorhabditis elegans. Sequence comparisons with homologs from insects (Drosophila and Anopheles) and mammals suggest that two paralogs were present within an ancestral metazoan; additional insect paralogs and both extant mammalian genes likely resulted from gene duplications that occurred after the split between the arthropods and chordates. Analyses of gene function using RNAi indicate that odd-1 and odd-2 play essential and distinct roles during gut development. Specific expression of both genes in the developing intestine and other cells in the vicinity of the gut was shown using GFP-reporters. These results indicate primary functions for both genes that are most like those of the Drosophila paralogs bowel and drumstick, and support a model in which gut specification represents the ancestral role for genes in this family.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The odd gene family is defined by a highly conserved set of zinc fingers that comprise a subgroup within the broader family of “classic” or Cys2His2 (C2H2) type fingers first identified in TFIIIA (Miller et al. 1985). The founding member of this family is the Drosophila odd-skipped gene, which was identified during the genetic screens of Nüsslein-Volhard and Wieschaus (1980) and later cloned and found to encode a protein with four contiguous fingers (Coulter et al. 1990). The existence of the odd family was subsequently revealed via the isolation of homologous cDNAs derived from the Drosophila paralogs, bowel (bowl) and sister of odd and bowel (sob; Hart et al. 1996; Wang and Coulter 1996), and the identification of two predicted genes, odd-1 and odd-2, in the genome of the nematode, Caenorhabditis elegans (Wilson et al. 1994; Waterston 1998). More recently, an additional, highly degenerate Drosophila paralog, drumstick (drm), has been genetically and molecularly characterized (Green et al. 2002), and two vertebrate homologs, Osr1 and Osr2, have been identified through the characterization of ESTs and/or predicted gene sequences of both mice (So and Danielian 1999; Lan et al. 2001) and humans (Debeer et al. 2002; Katoh 2002).
In contrast to the more stereotypical functions of other conserved families of transcription factors such as the Hox genes, substantial functional divergence within and between phyla is indicated by the expression patterns and/or developmental phenotypes of various genes in the odd family. The eponymous member, odd-skipped, was identified on the basis of its function as a pair-rule gene (Nüsslein-Volhard and Wieschaus 1980; Coulter and Wieschaus 1988). Within the segmentation network, it appears to function primarily as a transcriptional repressor that targets segment-polarity and/or pair-rule genes (DiNardo and O’Farrell 1987; Kuhn et al. 2000; Bouchard et al. 2000; Mullen and DiNardo 1995), although a possible role in activation of selected genes has also been suggested by gain-of-function experiments (Saulier-Le Drean et al. 1998). The developmental regulation and expression pattern of odd are very dynamic and complex (Coulter et al. 1990; Ward and Coulter 2000). In addition to a characteristic “pair-rule” pattern of seven transverse stripes that arise during cellularization, odd is subsequently expressed in additional patterns and tissues, which include a change from the initial pair-rule (7 stripe) mode to a persistent segment polarity-like pattern (14 stripes) during the blastoderm-gastrula transition, and expression in a wide variety of non-epidermal cells at subsequent stages. Expression in some of these cell types has proven to be useful as a marker (e.g., Skeath 1998; Ward and Skeath 2000), but in most cases the functional significance of the post-blastoderm expression remains uncertain.
Reverse genetic analyses of the Drosophila paralogs sob and bowl suggest that their developmental functions have diverged appreciably. The expression pattern and mutant phenotype of bowl indicate a key role in gut development and a primary function in the terminal, rather than the segmentation, hierarchy (Wang and Coulter 1996). A similar but distinct function during gut morphogenesis is also indicated for the drm gene (Green et al. 2002). In contrast, sob is expressed in patterns that are nearly equivalent to odd (Hart et al. 1996). Although this would be consistent with some degree of functional redundancy, RNAi experiments and analysis of deletions that remove both genes (Hart et al., submitted for publication) suggest that, beyond the pair-rule functions ascribed to odd, neither gene may play a significant role in other aspects of epidermal patterning during embryogenesis. Instead, it appears that sob has an essential function in internal tissues and/or post-embryonic stages.
Among the vertebrate representatives whose transcriptional profiles have been analyzed, the mouse Osr1 and Osr2 and human Osr1 genes, expression is seen in a very diverse range of tissues or cell types (So and Danielian 1999; Lan et al. 2001; Katoh 2002). While no mutations in vertebrate odd homologs have yet been reported, it has been suggested on the basis of the expression patterns of Osr1 and Osr2 that mutations in these loci might underlie some heritable human syndromes that exhibit combinations of distal limb deficiencies, micrognatia and renal hypoplasia (Debeer et al. 2002). A role for Osr-1 in human endodermal development and/or differentiation is consistent with the altered expression levels in some cancer cell lines (Katoh 2002).
Even among the small sample of insect and mammalian representatives that have been characterized, genes in the odd family reveal remarkably diverse expression patterns that include tissues from all three germ layers. While this promiscuity may reflect a highly plastic history, in which the regulation and/or functions of these zinc finger proteins have become significantly altered or extended during evolution, it is likely that at least some ancestral functions have been retained among many or most extant phyla. To further investigate the function and evolution of genes in the odd family, we have undertaken a functional characterization of the two homologs found in C. elegans, odd-1 and odd-2. Our results indicate that both genes are functionally more similar to the Drosophila bowl and drm genes than to odd-skipped, and that the developmental specification or elaboration of gut cells represents a primordial function for the ancestral “odd-like” gene(s).
Materials and methods
Sequences and alignments
Predicted amino acid sequences of odd, sob, bowl, drm, Osr1 and Osr2 have been published elsewhere (Coulter et al. 1990; Hart et al. 1996; Wang and Coulter 1996; Green et al. 2002; So and Danielian 1999; Lan et al. 2001). Accession numbers for additional sequences identified through database (BLASTP) searches with either the complete odd or bowl sequences, or the first finger of bowl alone, are as follows: AAA19082 (odd-1), AAF39763 (odd-2), XP_317495 (Ano-1), XP_306979 (Ano-2), and XP_317494 (Ano-3); note that the “Ano” (for Anopheles odd) designations for the mosquito genes are provisional. High-scoring genes that are not analyzed here include highly conserved homologs of the mouse Osr genes from other mammals, and distantly related sequences with numerous C2H2 repeats whose elevated BLAST scores resulted from the accumulation of multiple, phase-shifted alignments with the query sequence. Phylogenetic trees were generated using the SEQBOOT, PROTPARS, and CONSENSE programs in PHYLIP (Felsenstein 1993); unrooted majority-rule consensus trees and bootstrap confidence levels were calculated from 2000 randomized data sets. For the tree shown in Fig. 1, zinc finger domains were compared using the indicated amino acid alignments; the total number of fingers, and the presence/absence of an intron within the second finger, were modeled as single character differences as follows: (1) sequences with fewer than five C2H2 repeats were extended using a single stop codon followed by missing-data characters (“?”), and (2) a final “residue”, consisting of either an arbitrarily chosen amino acid (representing the intron found in bowl, sob, and Ano-1) or a gap character (“-”), was appended to all sequences in the alignment. The tree was rooted assuming a phylogeny in which nematodes diverged earlier than chordates and arthropods, which is supported by recent evidence (Blair et al. 2002) that contradicts the “new” phylogeny (in which the “ecdysozoans”—nematodes and arthropods—diverged later than chordates; Adoutte et al. 1999). Rooting under the “traditional” assumption also allows placement of the two nematode paralogs (odd-1 and odd-2) on different basal branches; because these branches include arthropod paralogs (Fig. 1) this minimizes the number of gene duplication events required (four, rather than the five needed under the ecdysozoan model).
RNAi
Templates for in vitro transcription of dsDNA that included convergent T7 and/or T3 promoters were generated by PCR amplification of genomic DNA sequences. For both genes, an around 700-bp “fingerless” region was amplified from corresponding plasmid clones that had been generated by PCR amplification from wild-type (N2) C. elegans genomic DNA. For odd-1, both primers (WrmbFRNAi: GCT CAT CTA ATA CGA CTC ACT ATA GGG AGA TGT ATT CGG TTG ACC TTT TTA GA, and WrmbRRNAi: GCT CAT CTA ATA CGA CTC ACT ATA GGG AGA GTA AAG TGA CGG GCA CAG T) were tagged with the T7 promoter sequence, while the primers used for odd-2 (WrmcFRNAi: GCT CAT CTA ATA CGA CTC ACT ATA GGG AGA TGT AGT AGA TTA CCC CAT AAG ACG A, and WrmcRRNAi: GCT CAT C AA TTA ACC CTC ACT AAA GGG AGA CAG TAG TCC TTC CAC GTC CAG) included a T7 and T3 promoter, respectively. For odd-1, a 1,175-bp full-length fragment including the entire predicted ORF and intron plus additional flanking sequences was amplified from a larger (3 kb) amplified genomic DNA fragment using two T7 tagged primers (WrmbZFRNAi: GCT CAT CTA ATA CGA CTC ACT ATA GGG AGA TGT TGT ACC ATC TGT TGA GG, and WrmbZRRNAi: GCT CAT CTA ATA CGA CTC ACT ATA GGG AGA AAA GGT TCA CGA AGC CC). In vitro transcription reactions were performed using a MEGAscript Kit (Ambion); for each PCR-generated template, 1 μg DNA was transcribed using either T7 polymerase alone (odd-1) or both T7 and T3 polymerases in separate reactions (odd-2). Following DNase treatment (Promega, RNAse free), combined sense and antisense products were heated to 94°C for 2 min, cooled to 70°C for 10 min and left at room temperature overnight. Following phenol/chloroform extraction and precipitation with isopropanol, dsRNA samples were resuspended in TE and aliquots were measured spectrophotometrically and electrophoresed to assess the reaction efficiency.
Wild-type (strain N2) L4 worms were injected in the gonads with dsRNA at concentrations from 1–3.5 mg/ml for odd-2 and 1–4 mg/ml for either of the two odd-1 samples. In double RNAi experiments equal concentrations of both odd-1 and odd-2 were injected at 3.5 mg/ml each. Injected hermaphrodites were maintained at 25°C and transferred twice daily to fresh plates. F1 progeny derived from self-fertilization were examined for defects. For each RNAi construct, two or more rounds of injections into at least 10 animals were performed on separate days, with a minimum of 20 affected larvae examined in each round.
GFP reporters
5′ sequences from odd-1 and odd-2 were fused to a Green Fluorescent Protein (GFP) reporter using a “2-fragment” protocol (Guhathakurta et al. 2002) in which a small PCR-generated, RE-tagged 5′ fragment that includes the first exon is directionally cloned (using HindIII and BamHI sites) into a promoterless GFP vector and subsequently coinjected with a large overlapping PCR fragment extending much further 5′. A full-length construct is generated by homologous recombination in vivo. For odd-1, the primers Wrmbreporter1 (CCC CAA GCT TGT GCA TAA GTC CAT CAT CTT C), and Wrmbreporter2 (CCC CGG ATC CCG CTG TTC AGG GTT CAG GAA C) were used for cloning into the GFP vector and Wrmbreporter3 (GAA GTT TTG GGA GCT CAT ATC), and Wrmbreporter4 (GTT CAG GGT TCA GGA ACC TG) were used to amplify the coinjected fragment. The cloned insert spans a region from 65 bp upstream to 348 bp downstream of the start codon, while the coinjected fragment (4,362 bp) includes from 3,885 bp upstream to 345 bp downstream of the start codon. The corresponding odd-2 primers, Wrmcreporter1 (CCC CAA GCT TCC GTG GCA ACG ACA AGT TC) and Wrmcreporter2 (CCC CGG ATC CCG TCC AGG GAT CAT GAA CC), and Wrmcreporter3 (CAG TGC AGT TTG GCT GAC AAG) and Wrmcreporter4 (CCA GGG ATC ATG AAC CTA AAA C) amplified a 413-bp cloned fragment from 3 bp downstream to 405 bp downstream of the start codon, and a 4,233 bp coinjected fragment from minus 3,961 to plus 402 bp. After cloning into the GFP vector, plasmid DNA was digested with HindIII, and mixed with the corresponding upstream PCR fragment plus a rol-6 marker construct (Park and Kramer 1994) at a 1:1:8 ratio. DNA was injected at a total concentration of 0.2 mg/ml into the gonads of adult worms, and F1 transformants were selected based on the “rolling” (rol-6) phenotype.
Expression patterns in living embryos and larvae (treated with levamisole to induce paralysis) were examined using a compound microscope with fluorescence and Nomarski optics. To isolate lines with a chromosomal insertion of the odd-1::GFP transgene, a transgenic line was subjected to gamma irradiation mutagenesis and F3 progeny scored for 100% transmission of the rol-6 marker phenotype. Antibody staining of embryos from these lines was performed as in Hresko et al. (1994), using rabbit polyclonal anti-GFP (Clontech) and mouse monoclonal antibody MH33 (Francis and Waterston 1991).
Results and discussion
Sequence conservation and divergence of odd-related genes
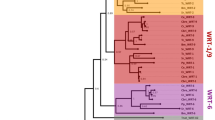
The predicted amino acid sequences of the zinc fingers derived from odd-related genes from insect (Drosophila and mosquito), vertebrate (mouse) and nematode (C. elegans) species are shown in Fig. 1. In addition to matching the consensus defined for the C2H2 family of DNA-binding domains (Rosenfeld and Margalit 1993), which includes the pairs of zinc-chelating cysteines and histidines plus additional hydrophobic residues that stabilize the core of the tertiary fold characteristic of this motif (Wolfe et al. 2000), the zinc fingers in this group comprise an obvious subfamily based upon the high degree of conservation at other “non-consensus” positions. These include the putative DNA contact residues and neighboring positions in the “recognition” helix of each finger, and the “linker” sequences that connect adjacent fingers, which may act to modulate protein stability and turnover during mitosis (Dovat et al. 2002). It should be noted that, because the serially-repeated fingers within each gene show considerable variability, this conservation is evident only when the fingers are aligned in the correct registration.
Zinc finger sequence alignments and taxonomy of odd-homologs from Drosophila melanogaster (bowl, sob, odd, drm), Anopheles gambiae (Ano-1, Ano-2, Ano-3), Caenorhabditis elegans (odd-1, odd-2), and Mus musculus (Osr1, Osr2). The top of the figure shows the consensus for two successive 28-residue C2H2 repeats; positions of amino acid residues predicted to adopt alpha or beta 2° or secondary structures and/or to form specific base contacts with DNA (Wolfe et al. 2000) are indicated by bars, zig-zag lines, and asterisks, respectively. Amino acid alignments for the finger domains of each protein are shown using the five zinc fingers of the bowl gene as a reference; dashes represent amino acid identities with bowl. Note that the zinc-finger repeats in each sequence are contiguous, and that drm and Ano-3 include an additional residue inserted between fingers 1 and 2. Ellipses represent adjacent sequences of variable length that extend to the N- and C-termini of each protein; these sequences include additional homologous stretches that are shared among particular pairs of homologs, including the two mouse genes (Osr1 and Osr2; So and Danielian 1999; Lan et al. 2001), odd and Ano-2, and drm and Ano-3 (data not shown). The boundaries delimiting individual fingers are based upon the limits of sequence homology within the odd family as a whole (Hart et al. 1996), and are shifted to a more N-terminal position than conventional representations of the C2H2 consensus. “Osr2(A)” designates the fourth and fifth fingers that are present in one of the two Osr2 isoforms. The cladogram was derived by parsimony analysis of the finger sequences of the indicated genes (see Materials and methods). Asterisks indicate branches that correspond to gene duplications, and the inferred reconstruction of gut function is traced in bold. Bootstrap values (in %) are indicated at each node, and the number of fingers in each sequence is indicated next to the corresponding symbol
While the remarkable degree of conservation within their zinc-finger domains suggests that the proteins in this group may have retained similar or overlapping binding properties, possible functional differences are suggested by variation in the number of fingers, and by the pattern of amino acid changes within the fingers that are present. A global comparison shows an N- to C-terminal gradient: amino-terminal fingers are more consistently present and show fewer amino acid substitutions. Thus, whereas the odd gene has a total of four C2H2 repeats, the vertebrate Osr1 gene and both of the nematode genes have only three fingers due to the absence of the most C-terminal finger found in odd. The Drosophila sob and bowl genes each include a fifth, C-terminal finger. The most divergent member of the family, the Drosophila drumstick gene, contains only two fingers, and the second histidine is not present in the C-terminal repeat. The transcriptional effects of drm appear to be mediated primarily through protein-protein, rather than protein-DNA, interactions (Green et al. 2002), and it is possible that the Drm protein has lost the capacity for specific DNA recognition. The absence of particular fingers from other homologs might reflect a similar trend, at least in cases where the C-terminal fingers have simply become dispensable. However, an alternative possibility is suggested by the mouse Osr-2 gene. In this case, alternative splicing can give rise to different isoforms with either three or five zinc-finger repeats, consistent with the idea that differences in finger number per se are sometimes advantageous.
It is possible that some of the variability in finger number reflects a gain, rather than loss, of C-terminal fingers. Considering the extensive similarity among the extant fourth and fifth fingers, and their presence in both chordate and arthropod representatives, it appears that such an event must have been unique and relatively ancient. Our phylogenetic reconstruction (Fig. 1) is consistent with a three-fingered ancestral condition that has persisted in the case of the two nematode genes (odd-1 and odd-2). However, homoplasy is indicated in the case of the remaining three-finger gene, Osr-1, which appears to have lost fingers from a five-fingered intermediate. Finger loss is also indicated for odd/Ano-2 (four fingers) and drm/Ano-3 (two fingers).
C-terminal divergence is also revealed by the patterns of amino acid substitutions among the extant fingers, especially within paralogous groups. For example, among the four fingers that are common to the Drosophila paralogs odd, sob and bowl, a majority of the accumulated changes in the least conserved member, odd, lie within the fourth finger. Similarly, most of the differences between sob and bowl fall within their common fifth finger. Similar tendencies are seen among the three fingers present in odd-1 and odd-2, but not those in Osr1 and Osr2, which have only one difference between them. It is uncertain whether these substitutions represent intermediate stages during the progressive loss of the finger from one paralog, as opposed to changes that are either advantageous or neutral with respect to domain function.
RNAi analysis of odd-1 and odd-2 in C. elegans
The odd-1 and odd-2 genes were identified in the C. elegans genome as predicted open reading frames encoding proteins of 159 and 254 amino acids, respectively. Despite the accumulation of several apparent indels, the two sequences exhibit similarities along their lengths, and each includes a single small intron that disrupts the ORF at an equivalent position upstream of the zinc fingers (Fig. 2). In contrast to the insect paralogs, which are clustered within a small region of the second chromosome (polytene bands 24A-C) of Drosophila and the third chromosome in Anopheles, odd-1 and odd-2 are not linked, mapping to chromosomes III and X, respectively.
Structures of odd-1 and odd-2. Open reading frames of odd-1 and odd-2 are indicated by rectangles, with shading used to indicate the zinc fingers (black) and additional regions with more limited amino acid conservation (gray). Putative indels involving >3 amino acid residues are depicted by lines connecting the two sequences, and narrow lines within each sequence indicate the intron and additional nucleotides upstream and downstream of the coding sequences. Although the introns are offset by two amino acid residues relative to the zinc-finger domains, each falls at an identical position within the second codon of a similar hexapeptide sequence (coding for PWFLNP and PWFMIP in odd-1 and odd-2, respectively). Bars shown above the odd-1 gene and below the odd-2 gene represent sequences used to generate full-length (odd-1) and fingerless (odd-1 and odd-2) RNAi constructs
To assess the developmental roles of odd-1 and odd-2, we used RNAi to inactivate one or both genes. The dsRNA sequences derived from each gene were injected into the gonads of L4 hermaphrodites (Fire et al. 1998; see Materials and methods). This procedure is expected to reduce or eliminate maternal as well as zygotic activity of the target gene or genes within the F1 progeny of injected worms. Because odd-1 and odd-2 share 73% identity at the nucleotide level within the finger domain, it is possible that dsRNA containing the finger region from one gene could result in inactivation of both loci. Therefore, experiments were initially performed for each locus using fingerless dsRNAs which were largely or entirely devoid of these sequences (Fig. 2).
Broods of eggs were collected daily from injected animals (see Materials and methods), and the progeny examined the following day for defects in morphology, behavior, and viability. Injection of fingerless dsRNA from odd-1 caused no detectable phenotype in the F1 progeny. Injection of odd-2 dsRNA, however, produced F1 larvae that hatched but failed to grow and mature. Most affected larvae arrested development at the late L1 to early L2 stage and did not exhibit normal rhythmic pumping of the pharynx. These phenotypes suggested that the arrest and subsequent lethality might be caused by an inability to feed. To test this, affected larvae were placed on ink-stained bacteria to feed, and then examined using Nomarski optics. Variable defects in digestive tract function were revealed, including failure to ingest bacteria or to move bacteria through the intestine (Fig. 3).
RNAi of odd-1 and odd-2 causes variable defects in the gastrointestinal tract. Nomarski images are shown with anterior to the left. a The anterior of a wild-type larva shows the muscular pharynx (Ph) extending from the anterior tip of the nose to the terminal bulb (Tb). The pharyngeal intestinal valve (PIV) connects the pharynx to the intestine, whose lumen is largest at its anterior (arrow) and compact along its remaining length (arrowhead). b The region of the terminal bulb, PIV, and anterior intestine is shown in higher magnification. c Injection of full-length odd-1 dsRNA resulted in cavities around the pharynx (arrowhead) and feeding defects in F1 larvae. Here, ink-stained bacteria have accumulated in the anterior intestine (arrow) just posterior to the PIV. d An odd-1 RNAi affected larva shows a putative apoptotic nucleus (arrow) in place of an absent or reduced PIV, and a morphologically abnormal anterior intestine (arrowhead). e RNAi of odd-2 caused accumulation of ink-stained bacteria in the expanded lumen of the posterior intestine (arrowhead); defective elimination is suggested by the normal appearance of the anterior lumen (arrow). f In some animals, reduced odd-2 activity caused an enlarged, empty intestinal lumen (arrowhead), here associated with a reduced PIV (arrow). g A feeding-defective larva resulting from double RNAi exhibits cavities around the pharynx and a novel phenotype in the anterior intestine. Although the rounded posterior border of the PIV (arrow) suggested that the PIV cells might have adopted a pharyngeal bulb fate, performing the double RNAi in animals expressing the pharyngeal marker myo-2::GFP (a gift from Aguan Wei) did not show ectopic expression of this marker in the PIV cells (data not shown). Instead, the novel appearance of the PIV/intestinal boundary may be due to the abnormally narrow lumen observed in the anterior intestine (arrowhead). Digestive tract morphology and function are described in Avery and Thomas (1997) and at http://www.wormatlas.org
In odd-2 RNAi-affected larvae, failure to feed was associated with variable morphological defects in the digestive tract (Fig. 3). Most commonly, animals exhibited morphological abnormalities in the pharyngeal intestinal valve (PIV), where the anterior muscular feeding organ is connected to the intestine. Although intestinal morphology appeared normal in many affected larvae, some animals exhibited a gross enlargement of the intestinal lumen. This phenotype appeared to be caused by constipation in a few cases, but was most often associated with an empty dilated lumen. Although a similar dilation is observed in wild-type larvae that have been starved for a long period of time, the rapid development of this phenotype in RNAi-affected larvae is most likely due to defects in gut specification or morphogenesis. These data indicate that odd-2 is specifically required for proper development or function of the digestive tract.
Because the failure of the fingerless odd-1 dsRNA to produce a phenotype could be due to problems with the dsRNA used, a second dsRNA that included the zinc finger region was prepared from the odd-1 gene (Fig. 2). As with the fingerless odd-2 sequences, injection of this extended odd-1 dsRNA also produced a larval arrest phenotype where affected animals failed to grow. Examination of affected larvae revealed morphological and functional defects in the vicinity of the PIV/intestine boundary (Fig. 3). In addition, the heads of odd-1 affected animals contained multiple cavities surrounding the isthmus of the pharynx (Fig. 3). These cavities resemble those found in aged wild-type worms (Garigan et al. 2002). However, because we observe these cavities in affected larvae that are 1–2 days old, and no cavities are detected in the odd-2 RNAi experiments, these cavities are likely to be a direct result of odd-1 inactivation. The presence of cavities suggests that inappropriate cell death is occurring within the head. This hypothesis is supported by the presence of button-like nuclei in cells within and/or adjacent to the terminal bulb and in the PIV (Fig. 3); these nuclei are similar to those in cell corpses that have not been engulfed (Ellis and Horvitz 1991; Ellis et al. 1991). Because the RNAi phenotypes of odd-1 and odd-2 are different, it is unlikely that inclusion of the zinc-finger region in the odd-1 dsRNA resulted in inactivation of the odd-2 gene. Together, these data suggest that odd-1 has a nonredundant function required for proper specification of cells that are in or associated with the pharynx.
To assess the possibility that the two genes have overlapping or redundant functions, injections were performed using both the full-length odd-1 dsRNA and the fingerless odd-2 dsRNA. As with either of the single dsRNA injections, the affected larvae showed a larval arrest phenotype, and most of the observed defects, including cavities in the head, an enlarged intestinal lumen, and feeding abnormalities, were consistent with an additive phenotype. The double RNAi did not provide evidence of additional tissues or structures where odd-1 and odd-2 might function redundantly. A possible exception is that the PIV and anterior intestine in some animals showed a novel morphology (Fig. 3). However, this phenotype may reflect either an additive effect or some form of regulatory or epistatic interaction between these genes within these structures, rather than functional redundancy.
Expression of odd-1 and odd-2 during C. elegans development
To assess the expression profiles of the two worm homologs during development, we established transgenic lines that carried GFP reporters driven by the promoter and around 3 kb of upstream regulatory sequences from each gene (Materials and methods). For each reporter, living animals from two or more independent transgenic lines were examined using Nomarski and fluorescence microscopy. Uniform expression of odd-1::GFP was detected in the intestine of embryos and larvae when the transgene was expressed from an extrachromosomal array (Fig. 4). To more carefully assess odd-1::GFP localization, an integrated transgenic line was isolated (see Materials and methods) and embryos from this line double-stained with antibodies specific for GFP and for the intestinal marker MH33 (Francis and Waterston 1991). Uniform odd-1::GFP staining was detected in presumptive gut cells prior to the onset of elongation (Fig. 5). However, unlike the more static pattern seen with the unstable extrachromosomal lines, differences in expression in the head and intestine were seen during elongation. As shown in Fig. 5, the GFP signal increased in the most anterior cells of the intestine, and showed a concomitant relative decrease throughout the rest of the intestine, during the early stages of elongation. As elongation proceeded, additional GFP-positive cells were detected in the head. The observed expression pattern correlates well with the RNAi data, which point to a role for odd-1 in gut function and in cell specification in the head (probably in or associated with the pharynx).
Green Fluorescent Protein (GFP) expression in living animals reveals that odd-1 and odd-2 are expressed in the intestine. Each row shows a single animal viewed by Nomarski (left) and fluorescence (right) microscopy. a, b An elongating embryo (2.5-fold stage) expressing odd-1::GFP from an extrachromosomal array shows uniform GFP expression in the intestine. c, d In contrast, the intestinal expression of odd-2::GFP appears brightest in the more anterior and posterior portions of the intestine (arrowheads), shown here in a hatched larva. Additional cells that appear to lie outside the posterior intestine also express GFP (arrow)
A Green Fluorescent Protein (GFP) reporter reveals odd-1 expression in the developing intestine and within unidentified cells in the head. Each row contains images from a single embryo. In Nomarski images (left column), anterior (A) and posterior (P) tips are indicated. Prior to elongation (a–c), odd-1::GFP (b) is detected in all presumptive intestinal cells, which are identified by MH33 staining (c), and within unidentified cells in the head (not shown). As elongation proceeds, GFP becomes prominent in the most anterior intestinal cell (arrowhead in e) and relatively dim in the remaining intestine, shown here in a 1.25-fold embryo (d–f). Signal in the anterior intestine remains high as additional cells become visible in the head (arrowhead in h), shown here in an embryo near the 2-fold stage
Transgenic lines expressing odd-2::GFP also contained signal within the intestine of embryos, larvae, and adults. At all stages, transgenic animals expressed the highest levels of GFP in the most anterior and most posterior portions of the intestine, as well as signal in cells that appeared to be outside the intestine, near the anus (Fig. 4). Unfortunately, lines expressing the odd-2 reporter were difficult to generate, and could only be maintained for a few generations, precluding a more careful examination of GFP localization using integrated lines and antibody staining.
Conclusions and evolutionary perspectives
Our observations from both the single and double RNAi experiments indicate that odd-1 and odd-2 have non-overlapping functions that are essential for normal gut development. Similarly, the expression data indicate that both genes are active during the development and differentiation of the intestine and other cells in the vicinity of the alimentary canal. It is possible that additional functions for these genes were not revealed due to limited effectiveness of RNAi, or that certain aspects of their expression patterns were not revealed by the GFP reporters. Nevertheless, these results indicate primary functions for both genes that are most reminiscent of the Drosophila paralogs, bowl and drm, and support the hypothesis that the developmental specification of gut cells is a primordial function of the odd gene family.
Our phylogenetic analysis (Fig. 1) indicates that two odd family genes were present in an ancestral metazoan prior to the divergence of the nematode and arthropod/chordate clades (note that an ancestral duplication preceding the arthropod/nematode split is indicated under any rooting method). Descendants of both genes have gut-specific functions in insects (i.e., bowl and drm). Additional paralogs in the odd/sob/bowl branch arose via duplications that occurred subsequent to the arthropod/chordate split. In contrast, the extant mammalian genes (Osr-1 and Osr-2) apparently arose via duplication of one of the ancestral genes, with the apparent loss of the other; compared to the insect and nematode paralogs, a relatively recent origin for these sequences is suggested by the remarkably high degree of conservation along the entire ORF (Lan et al. 2001). While the C. elegans genes show relatively poor bootstrap resolution, odd-1 appears to be orthologous to Drosophila drm. A similar inference for odd-2 and the bowl/odd/sob group of Drosophila is also supported under our rooting assumptions (see Materials and methods). However, the apparent degeneration and loss of zinc-finger sequences from drm (which has less than two intact C2H2 repeats) suggests that both nematode genes (which include three fingers each) may have retained functions that are more similar to those of Drosophila bowl. In any event, the functional divergence of these paralogs may have been important for the elaboration of the distinct, and increasingly complex, digestive systems that evolved in each lineage.
It has been suggested that the segmentation functions of pair-rule genes reflect a relatively recent adaptation whereby genes with distinct ancestral roles became co-opted into the segmentation hierarchy during the evolution of long germ band development (Patel et al. 1992). Although some studies have called this view into question (e.g., Schroder et al. 2000), the available data from flies and worms suggest that the segmentation functions of odd are indeed more recent than the terminal/endodermal functions associated with bowl and drm. However, the issue of odd and its evolution is complicated by the existence of the closely linked paralog, sob. Whereas sob has structural affinities with bowl that are consistent with a more recent evolutionary origin than odd (Fig. 1), sob is expressed during segmentation in a pattern that is almost identical to that of odd (Hart et al. 1996). This implies that striped expression, and a corresponding role in segmentation, may have evolved in an ancestral homolog prior to gene duplication; in the case of bowl, this would account for both the faint remnant stripes and occasional segmentation defects seen in mutants (Wang and Coulter 1996). Alternatively, the striped expression of sob may be an artifact of its proximity to odd. Since sob function appears to be largely insignificant for embryonic patterning (Hart et al., submitted for publication), it is possible that both genes respond to cis-regulatory elements that are only functionally relevant for odd. Notably, three odd-like genes that are orthologous to odd, drm and either bowl or sob (Fig. 1) have been identified in another dipteran insect (Anopheles) with a long germ pattern of embryogenesis very similar to that of Drosophila (Monnerat et al. 2002). Because a second mosquito gene in the sob/bowl group has not been reported to date, it appears that a sob ortholog may not be present within this genus, supporting the apparently recent sob/bowl split in Drosophila. Notwithstanding the remaining uncertainties about their evolutionary origins and functions, the diverse patterns exhibited by various odd homologs attest to the evolutionary adaptability of genes in this family.
References
Adoutte A, Balavoine G, Lartillot N, de Rosa R (1999) Animal evolution. The end of the intermediate taxa? Trends Genet 15:104–108
Avery L, Thomas JH (1997) Feeding and defecation. In: Riddle DL, Blumenthal T, Meyer BS, Priess JR (eds) C. elegans II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
Blair JE, Ikeo K Gojobori T, Hedges SB (2002) The evolutionary position of nematodes. BMC Evol Biol 2:7
Bouchard M, St-Amand J, Cote S (2000) Combinatorial activity of pair-rule proteins on the Drosophila gooseberry early enhancer. Dev Biol 222:135–146
Coulter DE, Wieschaus E (1988) Gene activities and segmental patterning in Drosophila: analysis of odd-skipped and pair-rule double mutants. Genes Dev 2:1812–1823
Coulter DE, Swaykus EA, Beran-Koehn MA, Goldberg D, Wieschaus E, Schedl P (1990) Molecular analysis of odd-skipped, a zinc finger encoding segmentation gene with a novel pair-rule expression pattern. EMBO J 9:3795–3804
Debeer P, de Ravel TJ, Devriendt K, Fryns JP, Huysmans C, Van de Ven WJ (2002) Human homologues of Osr1 and Osr2 are not involved in a syndrome with distal limb deficiencies, oral abnormalities, and renal defects. Am J Med Genet 111:455–456
DiNardo S, O’Farrell PH (1987) Establishment and refinement of segmental pattern in the Drosophila embryo: spatial control of engrailed expression by pair-rule genes. Genes Dev 1:1212–1225
Dovat S, Ronni T, Russell D, Ferrini R, Cobb BS, Smale ST (2002) A common mechanism for mitotic inactivation of C2H2 zinc finger DNA-binding domains. Genes Dev 16:2985–2990
Ellis RE, Horvitz HR (1991) Two C. elegans genes control the programmed deaths of specific cells in the pharynx. Development 112:591–603
Ellis RE, Jacobson DM, Horvitz HR (1991) Genes required for the engulfment of cell corpses during programmed cell death in Caenorhabditis elegans. Genetics 129:79–94
Felsenstein J (1993) PHYLIP (Phylogeny Inference Package) version 3.5c. Distributed by the author at http://evolution.genetics.washington.edu/phylip.html
Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806–811
Francis R, Waterston RH (1991) Muscle cell attachment in Caenorhabditis elegans. J Cell Biol 114:465–479
Garigan D, Hsu AL, Fraser AG, Kamath RS, Ahringer J, Kenyon C (2002) Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics 161:1101–1112
Green RB, Hatini V, Johansen KA, Liu XJ, Lengyel JA (2002) Drumstick is a zinc finger protein that antagonizes Lines to control patterning and morphogenesis of the Drosophila hindgut. Development 129:3645–3656
Guhathakurta D, Schriefer LA, Hresko MC, Waterston RH, Stormo GD (2002) Identifying muscle regulatory elements and genes in the nematode Caenorhabditis elegans. Proceedings of the 7th Pacific symposium on biocomputing, Lithue, Hawaii, 3–7 January 2002. pp 425–436
Hart MC, Wang L, Coulter DE (1996) Comparison of the structure and expression of odd-skipped and two related genes that encode a new family of zinc finger proteins in Drosophila. Genetics 144:171–182
Hresko MC, Williams BD, Waterston RH (1994) Assembly of body wall muscle and muscle cell attachment structures in Caenorhabditis elegans. J Cell Biol 124:491–506
Katoh M (2002) Molecular cloning and characterization of OSR1 on human chromosome 2p24. Int J Mol Med 10:221-225
Kuhn DT, Chaverri JM, Persaud DA, Madjidi A (2000) Pair-rule genes cooperate to activate en stripe 15 and refine its margins during germ band elongation in the D. melanogaster embryo. Mech Dev 95:297–300
Lan Y, Kingsley PD, Cho ES, Jiang R (2001) Osr2, a new mouse gene related to Drosophila odd-skipped, exhibits dynamic expression patterns during craniofacial, limb, and kidney development. Mech Dev 107:175–179
Miller J, McLachlan AD, Klug A (1985) Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J 4:1609–1614
Monnerat AT, Machado MP, Vale BS, Soares MJ, Lima JB, Lenzi HL, Valle D (2002) Anopheles albitarsis embryogenesis: morphological identification of major events. Mem Inst Oswaldo Cruz 97:589–596
Mullen JR, DiNardo S (1995) Establishing parasegments in Drosophila embryos: roles of the odd-skipped and naked genes. Dev Biol 169:295–308
Nüsslein-Volhard C, Wieschaus E (1980) Mutations affecting segment number and polarity in Drosophila. Nature 287:795–801
Park YS, Kramer JM (1994) The C. elegans sqt-1 and rol-6 collagen genes are coordinately expressed during development, but not at all stages that display mutant phenotypes. Dev Biol 163:112–124
Patel NH, Ball EE, Goodman CS (1992) Changing role of even-skipped during the evolution of insect pattern formation. Nature 357:339–342
Rosenfeld R, Margalit H (1993) Zinc fingers: conserved properties that can distinguish between spurious and actual DNA-binding motifs. J Biomol Struct Dyn 11:557–570
Saulier-Le Drean B, Nasiadka A, Dong J, Krause HM (1998) Dynamic changes in the functions of Odd-skipped during early Drosophila embryogenesis. Development 125:4851–4861
Schroder R, Jay DC, Tautz D (2000) Elimination of EVE protein by CALI in the short germ band insect Tribolium suggests a conserved pair-rule function for even skipped. Mech Dev 90:329
Skeath JB (1998) The Drosophila EGF receptor controls the formation and specification of neuroblasts along the dorsal-ventral axis of the Drosophila embryo. Development 125:3301–3312
So PL, Danielian PS (1999) Cloning and expression analysis of a mouse gene related to Drosophila odd-skipped. Mech Dev 84:157–160
Wang L, Coulter DE (1996) bowel, an odd-skipped homologue, functions in the terminal pathway during Drosophila embryogenesis. EMBO J 15:3182–3196
Ward EJ, Coulter DE (2000) odd-skipped is expressed in multiple tissues during Drosophila embryogenesis. Mech Dev 96:233–236
Ward EJ, Skeath JB (2000) Characterization of a novel subset of cardiac cells and their progenitors in the Drosophila embryo. Development 127:4959–4969
Waterston R (1998) Genome sequence of the nematode C. elegans: a platform for investigating biology. The C. elegans Sequencing Consortium. Science 282:2012–2018
Wilson R, Ainscough R, Anderson K, Baynes C, Berks M, et al (1994) 2.2 Mb of contiguous nucleotide sequence from chromosome III of C. elegans. Nature 368:32–38
Wolfe SA, Nekludova L, Pabo CO (2000) DNA recognition by Cys2His2 zinc finger proteins. Annu Rev Biophys Biomol Struct 29:183–212
Acknowledgements
We thank Larry Schriefer for instruction and assistance in generating the GFP reporter constructs and transgenic lines. We also thank Tim Schedl and lab members, particularly Sudhir Nayak and Dave Hansen, for scientific discussions and the use of the lab microscope, Aguan Wei for the myo-2::GFP plasmid, and an anonymous reviewer for helpful comments and suggestions. This research was supported by a Beaumont Faculty Development Award from Saint Louis University.
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by C. Desplan
Rights and permissions
About this article
Cite this article
Buckley, M.S., Chau, J., Hoppe, P.E. et al. odd-skipped homologs function during gut development in C. elegans . Dev Genes Evol 214, 10–18 (2004). https://doi.org/10.1007/s00427-003-0369-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00427-003-0369-x