Abstract
Main conclusion
This study reveals that mutations in BRIP1/2 subunits of the BAS complex disrupt root meristem development by decreasing PIN genes expression, affecting auxin transport, and downregulating essential root genes PLT.
Abstract
Switch defective/sucrose non-fermentable (SWI/SNF) chromatin remodeling complexes play vital roles in plant development. BRAHMA-interacting proteins1 (BRIP1) and BRIP2 are subunits of BRAHMA (BRM)-associated SWI/SNF complex (BAS) in plants; however, their role and underlying regulatory mechanism in root development are still unknown. Here, we show that brip1 brip2 double mutants have a significantly shortened root meristem and an irregular arrangement in a portion of the root stem cell niche. The mutations in BRIP1 and BRIP2 cause decreased expression of the PIN-FORMED (PIN) genes, which in turn reduces the transport of auxin at the root tip, leading to the disruption of the accurate establishment of normal auxin concentration gradients in the stem cells. Chromatin immunoprecipitation (ChIP) experiments indicated that BRIP1 and BRIP2 directly bind to the PINs. Furthermore, we found a significant down-regulation in the expression of key root development genes, PLETHORA (PLT), in brip1 brip2. The brip1 brip2 plt1 plt2 quadruple mutations do not show further exacerbation in the short-root phenotype compared to plt1 plt2 double mutants. Using a dexamethasone (DEX)-inducible PLT2 transgenic line, we showed that acute overexpression of PLT2 partially rescues root meristem defects of brip1 brip2, suggesting that BRIP1 and BRIP2 act in part through the PLT1/2 pathway. Taken together, our results identify the critical role and the underlying mechanism of BRIP1/2 in maintaining the development of root meristem through the regulation of auxin output and expression of PLTs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Roots are fundamentally crucial for plants to adapt to their living environments. The root growth of higher plants is sustained by multipotential stem cells in the root meristem (Scheres 2007). In Arabidopsis root meristem, the quiescent center (QC) and peripheral stem cells together form the root stem cell niche (Petricka et al. 2012). QC cells are seldom divided under normal conditions; however, it takes the activity of QC to sustain the root stem cells (van den Berg et al. 1997). Peripheral stem cells encircle QC, maintaining pluripotency in directly adjacent cells, thus providing a cellular source for generating different root tissue types (Dinneny and Benfey 2008). Therefore, the maintenance of stem cell niche and the proper development of meristem play an irreplaceable role in the normal growth of roots.
Auxin plays a pivotal role as a primary plant hormone in the regulation of root growth (Teale et al. 2006). The function of auxin depends on its differential distribution within the root meristem, forming an auxin maximum and concentration gradient in the root apex (Grieneisen et al. 2007; Petersson et al. 2009). The auxin efflux carriers PIN-FORMED (PIN) proteins, which consist of at least five members in Arabidopsis (PIN1, PIN2, PIN3, PIN4, and PIN7), play major roles in maintaining the proper auxin gradient for root growth (Grieneisen et al. 2007; Zazímalová et al. 2007; Petrásek and Friml 2009). Each PIN protein has been found to exhibit a unique expression pattern and localization depending on cell types (Blilou et al. 2005). PIN1 primarily localizes to the basal side of vascular cells (Blilou et al. 2005). PIN2 exhibits apical localization in epidermal and lateral root cap cells, while predominantly localizing basally in cortical cells (Müller et al. 1998). The localization of PIN3 can be observed in the second and third layers of the columella cells, as well as at the basal side of vascular cells and toward the lateral side of pericycle cells in the elongation zone (Michniewicz et al. 2007). PIN4 is detected in QC and its surrounding cells (Friml et al. 2002a). PIN7 is located on the lateral and basal surfaces of vascular cells in the meristem and elongation zone, while its distribution in columella cells is the same as that of PIN3 (Blilou et al. 2005). PIN1, PIN3, and PIN7 mediate the downward flow of auxin toward the root tip, resulting in higher concentrations of auxin in the root stem cell niche (Friml et al. 2002b, 2003b); whereas, PIN2 mediates the upward flow of auxin toward the elongation zone, maintaining a gradient of auxin concentrations within the root (Müller et al. 1998; Friml et al. 2003a). Studies have shown that single mutants of PIN1 and PIN2 display a slight decrease in root length and size of the root meristem (Müller et al. 1998; Sabatini et al. 1999). Single mutants of PIN3, PIN4, and PIN7 exhibit only minor defects in the QC and the columella root cap (Friml et al. 2002a, b, 2003b). However, most combinations of multiple mutants display more pronounced deficiencies in root length and size of the root meristem (Friml et al. 2003b; Blilou et al. 2005). This dynamic distribution network of PIN proteins effectively establishes and stores high levels of auxin in the root, ensuring the generation of maximal auxin levels in the QC and nearby positions.
Optimal development of root meristem necessitates the proper maintenance of stem cell niches. Two key transcriptional control pathways working in parallel are essential for maintaining the normal functions of the stem cell niche in Arabidopsis roots, namely the SHORT-ROOT (SHR)/SCARECROW (SCR) pathway independent of auxin and the PLETHORA (PLT) pathway that is related to auxin (Helariutta et al. 2000; Nakajima et al. 2001; Sabatini et al. 2003; Aida et al. 2004; Blilou et al. 2005). SHR and SCR encode transcription factors of the GRAS family to regulate the radial organization of root in order to provide location information along the radial axis (Di Laurenzio et al. 1996; Helariutta et al. 2000; Levesque et al. 2006; Cui et al. 2007). Mutations in either SHR or SCR lead to irregular morphology in the stem cell niche, ultimately resulting in the collapse of the root meristem (Benfey et al. 1993). On the other hand, the PLT gene family encodes APETALA2 transcription factors, consisting of four homologous proteins with functional redundancy (Aida et al. 2004; Santuari et al. 2016). PLT1, 2, 3, and BABY BOOM (BBM) (also known as PLT4) are primarily expressed in QC and its surrounding region (Galinha et al. 2007). plt1 plt2 double mutants fail to maintain QC correctly, and the primary root of plt1 plt2 exhibits significant shortening (Aida et al. 2004; Galinha et al. 2007). Both PLT genes and auxin demonstrate a graded distribution in the root; therefore, PLTs were considered a readout of the auxin gradient (Galinha et al. 2007; Grieneisen et al. 2007; Petersson et al. 2009; Band et al. 2014; Mähönen et al. 2014). However, existing studies also suggested that the PLT gradient is not a direct or proportional reading of the auxin gradient. Prolonged elevation of auxin levels leads to the formation of a restricted transcriptional domain for PLT genes, which serves as the source for generating a gradient of PLT proteins through gradual growth dilution and cell-to-cell transportation (Mähönen et al. 2014). Moreover, PIN genes maintain the normal development of the root stem cell niche by regulating the expression of PLT genes; in turn, PLT genes are needed to stabilize auxin maximum at the root tip to support the expression of PIN genes (Blilou et al. 2005). Hence, an interaction network consisting of auxin transport and PLT, the determinant of root fate, is indispensable for coordinating the normal growth and development of plant roots.
The switch defective/sucrose non-fermentable (SWI/SNF) complexes are chromatin remodeling complexes which utilize the energy generated by adenosine triphosphate (ATP) hydrolysis to alter the interaction between histone octamer and nucleosome DNA (Clapier and Cairns 2009; Ho and Crabtree 2010). In Arabidopsis thaliana, we and others recently reported that SWI/SNF complexes are divided into three distinct subcomplexes based on different ATPase subunits (Guo et al. 2022; Fu et al. 2023). BRAHMA (BRM) ATPase subunit forms one of these subcomplexes known as the BAS complex (BRM-associated SWI/SNF complex) (Guo et al. 2022; Fu et al. 2023). BRM-interacting proteins1 (BRIP1) and BRIP2 were identified as the subunits of the BAS complexes. BRIP1 and BRIP2 contain a conserved Glioma Tumor Suppressor Candidate Region (GLTSCR) domain, which interacts with BRM ATPase (Yu et al. 2020). Functionally, current research suggests that BRIP1 and BRIP2 are mainly involved in maintaining the stability of the BAS complexes (Yu et al. 2020).
The double mutants of BRIP1 and BRIP2 exhibit multiple phenotypes in the above-ground tissues, such as semi-dwarfness, curled leaves, early flowering, and shortened siliques. However, these abnormal phenotypes were much less severe than the phenotypes observed in the brm-1 null mutants (Yu et al. 2020). These genetic observations suggested that the function of BAS complexes in the above-ground tissues is only partially compromised in the absence of BRIP1 and BRIP2. Previous studies have indicated that BRM plays a significant role in root development through its influence on auxin distribution via regulating PINs and PLT1/2 expression (Yang et al. 2015). However, whether and to what extent BRIP1 and BRIP2 are required for the function of BAS complexes in the root development remain unknown. The potential for BRIP1/2 to play different roles in different parts of the plant is also unclear.
Here, we describe the critical functions of BRIP1 and BRIP2 in root development and demonstrate that they play an essential role in maintaining the root meristem by promoting the root stem cell niche. Our findings offer insights into the specific transcriptional regulatory mechanisms of BRIP1 and BRIP2 in root development.
Materials and methods
Plant materials and growth conditions
Arabidopsis thaliana ecotype Columbia-0 (Col-0) was used as the wild type in this study. The mutants brip1-1 (SALK_133464); brip2-1 (SALK_177513); brm-1 (SALK_030046); and brm-3 (SALK_088462) were previously described (Li et al. 2016; Yu et al. 2020). The mutants plt1-4 plt2-2 were previously described (Aida et al. 2004).
The following transgenic plants were previously described: pDR5:GFP and pPIN1:PIN1-GFP (Benková et al. 2003); pPIN2:PIN2-GFP; pPIN3:PIN3-GFP and pPIN7:PIN7-GFP (Blilou et al. 2005); pPLT1:PLT1-YFP; pPLT2:PLT2-YFP and p35S:PLT2-GR (Galinha et al. 2007); pBRIP1:BRIP1-GFP; and pBRIP2:BRIP2-GFP (Yu et al. 2020).
Arabidopsis seeds were surface sterilized in 20% NaClO for 15 min and washed five times with distilled water. Then, the seeds were stratified at 4 °C in the dark for 3 days and vertically germinated on MS medium (Murashige and Skoog) with 1.5% sucrose and 0.8% agar. All plants were grown in a greenhouse maintained at 22 °C under a 16-h light/8-h dark photoperiod.
Root length and root meristem size measurement
First, plants were allowed to grow vertically in a Petri dish. For the measurement of root length, plants with different genotypes were placed side-by-side in Petri dishes. After taking pictures with a camera, the root length was measured with Image J software. For the measurement of root meristem size, we first stained root tips by propidium iodide and photographed them under a confocal microscope. Root meristem size is determined by measuring the number of cortical cells from QC to the first elongated cell (Perilli and Sabatini 2010). The meristem size was measured by Image J. For DEX induction, following 3 days of vertical growth, DEX was applied to induce PLT2 expression. Two days post-induction, the root meristem size was measured.
Microscopy
The plants were first grown vertically and removed with tweezer. Roots were cleared in HCG solution (chloroacetaldehyde:water:glycerol = 8:3:1, by vol.) for several minutes before microscopy analysis. For confocal imaging, the plants were immersed in a staining solution containing 10 μg/mL propidium iodide for one minute. Confocal images were performed using a Zeiss 7 Duo NLO affiliated 880 laser scanning. The excitation/emission wavelengths are 561 nm/591–635 nm for PI, 488 nm/505–530 nm for GFP, and 514 nm/520–550 nm for YFP. For Lugol staining of roots, the tissues were treated with Lugol solution (Sigma-Aldrich, available at http://www.sigmaaldrich.com/) for a duration of 2 min. Following this incubation, the samples were rinsed once with double-distilled water and then mounted in HCG solution.
Gene expression analysis via RT-qPCR
Root tips (~ 5 mm) were quickly harvested and immediately frozen in liquid nitrogen. Total RNA was extracted with the RNA prep pure Plant kit (MAGEN, Guangzhou, China; Cat. No. R4151-02C). Reverse-transcription reactions were performed using 1 mg of total RNA with HiScript II Q RT SuperMix for qPCR (+ gDNA wiper) with gDNA eraser (Vazyme, Nanjing, China; Cat. No. R223-01). qRT-PCR analyses were conducted using SYBR Green SuperMix and processed with StepOne Software v.2.3 (Applied Biosystems) with 40 cycles. These experiments were replicated three times biologically. Data were analyzed with the − ΔΔCt (cycle threshold) method (Livak and Schmittgen 2001). The Arabidopsis UBQ10 gene was used as an internal control. Statistical significance was evaluated by one-way ANOVA analysis. Primer sequences are given in Supplemental Table S1.
Chromatin immunoprecipitation (ChIP)-qPCR
ChIP assays were performed as previously described with minor modifications (Li et al. 2016). In brief, 2 g of roots from 14 DAG seedlings grown on vertically oriented plates with MS medium was collected, fixed in water containing 1% formaldehyde, and then ground into fine power with liquid nitrogen. Chromatin was isolated and sheared into 200–800 base pair fragments by sonication. The chromatin was immunoprecipitated with specific antibodies including anti-GFP (Abcam, Cambridge, UK; Cat. No. ab290). Equal amount of the sonicated chromatin solution was set aside as the input sample. ChIP-qPCR was performed with three biological replicates. ChIP-qPCR results were calculated as a percentage of input DNA according to the Champion ChIP-qPCR user manual (SABioscience, Frederick, MD, USA). The primer sequences can be found in Supplemental Table S1. The exon region of retrotransposon TA3 was used as negative control.
Results
The brip1 brip2 double mutants show defective root growth and reduced root meristem size
To investigate whether BRIP1 and BRIP2 play roles in root development, we analyzed the primary root length. The single mutants, brip1 and brip2, showed no significant differences in primary root length compared to the wild-type (WT) plants (Fig. 1a). This lack of difference can be attributed to the functional redundancy between BRIP1 and BRIP2. The brip1 brip2 double mutants, on the contrary, displayed a strong reduction in primary root length compared to both the WT and single mutants (Fig. 1a and c). The differences in primary root growth between brip1 brip2 and WT or single mutants became evident starting from day 3 post imbibition and gradually increased as the seedlings grew (Fig. 1c). On the ninth day of germination (DAG), the primary root length of brip1 brip2 mutants was less than 25% of the WT root (brip1 brip2: ~ 0.7 cm, WT: ~ 3.2 cm) (Fig. 1c). Given the correlation between the size of the root meristem and the length of the root, we conducted a comparative observation of the root meristem size in WT, brip1, brip2, and brip1 brip2. The results indicated no significant differences in root meristem size between the WT and single mutants. However, the brip1 brip2 double mutants exhibited a significantly smaller root meristem size (Fig. 1b and d).
brip1 brip2 mutants show defective root growth and reduced root meristem size. a The root phenotypes of the wild-type (WT), brip1, brip2, brip1 brip2, brm-3, brm-1, brip1 brip2 brm-3, and brip1 brip2 brm-1 seedlings on 1/2 MS medium at 5 days after germination (DAG). Scale bar = 1 cm. b The root meristem size of the WT, brip1, brip2, brip1 brip2, brm-3, brm-1, brip1 brip2 brm-3, and brip1 brip2 brm-1 at 5 DAG. The white arrow indicates the beginning of root meristem, and the yellow arrow indicates the end of root meristem. The distance between the white arrow and the yellow arrow represents the length of the root meristem. Scale bar = 50 μm. c The primary root length of the WT, brip1, brip2, brip1 brip2, brm-3, brm-1, brip1 brip2 brm-3, and brip1 brip2 brm-1 from 3 to 10 DAG. The data represent mean ± SD (n = 15 plants). d The root meristem size was measured in the WT, brip1, brip2, brip1 brip2, brm-3, brip1 brip2 brm-3, and brip1 brip2 brm-1 at 5 DAG. The root meristem size is expressed as the length from cortex cells in Quiescent center (QC) to the transition zone. Data shown are means ± SD (n = 10 plants). Lower-case letters indicate significant differences, as determined by two-way ANOVA with Tukey’s multiple comparisons test
Our previous study showed that brip1 brip2 mutants exhibit relatively weak phenotypes in the above-ground parts of plants compared to those by the brm-1 null mutants, indicating that loss of BRIP1 and BRIP2 only partially compromised the role of BAS complexes in regulating the development of the above-ground plant organs (Yu et al. 2020). However, we found here that the defects in root length and root meristem of brip1 brip2 mutants were almost as strong as those of brm-1 loss of function mutants, suggesting that lack of BRIP1 and BRIP2 nearly completely abolished the function of BAS complexes in root tissues. We crossed the weak brm-3 mutant and the null brm-1 mutant with brip1 brip2. The primary root length of brip1 brip2 brm-3 was shorter than that of brm-3, but it was comparable to the primary root length of brip1 brip2 (Fig. 1a and c). The primary root length of brip1 brip2 brm-1 was similar to that of brm-1, indicating that loss of BRIP1/2 did not further enhance the brm-1 root phenotype (Fig. 1a and c). Additionally, the root meristem size of brip1 brip2 brm-3 closely resembled that of brip1 brip2, but was smaller than that of brm-3 (Fig. 1b and d). The root meristem sizes of brip1 brip2 brm-1 and brm-1 were almost identical (Fig. 1b and d). Together, these results suggest that BRIP1 and 2 are more essential for BAS-mediated root development than BAS-mediated above-ground tissue development.
BRIP1 and BRIP2 are necessary for the maintenance of stem cell niche
The above results indicated that BRIP1 and 2 are pivotal in maintaining root meristem size. To investigate whether loss of BRIP1/2 affects the QC and surrounding stem cells, we examined the root apical structure of WT, single, and double mutants under confocal microscopy (Fig. 2; Suppl. Fig. S1). At 3 DAG, both the WT and single mutants displayed a regular cellular arrangement, with the QC cells aligned parallelly, encircled by surrounding meristem cells (Fig. 2a) (van den Berg et al. 1997). Conversely, nearly 30–40% of the brip1 brip2 double mutants displayed an irregular cellular pattern in root tips, with the QC becoming unidentifiable (Fig. 2a and b). This defection was even more pronounced in brm-1, with up to four fifth showing disrupted QC identification (Fig. 2a and b). Furthermore, at 5 DAG, single mutants still maintained a standard cellular pattern, but about 25% of the brip1 brip2 double mutants showed pronounced irregularities, making the QC location hard to discern (Suppl. Fig. S1). Simultaneously, to rigorously validate our results and rule out the possibility of non-specific or unrelated phenotypic effects due to the T-DNA mutations examined, we assessed both the root length and the arrangement of the QC and its surrounding cells in pBRIP1:BRIP1-GFP brip1 brip2 and pBRIP2:BRIP2-GFP brip1 brip2 lines, both expressing GFP-tagged BRIP1 or BRIP2 under their respective native promoters. Remarkably, the short-root phenotype of brip1 brip2 was fully complemented (Suppl. Fig. S2), and there were no discernible defects in the arrangements of QC and its adjacent cells in the complement plants (Fig. 2a and b).
BRIP1 and BRIP2 are necessary for the maintenance of stem cell niche. a The arrangement of QC in the WT, brip1, brip2, brip1 brip2, pBRIP1:BRIP1-GFP brip1, pBRIP2:BRIP2-GFP brip2, pBRIP1:BRIP1-GFP brip1 brip2, and pBRIP2:BRIP2-GFP brip1 brip2 root tips by PI staining at 3 DAG. The white arrow points to the position of Quiescent center. Scale bar = 50 μm. b The percentage of QC irregularly arranged in the WT, brip1, brip2, brip1 brip2, pBRIP1:BRIP1-GFP brip1, pBRIP2:BRIP2-GFP brip2, pBRIP1:BRIP1-GFP brip1 brip2, and pBRIP2:BRIP2-GFP brip1 brip2 root tips at 3 DAG. c The expression pattern of pWOX5:GFP in the root tips of WT, brip1, brip2, and brip1brip2 at 3 DAG. Scale bar = 50 μm. d Lugol stain at root tip in the WT, brip1, brip2, and brip1 brip2 at 5 DAG. The yellow arrows indicate the position of QC in root tips. Scale bar = 50 μm
WUSCHEL-RELATED HOMEOBOX 5 (WOX5) encodes a key homeodomain transcription factor involved in maintaining stem cell identity in the root initials surrounding the QC (Sarkar et al. 2007). We crossed the pWOX5:GFP reporter, which encodes a green fluorescent protein (GFP) driven by the WOX5 promoter, into our brip1 brip2 mutants. In WT, brip1, and brip2, GFP signals were detected specifically in QC cells. Interestingly, in brip1 brip2, GFP signals were diffused and expanded to neighboring cells in approximately 30% of brip1 brip2 roots (15 out of 49 examined roots) (Fig. 2c). Lugol staining of the roots revealed that both brip1, brip2, and WT seedlings exhibited the same number of tiers of starch-containing columella root cap cells. Notably, however, there were fewer layers of starch-containing cells in the brip1 brip2 seedlings compared to those in the WT (Fig. 2d). These results suggest that BRIP1 and BRIP2 are essential for maintaining the stem cell niche.
The brip1 brip2 double mutants reduce auxin accumulation and the expression of PINs in the root tips
We previously reported that BRM maintains stem cells at the root tip by regulating the auxin signaling pathway (Yang et al. 2015). To explore whether the short-root phenotype of brip1 brip2 double mutants is related to auxin, we performed a genetic cross between brip1 brip2 and a pDR5:GFP marker line, which exhibits a characteristic expression pattern that reliably indicates the accumulation and distribution of auxin (Ulmasov et al. 1997). We found that, compared to WT, there was no change in GFP fluorescence intensity in the single mutants. However, the double mutants exhibited a substantial reduction in fluorescence intensity (Fig. 3a and b), suggesting that BRIP1 and 2 are required for the normal auxin accumulation in roots.
brip1 brip2 mutants decrease auxin content and lead to down-regulation of PIN genes. a The expression of pDR5:GFP in the root tips of WT, brip1, brip2, and brip1 brip2 at 5 DAG. Scale bar = 50 \(\upmu\)m. b The statistical result for relative fluorescence of pDR5:GFP in the root tips of WT, brip1, brip2, and brip1 brip2 at 5 DAG. The data represent mean ± SD (n = 15 plants). Lower-case letters indicate significant differences, as determined by two-way ANOVA with Tukey’s multiple comparisons test. c PIN1, PIN2, PIN3, PIN4 and PIN7 expression were determined by RT-PCR in the root tips of WT, brip1, brip2, brip1 brip2, brm-3, and brip1 brip2 brm-3 at 5 DAG. The data represent mean ± SD from three biological replicates. *P < 0.05, determined by the Student’s t-test
Next, we conducted experiments with exogenous auxin application to explore the possibility of complementing the root development defects of the brip1 brip2 double mutants. Upon treatment with exogenous auxin, we observed that at a concentration of 0.02 µM, there was no complementation on the short-root defect of brip1 brip2 (Suppl. Fig. S3). When the auxin concentration was elevated to 0.05 µM, due to the biphasic effect of auxin, root growth was suppressed not only in the mutants but also in WT (Suppl. Fig. S3). However, we found that the treatment with exogenous auxin partially compensates the defects in root meristem (Suppl. Fig. S4), suggesting the lower auxin levels causes, at least partially, the defects of the root meristem in brip1 brip2 double mutants. Considering the pivotal role of PIN proteins in shaping the maxima and minima of auxin that guide root development, we performed RT-qPCR to examine the relative expression levels of PINs and observed a significant decrease in PINs expression in brip1 brip2 root tips compared to WT (Fig. 3c). Moreover, we also investigated the expression levels of PINs in the root tip of brip1 brip2 brm-3 triple mutants. The results revealed that the triple mutants exhibited no further down-regulation in PINs expression compared to the double mutants (Fig. 3c). This finding further supports the notion that BRIP1 and BRIP2 indeed belong to the same subcomplex as BRM to cooperatively control root development through transcriptionally mediating auxin transporters.
Finally, we crossed brip1 brip2 with pPINs:PINs-GFP to assess any changes in the protein levels of PIN1, PIN2, PIN3, and PIN7. Compared to WT, the fluorescence signals of individual single mutants did not show significant alterations (Fig. 4). However, a significant decrease in fluorescence signals of PIN1, PIN2, PIN3, and PIN7 proteins were observed in brip1 brip2 (Fig. 4). Together, these results indicate that the short-root phenotype of the double mutants is likely caused by the reduction of auxin accumulation and polar transport-related gene expression.
The protein accumulation pattern of PINs-GFP in root tips of the brip1 brip2. a, c, e and g The protein accumulation of pPINs:PINs-GFP in the root tips of WT, brip1, brip2, and brip1 brip2 at 5 DAG. Scale bar = 50 μm. b, d, f and h The statistical result for relative fluorescence of pPINs:PINs-GFP in the root tips of WT, brip1, brip2, and brip1 brip2 at 5 DAG. The data represent mean ± SD (n ≥ 10 plants). Lower-case letters indicate significant differences, as determined by two-way ANOVA with Tukey’s multiple comparisons test
BRIP1 and BRIP2 directly regulate auxin transport-related genes
To detect whether BRIP1 and BRIP2 directly regulate the PIN genes, we first analyzed ChIP-seq data reported by Yu et al. (2020) and found that BRIP1 and BRIP2 exhibited significant binding signals at all the five PINs gene loci in seedlings (Fig. 5a). Visualization using the integrative genomics viewer (IGV) demonstrated the enrichment of BRIP2 near the transcription start site (TSS) of PIN1, at the transcription termination site (TTS) of PIN2, and near the TSS and in the gene body of PIN3, PIN4, and PIN7 (Fig. 5a). These data imply that BRIP1 and BRIP2 may also directly bind to PINs in the roots. To test this hypothesis, root materials from 14-day-old WT, pBRIP1:BRIP1-GFP brip1, and pBRIP2:BRIP2-GFP brip2 seedlings were collected for ChIP-qPCR experiments. Specific primers were designed according to the seedling ChIP-seq data for the binding sites and non-binding sites in each PIN loci. ChIP-qPCR experiments using roots as the materials were largely consistent with the ChIP-seq findings, confirming the enrichment of BRIP1 and BRIP2 in PINs (Fig. 5b and c). These results revealed that the PIN genes were the target genes of BRIP1 and BRIP2, and both play a direct role in regulating genes of the auxin output pathway.
BRIP1 and BRIP2 can target to PINs directly. a Integrative genomics viewer (IGV) views of BRIP2 occupancy at PINs using ChIP-seq data from Yu et al. (2020). The scale is identical for the different tracks, and gene structures are shown below each panel. P represents the primer designed for the detected regions. b, c ChIP-qPCR analysis of the enrichment of BRIP1and BRIP2 to the different regions of PINs loci in WT, pBRIP1:BRIP1-GFP, and pBRIP2:BRIP2-GFP Arabidopsis roots. The data represent mean ± SD from three biological replicates. *P < 0.05, determined by the Student’s t-test
BRIP1 and BRIP2 indirectly promote the expression of PLT1/2
The above results indicate that the root developmental defects in brip1 brip2 are associated with reduced auxin levels and impaired auxin polar transport. The PLT pathway, which is closely linked to the auxin gradient in the root tip, plays a crucial role in plant root development (Blilou et al. 2005; Mähönen et al. 2014). Therefore, we wondered whether mutations in BRIP1 and BRIP2 affect the gene expression of the PLT pathway in the root meristem. The results showed that, compared to WT, single mutants of brip1 and brip2 did not show significant changes in the transcription of PLT1 and PLT2 (Fig. 6a). However, brip1 brip2 mutants exhibited a significant decrease in the transcription levels of PLT1 and PLT2 (Fig. 6a). Additionally, the brm-3 hypomorphic mutation did not further enhance the down-regulation of PLT1 and PLT2 expression in the double mutants (Fig. 6a).
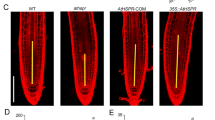
BRIP1/2 acts partially through the PLT1/2 pathway. a PLT1 and PLT2 expression were determined by RT-PCR in the root tips of WT, brip1, brip2, brip1 brip2, brm-3, and brip1 brip2 brm-3 at 5 DAG. The data represent mean ± SD from three biological replicates. *P < 0.05, determined by the Student’s t-test. b The protein levels of PLT2-YFP in the root tips of WT, brip1, brip2, and brip1 brip2 at 5 days after germination (DAG). Scale bar = 50 µm. c The statistical result for relative fluorescence of PLT2-YFP in the root tips of WT, brip1, brip2 and brip1 brip2 at 5 DAG. The data represent mean ± SD (n = 15 plants). Lower-case letters indicate significant differences, as determined by two-way ANOVA with Tukey’s multiple comparisons test. d Root phenotypes of the WT, brip1 brip2, plt1 plt2, and brip1 brip2 plt1 plt2 single, double, and quadruple mutants at 7 DAG. e The statistical result for primary root length of the WT, brip1 brip2, plt1 plt2, and brip1 brip2 plt1 plt2. The data represent mean ± SD (n = 10 plants). Lower-case letters indicate significant differences, as determined by two-way ANOVA with Tukey’s multiple comparisons test
Next, we investigated the impact of BRIP1 and BRIP2 on the protein level of PLT2 in the root meristem. We hybridized the brip1 brip2 double mutants with the transgenic plants carrying pPLT2:PLT2-YFP. As depicted in Fig. 6b and c, the fluorescence intensity of PLT2-YFP in the root tips of brip1 brip2 was significantly lower compared to the WT. We then designed primers targeting different regions of the PLT1/2 genes to investigate whether they are direct targets of BRIP1/2. ChIP-qPCR experiments revealed that BRIP2 did not directly bind to the PLT1 and PLT2 loci (Suppl. Fig. S5). Finally, to further confirm the involvement of BRIP1/2 in the PLT pathway, we performed cross between brip1 brip2 and plt1 plt2 double mutants. The root length of the brip1 brip2 plt1 plt2 quadruple mutants was similar to that of the plt1 plt2 double mutants (Fig. 6d and e). This indicates that the quadruple mutants did not exhibit an exacerbated short-root phenotype compared to the plt1 plt2, providing further evidence for the role of BRIP1/2 through partially the PLT pathway in regulating root development.
Overexpression of PLT2 partially rescues the stem cell niche defect of brip1 brip2
To determine whether overexpression of PLT genes can bypass the root meristem defects in brip1 brip2, a p35S:PLT2-GR construct was introduced (Galinha et al. 2007). Previous studies showed that dexamethasone (DEX) induction of p35S:PLT2-GR plants led to significant increases in root meristem cell number (Schena et al. 1991; Galinha et al. 2007). We performed genetic crosses between brip1 brip2 and the transgenic plant harboring the inducible construct p35S:PLT2-GR. After three days of vertical cultivation, dexamethasone (DEX) was added to acutely induce the expression of PLT2 in both p35S:PLT2-GR and p35S:PLT2-GR brip1 brip2 plants. Following two days of induction, it was observed that the growth of seedlings was not severely affected (Fig. 7a). However, upon induction with DEX, the short-term overexpression of PLT2 resulted in a significant increase in the number of cells in the proximal region of the wild-type root meristem (Fig. 7b and c). Importantly, in p35S:PLT2-GR brip1 brip2 plant, the size of the root meristem was also noticeably increased after short-term overexpression of PLT2 by dexamethasone (Fig. 7b and c). In addition, the SHR/SCR pathway is another key player in the regulation of root radial patterning and the maintenance of the stem cell niche (Di Laurenzio et al. 1996; Helariutta et al. 2000). Through ChIP-qPCR, we found that BRIP1/2 directly bound to the promoter of SCR, but not to SHR (Fig. 8a and b). One point of interest is whether the binding of BRIP1/2 to the SCR promoter leads to changes in SCR expression. We analyzed SCR transcription in the brip1 brip2 double mutants and found that SCR transcription in the brip1 and brip2 single mutants remains consistent with WT. In the brip1 brip2 double mutants, SCR transcription largely remained unchanged, displaying only a subtle, non-significant down-regulation (Fig. 8c). Importantly, this minor change in SCR expression contrasts with the more pronounced effects of BRIP1/2 on PIN genes. Overall, the partial rescue of root meristem growth in brip1 brip2 mutants by overexpression of PLT2 indicates the involvement of BRIP1/2 in the auxin-related PLT1/2 pathway during the regulation of root meristem development.
Overexpression of PLT2 partially rescues the root meristem of brip1 brip2. a The root phenotypes of the p35S:PLT2-GR and p35S:PLT2-GR brip1 brip2 seedlings after DMSO or 2 µM DEX (+ DEX) treatment on 1/2 MS medium at 5 DAG. Scale bar = 1 cm. b The statistical result for the number of cortex cells in the p35S:PLT2-GR and p35S:PLT2-GR brip1 brip2 seedlings after DMSO or 2 µM DEX (+ DEX) treatment at 5 days after germination (DAG). The data represent mean ± SD (n = 15 plants). *P < 0.05, determined by the Student’s t-test. c The root meristem of the p35S:PLT2-GR and p35S:PLT2-GR brip1 brip2 seedlings after DMSO or 2 µM DEX (+ DEX) treatment at 5 DAG. The white arrow indicates the beginning of root meristem, and the yellow arrow indicates the end of root meristem. The distance between the white arrow and the yellow arrow represents the length of the root meristem. The long white arrow indicates wild-type meristem beyond the microscopic field of view. Scale bar = 50 µm
The mutation in BRIP1/2 has no effects on the transcription of SHR and SCR. a, b ChIP-qPCR analysis of the enrichment of BRIP2 to the different regions of SHR and SCR loci in WT and pBRIP2:BRIP2-GFP Arabidopsis roots. The top of the diagram shows a schematic of the detected genes in the genome. The black thick frame, the black thin frame and the black line represent the exon, UTR, intron and promoter of genes, respectively. P/C represents the amplicon for the detected regions. The asterisk indicates that the transgenic plants have significant enrichment compared with the WT. *P < 0.05; NS, not significant as determined by Student’s t test. c SHR and SCR expression were determined by RT-qPCR in the root tips of WT, brip1, brip2, brip1 brip2, brm-3, and brip1 brip2 brm-3 at 5 DAG. The data represent mean ± SD from three biological replicates. *P < 0.05; NS, not significant as determined by Student’s t test
Discussion
BRIP1 and BRIP2 are essential factors for maintaining normal plant root growth
BRIP1 and BRIP2 were recently shown to be subunits of the BAS complexes and are involved in the development of the above-ground tissues. However, whether and to what extent BRIP1 and BRIP2 are required for the function of BAS complexes in the root development remain unknown. The potential for BRIP1/2 to play different roles in different parts of the plant is also unclear. In this study, we revealed the critical functions of BRIP1 and BRIP2 in root development. We observed that the primary root length of the brip1 brip2 double mutant was significantly shorter than that of WT, and the size of the root meristem was significantly smaller (Fig. 1). Normal growth and development of plants rely on the activity of undifferentiated cell populations, which serve as a cellular source for organ growth (van den Berg et al. 1997; Stahl and Simon 2005). Maintenance and homeostasis of the stem cell niche in Arabidopsis roots are crucial for the development of all types of root cells. Through microscopic observations, we found that BRIP1 and BRIP2 also play important roles in maintaining the root stem cell niche: compared to WT, the arrangement of the QC and surrounding cells in the brip1 brip2 double mutants was noticeably irregular, and the location of the QC could no longer be distinguished (Fig. 2). Hence, BRIP1/2 are essential proteins for maintaining the root stem cell niche in plants.
In the aerial parts of the plant, including leaves, stem, flowers, and siliques, the absence of BRIP1/2 only partially reduces the function of the BAS complexes, as evidenced by the relatively weak phenotypes observed in the brip1 brip2 double mutants compared to the brm-1 mutants (Yu et al. 2020). However, we show here that, in the root tissues, the absence of BRIP1/2 leads to a near complete disruption of the BAS complexes function, since the roots of the brip1 brip2 double mutants closely resemble those of the brm-1 mutants (Fig. 1). This differential contribution of BRIP1/2 to the functionality of the BAS complexes between the aerial parts and the roots underscores the intricacies of the molecular regulation of plant growth and development. The differential impact of BRIP1/2 mutations on the aerial and root tissues also highlights the adaptability of the plant system to cope with different environmental stimuli. One possibility to explain such a differential contribution by BRIP1/2 may be the existence of compensatory regulatory mechanisms from other subunits of the BAS complexes, such as BRD1/2/13 (Yu et al. 2021). Indeed, loss of BRD1/2/13 leads to significant impairments of above-ground development but does not cause significant phenotypes on roots (Yu et al. 2021). Thus, our finding is particularly valuable as it not only unravels the unique and essential roles of BRIP1/2 proteins in root development but also suggests the possibility of distinct regulatory mechanisms between BRIP1/2 and BRD1/2/13 in different parts of the plants. It will now be important to explore the molecular mechanisms and their implications for the differential contributions of BRIP1/2 observed in different plant tissues. Understanding the nuanced roles BRIP1/2 played in different tissues may enable the manipulation of these factors in a tissue-specific manner, potentially leading to innovative strategies for improving crop growth and productivity.
BRIP1 and BRIP2 contribute to the accumulation and transport of auxin
The normal development of plant roots relies on the action of plant hormones, with auxin playing a crucial role. Thus, the spatial distribution of auxin is essential for proper root development. In this study, we initially examined changes in auxin accumulation and found that the expression of pDR5:GFP was significantly reduced in the brip1 brip2 mutants (Fig. 3a and b), indicating that BRIP1/2 affect the auxin content in the root meristem. The primary mechanism controlling auxin distribution in plant roots is the active directional transport of auxin between cells. This transport is facilitated by membrane-based transporters of the AUX1/LIKE AUX1 (AUX1/LAX) family, which act as influx carriers, and by members of the PIN family and ATP-binding cassette subfamily B (ABCB) of the multidrug resistance/phosphoglycoprotein (ABCB/MDR/PGP) protein family, which serve as efflux carriers (Petrásek and Friml 2009). Auxin directional transport mediated by PIN efflux carriers plays a crucial guiding role in this regulation (Blilou et al. 2005; Adamowski and Friml 2015). Our results demonstrate that the root tips of brip1 brip2 plants exhibit a substantial decrease in both transcriptional levels of PIN1, PIN2, PIN3, PIN4, and PIN7 and fluorescence intensity from pPINs:PINs-GFP (Fig. 3c and 4). Additionally, our ChIP experiments showed that BRIP1/2 can directly target the PIN genes (Fig. 5b and c), indicating that BRIP1/2 directly act upstream of PINs and regulates their expression. Collectively, these data indicate that BRIP1/2 serve as key upstream regulators within the auxin hormone signaling pathway, significantly influencing the distribution and action of auxin within the plant. The BRIP1/2 proteins not only regulate hormone levels but also participate in orchestrating the cell-to-cell transport of these hormones, further enhancing the complexity of root development control. It would be interesting to understand the extent to which BRIP1/2 might mediate the interplay between auxin and gibberellins or abscisic acid (ABA), both of which are known to interact with auxin signaling during various developmental processes, including root development (Saini et al. 2013; Lee et al. 2013). ABA is known to stimulate the production of reactive oxygen species in mitochondria, diminish auxin accumulation, and ultimately inhibit primary root growth (He et al. 2012; Yang et al. 2014). Consequently, exploring the putative role of BRIP1/2 in these auxin-ABA interactions could potentially unlock new insights into plant hormone signaling dynamics, contributing to the regulation of plant root growth and stress responses.
BRIP1 and BRIP2 regulate root growth partially through the PLT pathway
Our findings suggest that mutations in BRIP1 and BRIP2 disrupt the establishment of normal auxin concentration gradients. The establishment of a dynamic gradient across the root meristem, mediated by PIN proteins, is a central mechanism in plant development (Grieneisen et al. 2007). Within this context, PLT proteins create a functional concentration gradient, reaching the elongation zone (Galinha et al. 2007). Within the basal embryo region, PIN proteins exert control over PLT expression, a regulation vital for initiating root primordium formation. Conversely, PLT genes actively maintain PIN transcription, thereby stabilizing the location of the distal stem cell niche (Blilou et al. 2005). In this study, we observed reduced expression of PLT2 in the brip1 brip2 mutants at both the transcriptional and protein levels, indicating that PLT2 level is regulated by BRIP1 and BRIP2 (Fig. 6a). Furthermore, the similarity in root length between the brip1 brip2 plt1 plt2 mutant and the plt1 plt2 double mutant suggests that BRIP1/2 and PLT1/PLT2 function within a common pathway (Fig. 6d and e). Finally, application of dexamethasone to p35S:PLT2-GR brip1 brip2 seedlings induced significant enlargement of the proximal meristem within a short period, partially rescuing the meristem defects in the brip1 brip2 mutant (Fig. 7). In conclusion, our results demonstrate that BRIP1 and BRIP2 participate in the maintenance of the root meristem by indirectly activating the expression of key genes, such as PLT1/2, involved in this process. However, our data also suggested that other pathways in addition to the PLTs or genes that act downstream of the PLTs may be controlled by BRIP1/2 in the regulation of root development.
Through ChIP-qPCR, we found that BRIP1/2 directly bound to the promoter of SCR, but not to SHR (Fig. 8). The SHR/SCR pathway is a key player in the regulation of root radial patterning and the maintenance of the stem cell niche (Di Laurenzio et al. 1996; Helariutta et al. 2000). However, RT-qPCR results showed that the brip1, brip2 single mutants, and brip1 brip2 double mutants did not show a significant down-regulation of SCR expression (Fig. 8). These findings support the notion that BRIP1/2 may not regulate SCR transcription in roots. Moreover, the lack of direct binding of BRIP1/2 to the SHR promoter does not exclude the possibility that BRIP1/2 might still influence SHR function indirectly. Further exploration could involve looking at how BRIP1/2 mutation or overexpression affect SHR translation, localization, or function, possibly through post-translational modifications or protein–protein interactions. The future investigation of the relationships between BRIP1/2 and the SHR/SCR pathway may further expand our understanding of the complex gene regulatory networks underlying root development.
The overexpression of PLT2-GR in brip1 brip2 mutants successfully restored the meristem but failed to rescue the primary root length. This intriguing observation suggests that while PLT1/2 and BRIP1/2 share a functional pathway, they may also have unique roles in root development. While PLT2 is pivotal in maintaining root meristem, the regulation of primary root growth might be mediated by the interplay of various factors. These include, but are not limited to, other genes in the auxin/PLT pathway, hormonal cross-talk, and environmental interactions, which may be misregulated in brip1/2 double mutants and may not be addressed by the modulation of PLT2 alone. The lack of complementation of primary root growth with exogenous auxin treatment underscores a multifaceted issue. On one hand, the application of exogenous auxin may indeed provide a seemingly adequate supply of the hormone. However, considering the pivotal role of PIN proteins in shaping the maxima and minima of auxin that guide root development, the diminished expression accumulation of PIN proteins, acting as crucial auxin efflux carriers, could potentially hinder the proper intracellular redistribution and maintenance of auxin within the cells. Consequently, even with a seemingly sufficient supply of auxin, the brip1 brip2 mutants might still manifest an inability to re-establish regular auxin gradients and distribution throughout the root tip, thus failing to rescue the short-root phenotype. On the other hand, it is critical to carefully consider the role of downstream auxin responses. The lack of complementation in terms of root length may also be associated with disruptions or alterations in auxin-responsive gene expression, or other auxin-related downstream processes, potentially inherently disordered in the brip1 brip2 mutants. Despite sufficient auxin levels, the dysfunctional or misregulated auxin response pathway might further contribute to the persistence of the observed root development anomalies, suggesting a complex network of regulatory mechanisms that extend beyond just auxin availability.
In conclusion, we demonstrated that BRIP1 and BRIP2 play critical roles in the BAS complexes to directly promote the expression of auxin efflux gene PINs. This leads to the proper transcription and translation of key genes, such as PLTs, during root development, ultimately resulting in normal elongation of the root and proper root development (Fig. 9). Finally, considering the conservation of BRIP1/2 proteins across plant species, our study suggests that BRIPs may play a crucial role in the root development process in other plants as well.
Accession numbers
Sequence data from this article can be found in the Arabidopsis Information Resource (TAIR; https://www.arabidopsis.org) under the following accession numbers: BRIP1 (AT3G03460), BRIP2 (AT5G17510), BRM (AT2G46020), PIN1 (AT1G73590), PIN2 (AT5G57090), PIN3 (AT1G70940), PIN4 (AT2G01420), PIN7 (AT1G23080), PLT1 (AT3G20840), PLT2 (AT1G51190), SCR (AT3G54220), SHR (AT4G37650), and TA3 (AT1G37110).
Data availability
BRIP2 ChIP-seq data were downloaded from GEO under accession no. GSE142369.
Abbreviations
- SWI/SNF:
-
Switch/sucrose non-fermenting
- BRM:
-
BRAHMA
- BRIP1/2:
-
BRM-interacting protein1/2
- QC:
-
Quiescent center
- PLT:
-
PLETHORA
- PIN:
-
PIN-FORMED
- SHR:
-
SHORT-ROOT
- SCR:
-
SCARECROW
- ChIP:
-
Chromatin immunoprecipitation
- GR:
-
Glucocorticoid receptor
- DEX:
-
Dexamethasone
- DAG:
-
Day of germination
References
Adamowski M, Friml J (2015) PIN-dependent auxin transport: action, regulation, and evolution. Plant Cell 27:20–32. https://doi.org/10.1105/tpc.114.134874
Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C et al (2004) The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119:109–120. https://doi.org/10.1016/j.cell.2004.09.018
Band LR, Wells DM, Fozard JA, Ghetiu T, French AP, Pound MP et al (2014) Systems analysis of auxin transport in the Arabidopsis root apex. Plant Cell 26:862–875. https://doi.org/10.1105/tpc.113.119495
Benfey PN, Linstead PJ, Roberts K, Schiefelbein JW, Hauser MT, Aeschbacher RA (1993) Root development in Arabidopsis: four mutants with dramatically altered root morphogenesis. Development 119:57–70. https://doi.org/10.1242/dev.119.1.57
Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115:591–602. https://doi.org/10.1016/S0092-8674(03)00924-3
Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J et al (2005) The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433:39–44. https://doi.org/10.1038/nature03184
Clapier CR, Cairns BR (2009) The biology of chromatin remodeling complexes. Annu Rev Biochem 78:273–304. https://doi.org/10.1146/annurev.biochem.77.062706.153223
Cui H, Levesque MP, Vernoux T, Jung JW, Paquette AJ, Gallagher KL et al (2007) An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science 316:421–425. https://doi.org/10.1126/science.1139531
Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G et al (1996) The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86:423–433. https://doi.org/10.1016/s0092-8674(00)80115-4
Dinneny JR, Benfey PN (2008) Plant stem cell niches: standing the test of time. Cell 132:553–557. https://doi.org/10.1016/j.cell.2008.02.001
Friml J, Benková E, Blilou I, Wisniewska J, Hamann T, Ljung K et al (2002a) AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108:661–673. https://doi.org/10.1016/s0092-8674(02)00656-6
Friml J, Wiśniewska J, Benková E, Mendgen K, Palme K (2002b) Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415:806–809. https://doi.org/10.1038/415806a
Friml J, Benková E, Mayer U, Palme K, Muster G (2003a) Automated whole mount localisation techniques for plant seedlings. Plant J 34:115–124. https://doi.org/10.1046/j.1365-313x.2003.01705.x
Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T et al (2003b) Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426:147–153. https://doi.org/10.1038/nature02085
Fu W, Yu Y, Shu J, Yu Z, Zhong Y, Zhu T et al (2023) Organization, genomic targeting, and assembly of three distinct SWI/SNF chromatin remodeling complexes in Arabidopsis. Plant Cell 35:2464–2483. https://doi.org/10.1093/plcell/koad111
Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R et al (2007) PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449:1053–1057. https://doi.org/10.1038/nature06206
Grieneisen VA, Xu J, Marée AF, Hogeweg P, Scheres B (2007) Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 449:1008–1013. https://doi.org/10.1038/nature06215
Guo J, Cai G, Li YQ, Zhang YX, Su YN, Yuan DY et al (2022) Comprehensive characterization of three classes of Arabidopsis SWI/SNF chromatin remodelling complexes. Nat Plants 8:1423–1439. https://doi.org/10.1038/s41477-022-01282-z
He J, Duan Y, Hua D, Fan G, Wang L, Liu Y et al (2012) DEXH box RNA helicase-mediated mitochondrial reactive oxygen species production in Arabidopsis mediates crosstalk between abscisic acid and auxin signaling. Plant Cell 24:1815–1833. https://doi.org/10.1105/tpc.112.098707
Helariutta Y, Fukaki H, Wysocka-Diller J, Nakajima K, Jung J, Sena G et al (2000) The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101:555–567. https://doi.org/10.1016/s0092-8674(00)80865-x
Ho L, Crabtree GR (2010) Chromatin remodelling during development. Nature 463:474–484. https://doi.org/10.1038/nature08911
Lee Y, Lee WS, Kim SH (2013) Hormonal regulation of stem cell maintenance in roots. J Exp Bot 64:1153–1165. https://doi.org/10.1093/jxb/ers331
Levesque MP, Vernoux T, Busch W, Cui H, Wang JY, Blilou I et al (2006) Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol 4:e143. https://doi.org/10.1371/journal.pbio.0040143
Li C, Gu L, Gao L, Chen C, Wei CQ, Qiu Q et al (2016) Concerted genomic targeting of H3K27 demethylase REF6 and chromatin-remodeling ATPase BRM in Arabidopsis. Nat Genet 48:687–693. https://doi.org/10.1038/ng.3555
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Mähönen AP, Ten Tusscher K, Siligato R, Smetana O, Díaz-Triviño S, Salojärvi J et al (2014) PLETHORA gradient formation mechanism separates auxin responses. Nature 515:125–129. https://doi.org/10.1038/nature13663
Michniewicz M, Brewer PB, Friml J (2007) Polar auxin transport and asymmetric auxin distribution. Arabidopsis Book 5:e0108. https://doi.org/10.1199/tab.0108
Müller A, Guan C, Gälweiler L, Tänzler P, Huijser P, Marchant A et al (1998) AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J 17:6903–6911. https://doi.org/10.1093/emboj/17.23.6903
Nakajima K, Sena G, Nawy T, Benfey PN (2001) Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413:307–311. https://doi.org/10.1038/35095061
Perilli S, Sabatini S (2010) Analysis of root meristem size development. Methods Mol Biol 655:177–187. https://doi.org/10.1007/978-1-60761-765-5_12
Petersson SV, Johansson AI, Kowalczyk M, Makoveychuk A, Wang JY, Moritz T et al (2009) An auxin gradient and maximum in the Arabidopsis root apex shown by high-resolution cell-specific analysis of IAA distribution and synthesis. Plant Cell 21:1659–1668. https://doi.org/10.1105/tpc.109.066480
Petrásek J, Friml J (2009) Auxin transport routes in plant development. Development 136:2675–2688. https://doi.org/10.1242/dev.030353
Petricka JJ, Winter CM, Benfey PN (2012) Control of Arabidopsis root development. Annu Rev Plant Biol 63:563–590. https://doi.org/10.1146/annurev-arplant-042811-105501
Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malamy J et al (1999) An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99:463–472. https://doi.org/10.1016/s0092-8674(00)81535-4
Sabatini S, Heidstra R, Wildwater M, Scheres B (2003) SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev 17:354–358. https://doi.org/10.1101/gad.252503
Saini S, Sharma I, Kaur N, Pati PK (2013) Auxin: a master regulator in plant root development. Plant Cell Rep 32:741–757. https://doi.org/10.1007/s00299-013-1430-5
Santuari L, Sanchez-Perez GF, Luijten M, Rutjens B, Terpstra I, Berke L et al (2016) The PLETHORA gene regulatory network guides growth and cell differentiation in Arabidopsis roots. Plant Cell 28:2937–2951. https://doi.org/10.1105/tpc.16.00656
Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, Nakajima K et al (2007) Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446:811–814. https://doi.org/10.1038/nature05703
Schena M, Lloyd AM, Davis RW (1991) A steroid-inducible gene expression system for plant cells. Proc Natl Acad Sci USA 88:10421–10425. https://doi.org/10.1073/pnas.88.23.10421
Scheres B (2007) Stem-cell niches: nursery rhymes across kingdoms. Nat Rev Mol Cell Biol 8:345–354. https://doi.org/10.1038/nrm2164
Stahl Y, Simon R (2005) Plant stem cell niches. Int J Dev Biol 49:479–489. https://doi.org/10.1387/ijdb.041929ys
Teale WD, Paponov IA, Palme K (2006) Auxin in action: signalling, transport and the control of plant growth and development. Nat Rev Mol Cell Biol 7:847–859. https://doi.org/10.1038/nrm2020
Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9:1963–1971. https://doi.org/10.1105/tpc.9.11.1963
van den Berg C, Willemsen V, Hendriks G, Weisbeek P, Scheres B (1997) Short-range control of cell differentiation in the Arabidopsis root meristem. Nature 390:287–289. https://doi.org/10.1038/36856
Yang L, Zhang J, He J, Qin Y, Hua D, Duan Y et al (2014) ABA-mediated ROS in mitochondria regulate root meristem activity by controlling PLETHORA expression in Arabidopsis. PLoS Genet 10:e1004791. https://doi.org/10.1371/journal.pgen.1004791
Yang S, Li C, Zhao L, Gao S, Lu J, Zhao M et al (2015) The Arabidopsis SWI2/SNF2 chromatin remodeling ATPase BRAHMA targets directly to PINs and is required for root stem cell niche maintenance. Plant Cell 27:1670–1680. https://doi.org/10.1105/tpc.15.00091
Yu Y, Liang Z, Song X, Fu W, Xu J, Lei Y et al (2020) BRAHMA-interacting proteins BRIP1 and BRIP2 are core subunits of Arabidopsis SWI/SNF complexes. Nat Plants 6:996–1007. https://doi.org/10.1038/s41477-020-0734-z
Yu Y, Fu W, Xu J, Lei Y, Song X, Liang Z et al (2021) Bromodomain-containing proteins BRD1, BRD2, and BRD13 are core subunits of SWI/SNF complexes and vital for their genomic targeting in Arabidopsis. Mol Plant 14:888–904. https://doi.org/10.1016/j.molp.2021.03.018
Zazímalová E, Krecek P, Skůpa P, Hoyerová K, Petrásek J (2007) Polar transport of the plant hormone auxin—the role of PIN-FORMED (PIN) proteins. Cell Mol Life Sci 64:1621–1637. https://doi.org/10.1007/s00018-007-6566-4
Acknowledgements
We thank the Arabidopsis Biological Resource Center (ABRC) for seeds of T-DNA insertion lines, Songguang Yang (Guangdong Academy of Agricultural Sciences) for providing p35S:PLT2-GR, pPINs:PINs-GFP, and p35S:PLT2-GR seeds. This work was supported by the National Natural Science Foundation of China to C.L. (32070212, 32270322, and 31870289).
Author information
Authors and Affiliations
Contributions
CL conceived the project. XS performed most of the experiments. XS, YY, and JZ analyzed data. CL wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of financial interest.
Additional information
Communicated by Dorothea Bartels.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplemental Fig. S1: a
The arrangement of QC in the WT, brip1, brip2, and brip1 brip2 root tips by PI staining at 5 DAG. The white arrow points to the position of QC. Scale bar = 50 μm. b The percentage of QC irregularly arranged in the WT, brip1, brip2, and brip1 brip2 root tips. At least 50 root tips are viewed under a microscope for each background (PPTX 410 KB)
Supplemental Fig. S2:
Root phenotype analysis in plants expressing GFP-tagged BRIP1 or BRIP2 in corresponding single/double mutant backgrounds. a The root phenotypes of the WT, brip1, brip2, brip1 brip2, pBRIP1:BRIP1-GFP brip1, pBRIP2:BRIP2-GFP brip2, pBRIP1:BRIP1-GFP brip1 brip2, and pBRIP2:BRIP2-GFP brip1 brip2 seedlings on 1/2 MS medium at 7 DAG. Scale bar = 1 cm. b Statistical result for primary root length of the WT, brip1, brip2, brip1 brip2, pBRIP1:BRIP1-GFP brip1, pBRIP2:BRIP2-GFP brip2, pBRIP1:BRIP1-GFP brip1 brip2, and pBRIP2:BRIP2-GFP brip1 brip2. The data represent mean ± SD (n = 10 plants). Lower-case letters indicate significant differences, as determined by two-way ANOVA with Tukey’s multiple comparisons test (PPTX 546 KB)
Supplemental Fig. S3:
Exogenous application of auxin fails to rescue the short-root phenotype of brip1 brip2. a The root phenotypes of the WT, brip1, brip2, and brip1 brip2 seedlings on 1/2 MS medium or 1/2 MS medium supplemented with 0.02 μM IAA, 0.05 μM IAA, 0.1 μM IAA, and 0.2 μM IAA at 5 DAG. Scale bar = 1 cm. b Statistical result for primary root length of the WT, brip1, brip2, and brip1 brip2 seedlings on 1/2 MS medium or 1/2 MS medium supplemented with 0.02 μM IAA. The data represent mean ± SD (n = 10 plants). NS, not significant as determined by Student’s t test (PPTX 599 KB)
Supplemental Fig. S4:
Exogenous application of auxin partially rescues the root meristem of brip1 brip2. a The root meristem of the WT and brip1 brip2 seedlings on 1/2 MS medium or 1/2 MS medium supplemented with 0.02 μM IAA at 5 DAG. Scale bar = 50 μm. b Statistical result for root meristem size of the WT and brip1 brip2 seedlings on 1/2 MS medium or 1/2 MS medium supplemented with 0.02 μM IAA. The data represent mean ± SD (n ≥ 10 plants). *P < 0.05; NS, not significant as determined by Student’s t test (PPTX 637 KB)
Supplemental Fig. S5:
BRIP2 does not target to PLT1 and PLT2 directly. a, b ChIP-qPCR analysis of the enrichment of BRIP2 to the different regions of PLT1and PLT2 loci in WT and pBRIP2:BRIP2-GFP Arabidopsis roots. The top of the diagram shows a schematic of the detected genes in the genome. The black thick frame, the black thin frame and the black line represent the exon, UTR, intron and promoter of genes, respectively. P/C represents the primer designed for the detected regions. *P < 0.05; NS, not significant as determined by Student’s t test. (PPTX 85 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Song, X., Yu, Y., Zhu, J. et al. BRIP1 and BRIP2 maintain root meristem by affecting auxin-mediated regulation. Planta 259, 8 (2024). https://doi.org/10.1007/s00425-023-04283-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-023-04283-0