Abstract
Main conclusion
Lysin motif (LysM)-receptor-like kinase (RLK) and leucine-rich repeat (LRR)-RLK mediated signaling play important roles in the development and regulation of root nodule symbiosis in legumes.
Abstract
The availability of water and nutrients in the soil is a major limiting factor affecting crop productivity. Plants of the Leguminosae family form a symbiotic association with nitrogen-fixing Gram-negative soil bacteria, rhizobia for nitrogen fixation. This symbiotic relationship between legumes and rhizobia depends on the signal exchange between them. Plant receptor-like kinases (RLKs) containing lysin motif (LysM) and/or leucine-rich repeat (LRR) play an important role in the perception of chemical signals from rhizobia for initiation and establishment of root nodule symbiosis (RNS) that results in nitrogen fixation. This review highlights the diverse aspects of LysM-RLK and LRR receptors including their specificity, functions, interacting partners, regulation, and associated signaling in RNS. The activation of LysM-RLKs and LRR-RLKs is important for ensuring the successful interaction between legume roots and rhizobia. The intracellular regions of the receptors enable additional layers of signaling that help in the transduction of signals intracellularly. Additionally, symbiosis receptor-like kinase (SYMRK) containing the LRR motif acts as a co-receptor with Nod factors receptors (LysM-RLK). Cleavage of the malectin-like domain from the SYMRK ectodomain is a mechanism for controlling SYMRK stability. Overall, this review has discussed different aspects of legume receptors that are critical to the perception of signals from rhizobia and their subsequent role in creating the mutualistic relationship necessary for nitrogen fixation. Additionally, it has been discussed how crucial it is to extrapolate the knowledge gained from model legumes to crop legumes such as chickpea and common bean to better understand the mechanism underlying nodule formation in crop legumes. Future directions have also been proposed in this regard.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Crop production is influenced by the availability of water and nutrients in the soil. Therefore, cultivated soil needs a continuous supply of nutrients. Nitrogen fertilizers have been used as a source of nutrients and have significantly contributed to the Green Revolution. However, their continuous use is costly and detrimental to soil health (Schroeder et al. 2013). The symbiotic relationship between fungi and bacteria helps plants absorb nutrients. Plants of the Leguminosae family form a mutualistic interaction with nitrogen-fixing Gram-negative soil bacteria called rhizobia. These rhizobia convert inert atmospheric dinitrogen into ammonia. In return, the rhizobia receive photosynthate-derived carbon from plants for their survival. This symbiotic relationship enhances legume production and subsequent cereal production by improving soil fertility. In addition, this symbiosis can significantly reduce global warming, the use of chemical fertilizers, and water contamination (Ouma et al. 2016).
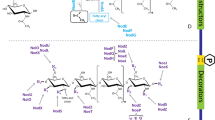
Establishing a successful symbiotic relationship requires the exchange of chemical signals between the legume and its partner rhizobia. Legumes exude flavonoids and isoflavonoids into the rhizosphere, which may either directly associate with the rhizobial NodD protein or induce nod gene expression in the rhizobia (Göttfert 1993; Kelly et al. 2018). The expression of rhizobial nod genes triggers the synthesis of lipo-chitooligosaccharides (LCO) known as Nod factors (NFs) (Göttfert 1993; Harris et al. 2018). Further, the lysin motif (LysM)-type receptors, in co-ordination with leucine-rich repeat (LRR)-type receptors, perceive rhizobial NFs, trigger infection thread formation, and further initiate nodule formation (Oldroyd et al. 2011; Harris et al. 2018). This review explores the contribution of LRR-receptor-like kinases (RLKs) and LysM-RLKs, and their molecular structure, functions, and regulation via interactions with different downstream signaling molecules, for the establishment of root nodule symbiosis (RNS) (Fig. 1).
Different steps involved in Nod factors (NF) signaling in the legume–rhizobial interactions for root nodule symbiosis. (1) Legume roots secrete flavonoids in nitrogen-deficient soil. Flavonoids are sensed by rhizobia, and rhizobia release NF. (2) NF induces curling, and rhizobia get entrapped inside the curled structure. As infection thread formation starts, inner cortical cells begin to divide. (3) Infection thread progresses into inner cortical cells. (4) Rhizobia reach the nodule primordial and bacteroid differentiation begins. (5) Nodule matures and fixes nitrogen
Role of Nod factors (NFs) receptors in nodule symbiosis
The perception of rhizobial NFs by legume roots is the first important step in nodule formation for nitrogen fixation (Oldroyd et al. 2011). In the model legumes, Lotus japonicus and Medicago truncatula, the plasma membrane-specific receptors, NF RECEPTOR 1 (LjNFR1), LjNFR5, LysM-RLK 3 (MtLYK3), and NF PERCEPTION (MtNFP) perceive the NFs (Arrighi et al. 2006; Radutoiu et al. 2003; Smit et al. 2007; Madsen et al. 2003; Limpens et al. 2003). In addition, the L. japonicus epidermal LysM receptor (NFRe) has also been reported to be involved during the infection process by amplifying signals in root epidermal cells (Murakami et al. 2018). LjNFR1/MtLYK3, LjNFR5/MtNFP, and LjNFRe contain the LysM domain in the extracellular region of the protein, and hence they are known as LysM-RLKs. The LysM-RLKs contain an intracellular cytoplasmic kinase domain, extracellular LysM, and a single-pass transmembrane domain that joins the ectodomain and cytosolic domain. NFR5 lacks a functional kinase domain, and hence it is termed a pseudo-kinase. In contrast, NFR1 comprises a functional cytoplasmic kinase domain that is required for NF signaling. The NFR1 cytoplasmic kinase domain is capable of auto-phosphorylation as well as trans-phosphorylation. During trans-phosphorylation, the NFR1 kinase domain phosphorylates the cytoplasmic pseudo-kinase domain of NFR5 (Madsen et al. 2011). Furthermore, the results indicated that the kinase domains of different RLKs are important for the initiation of downstream signaling through a series of trans-phosphorylation events (Oldroyd et al. 2011). Plant-based expression systems and insect cell expression system were used to express and purify NFR5, NFR1, and NFRe that were further used to show the binding affinity of the ectodomain of NFR5, NFR1, and NFRe with NFs in the nanomolar range (Broghammer et al. 2012; Murakami et al. 2018).
The formation of a functional root nodule is the result of two coordinated programs: bacterial infection of the root epidermis, followed by nodule primordia inception in the root cortex (Gamas et al. 2017). In the epidermis, activation of this signaling pathway leads to localized growth inhibition at the tip of root hairs, inducing their curling (Gage 2004). Rhizobia entrapped in the curl enter the root hair by local hydrolysis of cell wall and invagination of the plasma membrane. Growing the tip toward the base of the root hair results in an intracellular tube called the infection thread. NF-induced pectate lyase is required for the localized degradation of the plant cell wall at the site of infection thread initiation (Xie et al. 2012). The infection thread then grows and branches through the cortex. These processes depend on plant cell wall degradation, synthesis of infection thread wall components, and cytoskeletal rearrangements forming the preinfection structure (Monahan-Giovanelli et al. 2006). Simultaneously with the rhizobial infection, cortical cells reinitiate meristematic activity to form the nodule primordia. Activation of such a program is mediated by a mobile unidentified signal (possibly cytokinins or, less likely, NSP1 and NSP2) originating in the epidermis, which triggers a cortical signaling cascade (Frugier et al. 2008; Hayashi et al. 2014). A role in coordinating epidermal and cortical responses has also been attributed to nodule inception (NIN). This protein displays a complex and tissue-specific role during nodulation. It activates bacterial infection in the epidermis and nodule organogenesis in the cortex while negatively regulating the number of nodules that are formed (Vernié et al. 2015).
Concurrently with the rhizobial infection, the fate of differentiated cortical root cells changes, and they revive their meristematic activity. Activated cells start dividing faster and a tumor-like nodule primordium forms. Furthermore, in response to rhizobial infection, the nuclei of the cortical cells re-localize themselves to the center of the cell and cytoplasmic bridges form between adjacent cortical cells to guide the infection thread. The formation of these cytosolic bridges in the cortex is also known as preinfection threads, which is a preparatory step for cell division (Van Brussel et al. 1992). The nodule primordium subsequently develops into a nodule by facilitating the invasion of rhizobia. Both NF receptors, LjNFR1/MtLYK3 and LjNFR5/NFP, are required to develop the nodule primordium.
The final stage of infection involves the release of rhizobia into the newly divided cortical cells. Both NF receptors control the release of rhizobia into the cytoplasm of the cells, which leads to nodule differentiation and the induction of endoreduplication in the nodules. As a result, the infected cells get enlarged. The role of NFP in the release of rhizobia and endoreduplication has been demonstrated by utilizing an NFP knockdown mutant. The NFP mutants are characterized by impaired release of rhizobia from infection threads (Bensmihen et al. 2011). The cells devoid of the infection thread remained small due to the abortion of endoreduplication (Moling et al. 2014).
Further, bacterial differentiation inside the nodule depends upon the secretion of nodule-specific cysteine-rich (NCR) peptides, which are processed by defective in nitrogen fixation 1 (DNF1), a predicted module of the nodule-specific signal peptidase complex (Wang et al. 2010). The NCR peptides are a characteristic feature of legumes belonging to the inverted repeat-lacking clade (IRLC). IRLC members include the Cicer arietinum, Medicago spp., Pisum sativum, Vicia spp., and Trifolium repens. However, the NCR peptides are not found in non-IRLC legumes such as L. japonicus, Phaseolus vulgaris, and Glycine max. Legumes of the IRLC clade form “indeterminate” nodules that are characterized by persistent nodule apical meristems, which result in elongated or multi-lobed shapes. Non-IRLC legumes form “determinate” nodules that are characterized by transiently active apical meristems, which result in a spherical shape. In indeterminate nodules, rhizobia undergo terminal differentiation and are not able to revert to the free-living form (Singh and Valdés-López 2022a). Different developmental zones in determinate nodules are not easily differentiated; rhizobia-infected cells are intermixed with non-infected cells. No terminal differentiation has been observed for rhizobia inside determinate nodules. In comparison to indeterminate nodules, rhizobia inside determinate nodules maintain standard genome content, average bacterial size, and reproductive capacity (Brewin 1991; Mergaert et al. 2006). Downstream of NF signaling, calcium spiking occurs, which leads to the activation of a calcium/calmodulin-dependent protein kinase (CCaMK) (Lévy et al. 2004). The activated CCaMK interacts with and phosphorylates the coiled-coil protein, CYCLOPS, which is a transcription factor (Singh et al. 2014). This phosphorylation leads to the stimulation of a series of transcription factors for symbiosis. A constitutively active form of CCaMK results in spontaneous nodule formation (Takeda et al. 2012).
Structural characterization of NF receptor binding sites in legumes is essential as it provides a basis for understanding the evolution of the specificity of NF recognition. Furthermore, the characterized structures may be used as a blueprint for engineering NF receptors that can be used to improve the nodulation trait or even to transfer the trait to non-legume crops. Such engineering would enable non-legume crops to be grown without nitrogen fertilizer.
Role of LysM1 domain in discriminating between symbiotic and pathogenic ligands
In nature, plants live and interact with diverse microbes, including symbionts and pathogens and plant adaptation relies on an appropriate response to symbiotic and pathogenic microbes. Recently, LysM1 of LysM-RLK was reported to be involved in recognition of symbiotic partners and the discrimination of pathogenic microbes (Bozsoki et al. 2020). The researchers selected two receptors, NFR1 (specific for symbiosis) and CERK6 (specific for immunity), and created chimeras in different combinations to identify determinants specific for Mesorhizobium loti (symbiotic partner of Lotus) and chitooligosaccharide 8 (CO8) for activation of NF signaling and immune signaling. All chimeras containing LysM1 domain of NFR1 restored nodulation in the nfr1 background, while chimeras comprising LysM1 of CERK6 failed to do so. Further, a mutation in LysM1 that converted isoleucine, a conserved amino acid residue, to tryptophan, a bulky residue, disrupted the potential binding pocket. This mutation in LysM1 resulted in impaired symbiosis. These results are in line with previous observations of the pea Sym37 mutant and Medicago Lyk3, where a mutation in the LysM1 of RLK resulted in defective symbiosis. Bozsoki et al. (2020) identified four structural regions in LysM1 and labeled them I, II, III, and IV regions based on sequence differences between NFR1 and CERK6 and tested whether they were required for NF recognition (Bozsoki et al. 2020). It was found that swapping regions I and III between NFR1 and CERK6 produced no detrimental effect on nodulation. However, swapping regions II and IV resulted in a “no nodule” phenotype. Further, NFR1 with regions II and IV from CERK6 showed no binding to Mesorhizobium loti NF. These results indicated that two motifs (regions II and IV) in LysM1 domains in NFR1 are ligand-binding sites and provide specificity for recognizing symbiotic ligands. These motifs constitute the most structurally divergent regions in NF receptors. Further, embedding regions II and IV of LYK3 into NFR1 abolished their capacity to induce nodulation in nfr1 by M. loti. In the same way, in Mtlyk3, where II and IV of NFR1 were embedded into LYK3, revealed a similar lack of nodulation in by S. meliloti. Together, these results provide support for the presence of molecular determinants for Nod factor signaling specificity in LysM1 of NFR1 and LYK3.
Interacting partners of NF receptors
To enable successful symbiosis, NF receptors interact with downstream signaling components. NF perception leads to a change in the stability and localization of LysM-RLKs through the compartmentalization of the receptors into nanodomains or microdomains at the plasma membrane. This compartmentalization, partitions RLKs into functionally well-defined signaling units (Ott 2017). Before exposure to NF or rhizobia, MtLYK3 shows high lateral motion at the plasma membrane. Rhizobial NF application leads to the immobilization of MtLYK3 in nanodomains along with MtFLOTILLIN4 (MtFLOT4), which is a scaffolding protein required for spatial regulation of signaling complexes during these symbiotic interactions (Haney et al. 2011). Symbiosis-specific remorin from Medicago (MtSYMREM1) is another scaffold protein that mediates the stabilization and immobilization of MtLYK3. MtSYMREM1 interacts with MtLYK3, MtNFP, and does not make infections 2 (MtDMI2) (Tóth et al. 2012; Liang et al. 2018). PLANT U-BOX PROTEIN1 (PUB1) was identified as an interacting protein of the MtLYK3 kinase domains by using the kinase domain of MtLYK3 as bait in yeast two-hybrid (Y2H) cDNA library screenings (Mbengue et al. 2010). It has been reported that PUB1 interacts with, and gets phosphorylated by, MtLYK3. The RNA interference (RNAi) knockdown of MtPUB1 has been shown to result in an increased number of nodules and infection threads. Similarly, overexpression of MtPUB1 delayed nodulation. Therefore, the overexpression and RNAi phenotypes indicated that MtPUB1 negatively regulates rhizobial infection (Mbengue et al. 2010). PUB1 interacts with the receptor kinase DMI2 and negatively regulates rhizobial and arbuscular mycorrhizal symbioses in M. truncatula via its ubiquitination activity (Vernié et al. 2016). The second interacting partner of LjNFR5 kinase domain identified in L. japonicus was Rho-like small GTPase 6 (LjROP6) (Ke et al. 2012). The down-regulation of LjROP6 in transgenic hairy root cells inhibits infection thread growth through the cortical region of the root, resulting in fewer nodules. Thus, LjROP6 seems to positively regulate nodulation and formation of infection thread (Ke et al. 2012). Recently, by utilizing a proteomics approach, Wong et al. (2019) identified NFR5-interacting cytoplasmic kinase 4 (NiCK4) as an interacting partner of LjNFR5. It has been shown that NiCK4 phosphorylates NFR5/NFR1 kinase domains in vitro. The NFR5 and NiCK4 proteins are expressed in identical cellular region of root hair and nodule cells. Functional characterization using retrotransposon insertion mutants (Tnt1) revealed that NiCK4 has a role in nodule organogenesis. Interestingly, it has been seen that the NiCK4 protein relocates to the nucleus in response to NF treatment in an NFR1/NFR5-dependent manner. This indicates that NiCK4 is a missing link between NF perception and nodule organogenesis (Wong et al. 2019).
Exopolysaccharide receptor 3 (EPR3)
In legumes, rhizobial surface-exposed carbohydrates such as lipo- and exo-polysaccharides (EPS) are used as determinants to either block or promote symbiosis with rhizobia depending on their molecular composition. In L. japonicus, exopolysaccharide receptor 3 (EPR3) have been shown to be involved in EPS recognition. EPR3 binds EPS directly and distinguishes compatible and incompatible bacterial EPS. This protein is an LysM-RLK belonging to the NFR1 protein phylogenetic branch, and its expression in root hairs and epidermal cells is induced by NFs. EPR3 contains an intracellular kinase domain predicted to be active. EPR3 is likely assisted by a co-receptor containing an inactive pseudokinase domain to recognize EPS or other microbial surface carbohydrates, possibly for monitoring associated microbiota (Wong et al. 2020).
Plant sensing of bacterial EPS by EPR3 occurs at the stage of colonization and infection of epidermal cells. Using bacterial mutants affected in EPS production, it was observed that the EPR3 receptor controls the infection, irrespective of the way the L. japonicus microsymbiont invades epidermal cells (Kawaharada et al. 2015). Ortholog genes are present in other legumes and non-legume species, suggesting that the mechanism for bacterial EPS recognition is widespread among plants. EPR3 advances the intracellular infection mechanism that mediates infection thread invasion of the root cortex and nodule primordia. At the cellular level, Epr3 expression delineates progression of infection threads into nodule primordia, and cortical infection thread formation is impaired in epr3 mutants. Genetic dissection of this developmental co-ordination showed that Epr3 is integrated into the symbiosis signal transduction pathways. Further analysis showed differential expression of Epr3 in the epidermis and cortical primordia and identified key transcription factors controlling this tissue specificity (Kawaharada et al. 2017b). These results suggest that exopolysaccharide recognition is reiterated during the progressing infection and that EPR3 perception of compatible exopolysaccharide promotes an intracellular cortical infection mechanism maintaining bacteria enclosed in plant membranes.
To establish a deeper understanding of EPR3 involved in EPS recognition, Wong et al. (2020) determined the structure of the L. japonicus EPR3 ectodomain. EPR3 forms a compact structure built of three putative carbohydrate-binding modules (M1, M2, and LysM3). Unique βαββ and βαβ folds present in M1 and M2 motifs have been shown to be important for carbohydrate binding. Altogether, EPR3 is a defining member of a large and conserved unique class of plant receptors capable of directly perceiving EPS from different bacterial species (Wong et al. 2020). Future studies in different plant species will help us to better understand this class of receptor and its downstream signaling mechanisms.
SYMRK: a co-receptor with NF receptors
SYMRK is an indispensable central component for development of N2-fixing RNS and phosphate-acquiring arbuscular mycorrhizal (AM) symbiosis (Ried et al. 2014). SYMRK is positioned immediately downstream or at the same hierarchical level as NF receptors. In M. truncatula, SYMRK is known as DMI2. The DMI2/SYMRK protein acts as a co-receptor of rhizobial signals to initiate a rhizobial infection, and nodule development, for the establishment of a nitrogen-fixing symbiosis (Gherbi et al. 2008). DMI2/SYMRK was identified by map-based cloning as one of the genetic determinants of the non-nodulation traits of L. japonicus and Medicago sativa mutants and, by genetic analogy, was determined to be important for the related legumes, M. truncatula and P. sativum (Endre et al. 2002). Further studies revealed that SYMRK harbors an intracellular cytoplasmic kinase domain, a transmembrane domain, and the extracellular ectodomain with one LRR and malectin-like domain (MLD) (Markmann et al. 2008). Each structural component of SYMRK is further discussed below (Fig. 2A).
A Functional domains of SYMRK. SYMRK contains a signal peptide (SP), a malectin-like domain (MLD), a tetrapeptide Gly-Asp-Pro-Cys motif (GDPC), three Leu-reach repeats (LRR), a transmembrane domain (TM), and kinase domain (KD). B SYMRK regulation via cleavage of the MLD domain. MLD domain in the ectodomain in SYMRK is connected to LRR through the conserved GDPC motif. The MLD domain undergoes proteolytic cleavage at the GDPC motif. The resulting membrane-bound SYMRK version without the MLD domain containing the LRR is referred to as SYMRK-ΔMLD. SYMRK-ΔMLD strongly and specifically associates with NFR5, and both kinase domain and the LRR are found to be responsible for this association
Structural features of SYMRK
MLD
MLD gene families are greatly expanded in the plant kingdom, although knowledge of the exact function of MLDs in plants is still elusive (Pan et al. 2018). MLDs are generally present in the ectodomain of RLKs, which suggests that they play a role in the binding of extracellular ligands for the activation or deactivation of the intracellular kinase domain (Schallus et al. 2008).
Interestingly, it has been shown in L. japonicus that the ectodomain of SYMRK undergoes constitutive proteolytic cleavage to release MLD, which is independent of the presence or absence of rhizobia. Surprisingly, MLD-cleaved SYMRK protein outcompetes full-length SYMRK for its interaction with the NF receptor, NFR5. This suggests that MLD has a negative effect on complex formation with NF receptors (Antolín-Llovera et al. 2014). It was observed that when conserved amino acids of the SYMRK MLD were mutated, mutant SYMRK failed to restore nodule development in dmi2 mutants (Pan et al. 2018). Additionally, point mutations in MLDs have been shown to result in the constitutive degradation of SYMRK. These two analyses suggest that MLDs play a vital role in accumulation of DMI2 (Kosuta et al. 2011). In this way, legumes modulate protein level of SYMRK during establishment of nitrogen-fixing symbiosis, controlling nodule development. The GDPC motif present between MLD and the LRR regulates the abundance of the SYMRK protein in L. japonicus. Antolín-Llovera et al. (2014) revealed that GDPC is critical for MLD cleavage (MLD release) in planta, which is vital for successful symbiotic interactions. In transformed N. benthamiana leaf cells that were transiently expressing LjSYMRK, cleavage of the ectodomain to release the MLD was observed. This demonstrates that this cleavage is independent of rhizobial inoculation. SYMRK cleavage was thus postulated either to be mediated by proteases present in both N. benthamiana leaves and L. japonicus roots or to be an inherent property of the ectodomain. Even under the control of a strong promoter, the gene encoding the truncated SYMRK protein without MLD (SYMRK-ΔMLD) synthesized an insufficient amount of protein in N. benthamiana or L. japonicus. Therefore, it is evident that after MLD release, the remaining SYMRK-ΔMLD protein undergoes rapid degradation to maintain an optimum concentration of SYMRK for full symbiotic function (Antolín-Llovera et al. 2014). The GDPC was found to be an important feature of almost one-quarter of the predicted LRR-RLKs in Arabidopsis thaliana. This indicates that GDPC sequence plays an essential role in non-symbiotic function (Fig. 2B).
On the other hand, the symrk-14 har1-1 mutant, which showed an interesting symbiotic phenotype, was characterized by substitution from Pro to Lys in the conserved Gly-Asp-Pro-Cys (GDPC) domain in the extracellular domain upstream of first LRR in SYMRK protein. This symrk-14 har1-1 mutant showed few wild-type nodules along with many nodule primordia and/or small uncolonized nodules. Furthermore, symrk-14 mutant was defective in its epidermal responses to bacterial signaling in the form of calcium spiking, NIN gene expression, and infection thread formation (Kosuta et al. 2011).
LRR
LRR are present in several proteins and have diverse functions. The length of an LRR motif can range from 22 to 28 amino acids (Kobe and Deisenhofer 1994). When LRR are present in series, they form non-globular, crescent-shaped structures. The crescent shape is created through a solvent-exposed extended concave surface of parallel beta-strands that acts as a scaffold for protein–protein, and protein–ligand interactions. Each LRR crescent structure performs a specific function. RLKs containing the LRR motif represent most extensive group of receptor kinases in plants and are implicated in diverse plant signaling pathways (Chakraborty et al. 2019). SYMRK has been demonstrated to associate with both NFR1 and NFR5 in both N. benthamiana and L. japonicus independent of rhizobial inoculation (Antolín-Llovera et al. 2014; Ried et al. 2014). It remains to be explored whether the LRR of SYMRK is essential for this communication, leading to the assembly of the receptor complexes, or whether they have a more direct role via interactions with a signaling ligand.
The intracellular kinase domain of SYMRK
Biochemical analyses of the cytosolic domain of LjSYMRK revealed that it acts as a functional kinase domain with intermolecular auto-phosphorylation activity (Yoshida and Parniske 2005). Mutations in conserved amino acids of kinase domain have been shown to result in defective symbiosis in L. japonicus and P. sativum. This indicates that SYMRK kinase activity is essential for symbiosis. It has also been reported that non-phosphorylated SYMRK is less active than the phosphorylated form, which suggests that the phosphorylation status plays a vital role in SYMRK activity (Saha et al. 2016). Targeted mutagenesis revealed that three Ser/Thr residues are essential for full kinase activity. Quadrupole time-of-flight mass spectrometry analysis revealed that Ser-754 and Thr-760, which are located in the activation loop of the kinase domain, are phosphorylated, whereas Thr-593, which is located in the juxtamembrane region, remains non-phosphorylated. In SYMRK loss-of-function mutants, signaling was arrested at the very initial stages of nodule development. For all tested mutants of LjSYMRK, root hairs failed to form a curled structure upon inoculation with a rhizobial partner for the entrapment of bacteria in infection pockets (Li et al. 2018). Hence, no infection threads were observed and, in turn, the root hair cells responded with exaggerated swelling and branching, indicating that LjSYMRK was required for infection thread initiation (Stracke et al. 2002). The complete abortion of infection thread formation associated with SYMRK null mutants makes it difficult to evaluate the function of SYMRK at later stages of the symbiosis. RNAi generally results in the down-regulation of the targeted gene. Hence, RNAi-down-regulated plants were found to be suitable for assessing different weak phenotypes of SYMRK and its various orthologs in the later stages of nodule development. This technique has been used for legumes such as M. truncatula, Sesbania rostrata, P. vulgaris, and G. max, and actinorhizal plants such as Casuarina glauca and Datisca glomerata (Capoen et al. 2005; Limpens et al. 2005; Gherbi et al. 2008; Markmann et al. 2008; Sánchez-López et al. 2011; Indrasumunar et al. 2015). The GmSYMRK RNAi plant roots exhibit diminished nodule numbers and multiple other nodulation defects, ranging from highly branched infection threads to abnormal cell wall depositions. Further, infection threads with numerous bacteria embedded in a matrix bound by a cell wall and a membrane, zones of differentiated cells in nodules devoid of rhizobia, and impaired symbiosome formation have also been observed (Capoen et al. 2005; Limpens et al. 2005). The analysis of the functions of SYMRK orthologs from C. glauca and D. glomerata using transgenic (RNAi) roots of the respective plants showed a ubiquitous reduction in nodule numbers. Additionally, rhizobia-inoculated CgSYMRK transgenic (RNAi) roots developed pseudo-nodules concomitant with the accumulation of phenolic compounds (Gherbi et al. 2008). Thus, it can be concluded that SYMRK plays a crucial role in NF-induced signal transduction that leads to rhizobial infection of the root epidermis and nodule organogenesis.
Interesting observations were made during a genetic screening of suppressors of the L. japonicus har1-1 hypernodulation phenotype. Two recessive SYMRK mutant alleles, symrk-13 and symrk-14, were identified. However, both conferred different nodulation phenotypes (Kosuta et al. 2011). The symrk-13 mutation resulted in an A to T substitution in predicted ATP binding site of protein kinase domain. This resulted in a complete block of nodulation in the symrk-13 har1-1 double mutant.
Interacting partners of SYMRK
Besides regulation through the GDPC, different proteins interact with SYMRK and regulate it at post-translational levels. Several interacting partners have been identified for SYMRK via Y2H-based screening, using the kinase domain of SYMRK as bait. These identified interacting partners of SYMRK have proven to be essential for an improved understanding of the molecular and biochemical functions of SYMRK in symbiotic signaling. Some of these interacting partners of SYMRK regulate RNS positively, while others regulate RNS negatively. The 3-hydroxy-3-methylglutaryl coenzyme A reductase 1 (HMGR1) protein was one of the first proteins identified as an SYMRK-interacting protein in M. truncatula (Kevei et al. 2007). HMGR1 catalyzes a rate-limiting step in the production of isoprenoid compounds via mevalonate pathway. Down-regulation of HMGR1 results in a substantial reduction of nodulation, indicating its positive role in RNS (Kevei et al. 2007). SYMRK-interacting protein 1 (SIP1), a DNA-binding protein that contains a conserved AT-rich interaction domain (ARID), was identified as a second novel SYMRK interactor in L. japonicus. SIP1 binds to the LjNIN promoter to regulate NIN expression for the establishment of early dialogue between the rhizobia and host root cells (Zhu et al. 2008). A Y2H library screen using the SYMRK kinase domain identified a mitogen-activated protein kinase kinase (MAPKK) protein termed SYMRK-interacting protein 2 (SIP2). This interaction was found to be conserved at least between L. japonicus and M. sativa. Down-regulation of SIP2 expression via RNAi has been shown to result in a drastic reduction in nodule formation. This result indicates the positive role of SIP2 in signaling and nodule organogenesis during the initial stages of symbiosis (Chen et al. 2012). SYMRK-interacting E3 ubiquitin ligase (SIE3) interacts with and ubiquitinates SYMRK in planta and in vitro. A significant reduction in infection thread formation and nodule organogenesis was observed in roots with RNAi-down-regulated SIE3. The overexpression of SIE3 increased nodulation in transgenic hairy roots. These results suggest that SIE3 acts as a positive regulator of SYMRK for rhizobial infection and nodulation in L. japonicus (Yuan et al. 2012). The E3 ubiquitin ligase family protein, seven in absentia (SINA4), interacts with SYMRK in N. benthamiana leaves in small punctae at the cytosolic interface of the plasma membrane. The co-localization study of fluorescence-tagged SYMRK and SINA4 showed that SINA4 lead to the disappearance of SYMRK from the plasma membrane. This indicates that SYMRK abundance is negatively regulated by SINA4, which consequently modulates symbiosis signaling. Therefore, it can be concluded that SINA4 is a negative regulator of SYMRK (den Herder et al. 2012) (Fig. 3).
Nod factors are perceived at the plasma membrane by receptor-like kinase proteins (NFR1, NFR5, and SYMRK). Receptors at the plasma membrane interact with different proteins in the cytosol through the kinase domain and form a complex for the transduction of signaling to downstream components. SYMRK interacts with symbiosis-specific remorin1 (SYMREM1), 3-hydroxy-3-methylglutaryl coenzyme reductase1 (HMGR1), SYMRK-interacting protein 1(SIP1), SYMRK-interacting protein 2(SIP2), SEVEN IN ABSENTIA 4(SINA4), and SYMRK-interacting E3 ubiquitin ligase (SIE3). NFR5 interacts with symbiosis-specific remorin (SYMREM1) and NFR5-interacting cytoplasmic kinase 4 (NiCK4). NFR1 interacts with symbiosis-specific remorin (SYMREM1), Rho-like small GTPase 6 (ROP6), U-box-dependent E3 ubiquitin ligase (PUB1), and FLOTILLIN4 (FLOT4). Rhythmic calcium spiking, especially around and possibly within the nucleus, may activate a nuclear calcium/calmodulin-dependent protein kinase (CCaMK), which further activates the GRAS-type transcriptional regulators NSP1 and NSP2. NSP1, NSP2 along with DELLA and ERN1 induce the transcription of early nodulation genes, leading to root hair deformation and formation of nodules. CYC-box, cis-regulatory element recognized by the transcription factor CYCLOPS; NRE-box, cis-regulatory element recognized by the transcription factor NSP1
Regulation of SYMRK/DMI2
In legumes, an abundance of the SYMRK/DMI2 protein is regulated to ensure proper response toward rhizobia. In rhizobia-free environments, SYMRK/DMI2 protein is degraded by proteasomal degradation pathways. Degradation of SYMRK/DMI2 via the proteasome became more evident when a proteasomal inhibitor was shown to result in DMI2 protein accumulation in Medicago roots (Pan et al. 2018). Rhizobial inoculation induces SYMRK/DMI2 protein accumulation in the nodule as SYMRK/DMI2 is protected from the proteasome during rhizobial infection. Fine-tuning the abundance of SYMRK/DMI2 at the protein level is essential for maximizing the benefits of RNS for legumes as overexpression of kinase domain of SYMRK/ DMI2 causes spontaneous nodulation (Ried et al. 2014).
Rhizobial infection receptor-like kinase1 (RINRK1) and RINRK2
Analysis of infection thread-defective mutant itd4 (SL3055-2) created by ethyl methanesulfonate (EMS) mutagenesis led to the identification of new LRR-RLK rhizobial infection receptor-like kinase1 (RINRK1) (Li et al. 2019). Specifically, RINRK1 is induced by NFs and is involved in IT formation. RINRK1 displayed an infection-specific expression pattern, and NIN bound to the RINRK1 promoter, inducing its expression. RINRK1 was found to be an atypical kinase localized to the plasma membrane and did not require kinase activity for rhizobial infection. Being a pseudokinase, RINRK1 most probably acts to produce a signaling output in response to NF to promote the initiation and maintenance of infection threads along which rhizobia can grow. This could facilitate the spatial and temporal co-ordination of infection-related signaling by two different mechanisms. One possibility is that RINRK1 could interact with component(s) of the NF recognition complex and function along with an active kinase to phosphorylate target proteins. Another possibility is that RINRK1 may be involved in perceiving an unknown rhizobial signal that activates the formation of infection threads once the nodulation signaling pathway has been activated by NF. The potential positive feedback loop, resulting from NIN induction of RINRK1 expression and RINRK1 promoting NIN expression, could facilitate the commitment required by the plant to promote a structure that enables root infection by rhizobia. A paralog of RINRK1 was identified in L. japonicus and it is named RINRK2. The rinrk2 mutant produced similar infection threads as wild type. Quantification of infection events revealed that rinrk1-1 had fewer ITs than wild type and that rinrk2-1 was similar to wild type, which suggests that RINRK2 plays a relatively minor role in infection (Li et al. 2019). RINRK1 is required for full induction of early infection genes, including Nodule Inception NIN and NPL; on the other hand, RINRK2 is not required for induction of these genes. It is proposed that RINRK1 is an infection-specific RLK, which may specifically coordinate output from NF signaling or perceive an unknown signal required for rhizobial infection.
NF signaling in model legumes
Following NF perception by NF receptors, the symbiotic signal is transduced to the common symbiosis signaling pathway (CSSP). Several mutants impaired in nodulation pathway components were found to be compromised in AM symbiosis (AMS), which led to the notion of the CSSP. SYMRK activity is required for proper progression of rhizobia, even in late stage of infection (Limpens et al. 2005). SYMRK acts upstream of NF-induced calcium spiking (Wais et al. 2000). Activation of calcium spiking inside the nucleus requires the nuclear envelope-localized potassium-permeable channels (Ané et al. 2004; Peiter et al. 2007; Charpentier et al. 2008) and cyclic nucleotide-gated channels (Charpentier et al. 2016). Calcium spiking activates CCaMK inside the nucleus (Lévy et al. 2004). CCaMK interacts with and phosphorylates CYCLOPS, a coiled-coil protein (Singh et al. 2014), which further activates a series of transcription factors. It has been proposed that CCaMK and CYCLOPS probably interact with and activate key transcription regulators like NODULATION SIGNALING PATHWAY 1 (NSP1) and NSP2. These activated transcription regulators further influence the expression of several genes like NIN, ERF Required for Nodulation 1 (ERN1), and ERN2, which are related to symbiosis development (Kalo et al. 2005; Smit et al. 2005; Kawaharada et al. 2017a; Cerri et al. 2017; Singh and Verma 2023). NIN was one of the first genes identified for nodulation in L. japonicus (Schauser et al. 1999). NIN was found to regulate rhizobial infection and nodule organogenesis (Soyano et al. 2013). These activated transcription factors regulate the expression level of early nodulin genes such as ENOD11 and ENOD12, which encode regulators of infection thread growth and nodule primordium formation (Vernié et al. 2015).
DELLA proteins, which are regulators of gibberellic acid signaling, have been shown to regulate RNS by forming a complex with CCaMK–CYCLOPS (IPD3 in Medicago). DELLA proteins not only interact with CCaMK–CYCLOPS (IPD3) to increase the phosphorylation state of CYCLOPS (IPD3), but also interact with NSP2–NSP1 and nuclear factor Y subunit A1 (NF-YA1) to enhance the expression of ENOD11, which is an NF-inducible gene (Jin et al. 2016; Fonouni-Farde et al. 2016). Following these events, up-regulation of RNS-specific cytokinin receptors is observed, which leads to changes in the cytokinin status of the plant (Gonzalez-Rizzo et al. 2006; Tirichine et al. 2007).
Recent studies showed that abiotic factors such as soil salinity has a negative impact on root nodule symbiosis, mainly by regulating the expression of NIN gene downstream of the NF signaling pathway (Singh and Valdés-López 2022b). Current knowledge can only lead us to speculate that the reduced rhizobial infection process and nodule development under salt-stress conditions are caused by either a disruption in the formation of the CYCLOPS–DELLA–NSP2/1 complex, which inhibits NIN transcriptional activation (Singh and Verma 2021) or a failure of the phosphorylated form of NSP1a to activate the expression of ERN1, which could either directly or indirectly inhibit NIN expression (He et al. 2021). The root nodule symbiosis appears to be shaped by spatiotemporal NIN expression and activity, which are modulated in response to soil salt levels.
Autoregulation of nodulation
RNS is a costly and complicated process for the plant. Thus, there is a need for a balance between hosting rhizobia in nodules for their nitrogen benefits and the costs in the form of the nutrients provided to the rhizobia. To achieve this balance, another important signaling process takes place in RNS called autoregulation of nodulation (AON). AON acts systemically via another set of RLKs. External and internal cues have been reported to control the number of nodules. The concentration of nitrate in the soil acts as a major external cue. Meanwhile, a regulatory feedback system acts as an internal cue; nodules that have already been formed suppress further nodule formation through two-way communication: root to shoot and shoot to root. This regulatory feedback system is known as AON (Ferguson et al. 2010) (Fig. 4).
A conceptual model representing autoregulation of nodulation (AON). Legumes regulate nodulation either in response to pre-existing rhizobial infections or soil nitrate levels. Nitrate induces the production of an MtNLP4/LjNLP4, which induces CLE peptide (CLE RS1/2); CLE RS1/2 transported via the xylem to the shoot acts as ascending signal. In the shoot, MtSUNN/LjHAR1 perceives CLE RS1/2 peptide. MtSUNN/LjHAR1 acts as descending signal and is transported via the phloem to the roots where it inhibits further nodule progression and cell divisions by inhibiting TOO MUCH LOVE1/2 (TML1/2)
Recent reports on AON suggest that NFs perception activates the NIN/NIN-like protein (NLP) family, which further activates the expression of CLAVATA3/embryo surrounding region (CLE) peptides in the root as ascending signals. CLE peptides move from root to shoot and activate the LRR-RLK, HAR1, in L. japonicus, and its homologs super numeric nodules 1 (MtSUNN1) and nodule autoregulation receptor 1 (GmNARK1) in M. truncatula and soybean, respectively. Conversely, CEP (C-terminally Encoded Peptide) signaling peptides produced in nitrogen-starved roots act in shoots through the CRA2 (Compact Root Architecture 2) receptor to promote nodulation even in the absence of rhizobia. The shoot-produced miR2111 microRNA acts as a signaling effector of downstream shoot-to-root of these systemic pathways that negatively regulate TOO MUCH LOVE 1 (TML1) and TML2 transcripts accumulation in roots, ultimately promoting nodulation. Low nitrogen conditions and CEP1 signaling peptides induce the production of miR2111 in CRA2-dependent manner in shoots, thus favoring root competence for nodulation. The SUNN pathway along with miR2111 systemic effector regulates dynamic fine-tuning of the nodulation capacity of legume roots. In Pisum sativum (pea), Sym28 locus which encodes PsClv2 gene involved in nodulation (Krusell et al. 2011). Inactivation of the PsClv2 gene results in hypernodulation of the root and changes to the shoot architecture. Mutational substitution of an amino acid in one leucine-rich repeat of the corresponding Lotus LjCLV2 protein results in increased nodulation. Similarly, down-regulation of the Lotus Clv2 gene by RNAi-mediated reduction of the transcript level also resulted in increased nodulation (Krusell et al. 2011).
In addition, MtSUNN1/LjHAR1/GmNARK1 has been shown to play an essential role in maintaining plant status in the presence of nitrate. Forward genetic screening for loss of nitrate suppression of nodulation in L. japonicus resulted in identifying the NITRATE UNRESPONSIVE SYMBIOSIS 1 (LjNRSYM1)/NLP4 protein. A reverse genetic screening of NLP mutants in Medicago resulted in MtNLP1 identification. MtNLP1 and LjNRSYM1/NLP4 move to the nucleus in the presence of nitrate to induce their downstream CLE-root signal (RS) peptides. The induction of CLE-RS peptide expression by NLPs in the root nodule for the activation of CLAVATA1-like RLKs in the shoot is used by legumes to control the number of nodules in nitrate-and rhizobia-dependent fashion (Nishida and Suzaki 2018).
Recent evidence indicates that phosphate deficiency can also activate the AON pathway leading to a reduction in the number of nodules (Isidra‐Arellano et al. 2020). As discussed above, soil salinity reduces the number of nodules in diverse legumes, it is tempting to speculate that salinity might activate the AON pathway to restrict nodule formation under this abiotic condition. To prove this, it will be necessary to evaluate whether salt stress activates the main AON genetic components (i.e., CLE-RS1/2 and TML) as observed in phosphate deficiency (Isidra‐Arellano et al. 2020). It will also be interesting to see if the reduction in the number of nodules is observed in the AON mutant plants.
NFs signaling for RNS in non-legume plants
Rhizobium–root nodule symbiosis is thought to be unique to legumes. There is, however, one exception, Parasponia. The rhizobial nodule symbiosis evolved independently in this non-legume and is induced by rhizobium NFs, as in legumes (van Velzen et al. 2018). The Parasponia genus of the Cannabaceae family includes five tropical tree species associated with Lipochito-oligosaccharide (LCO) producing rhizobium and forming nitrogen-fixing nodules. Parasponia perceives NF by orthologous LysM-type receptors: PanNFP1, PanNFP2 PanLYK1, and PanLYK3 (Rutten et al. 2020). Two copies of NFP in Parasponia are a result of an ancient duplication event. Parasponia PanLYK3 gene encodes two protein variants, PanLYK3.1/3.2, due to duplication of the first exon in the extracellular domain. The first exon codes for an extracellular domain that contains three LysM motifs and signal peptide. CRISPR-Cas9 knockout mutants of PanNFP1/2 and PanLYK1/3 were used to determine their role in nodulation. Pannfp2 knockout mutant lines failed to develop nodules, indicating the requirement of PanNFP2 for nodulation. On the other hand, reduced nodulation efficiency was observed in Pannfp1 mutant lines demonstrating that Pannfp1 controls nodulation efficiency, but is not essential for rhizobium intracellular infection. Reduced nodulation was found in Panlyk3 Parasponia knockout mutants but not in Panlyk1. Panlyk3 mutant nodules were relatively smaller and showed a diverse range of phenotypes, half of the nodules were similar in appearance to the wild type, a few nodules contained infection threads only, but no fixation threads, while others showed a halfway phenotype having a limited number of infected cells. Furthermore, the expression of PanNFP2 was found to be nodule specific. Based on the expression pattern and phenotype of nodules in knockout mutants, PanLYK3 is shown to be a putative ortholog of LjNFR1/MtLYK3, while PanNFP2 of LjNFR5/MtNFP. Parasponia and legumes diverged at the root of the nitrogen-fixing clade > 100 million years ago (van Velzen et al. 2018) and provide a unique comparative system to obtain insight into evolutionary trajectories of LCO receptors that are essential for nodulation in legume and non-legume plants.
Very recently, He et al. (2019) provided a significant advancement in our understanding of how NF receptors can be engineered to improve the nodulation trait or even to transfer it to non-legume crops. Such engineering would enable the growing of non-legume crops without nitrogen fertilizer. The researchers produced transgenic rice expressing chimeric receptors with different combinations of intracellular and extracellular domains. The most interesting transgenic line involved the extracellular domains of the rice Myc factor receptors, OsMYR1 or OsCERK1, being replaced by the equivalent domains of the M. truncatula NF receptors, MtNFP or MtLYK3. The resulting rice plants responded to NF treatments by activating their CSSP with intense nuclear calcium oscillations, as they do in response to Myc factors (COs). This result has a wide implication in agriculture and can be used as a starting point for the most ambitious goals of the scientific community studying plant symbioses to introduce symbiotic nitrogen fixation in cereals.
Concluding remark
The legume–rhizobia symbiosis is a unique model for investigating the evolution of mutualistic plant–microbe interactions. The ability of an NF receptor to correctly distinguish between the NFs of compatible and non-compatible rhizobial partners allows the evolution of both symbiotic partners at the same time. The structural characterization of NF receptors, and especially their binding motifs, has proven to be a great advancement in understanding NF receptor evolution. Many regulatory events, from NF perception to downstream signaling processes, have been well characterized and explored in model leguminous plants such as L. japonicus and M. truncatula. However, such knowledge is limited in the case of legume crops. To translate current research from model legumes to crop legumes, it will be vital to gain a better evolutionary and structural understanding of the mechanism by which NFs bind to LysM-RLK and how LysM-RLK is regulated at the protein level. To do this, it will be necessary to quantitatively analyze LysM-RLK at the subcellular and tissue-specific levels. The further unearthing of genes involved in the transduction of symbiotic signals downstream of NF perception will help us to create more efficient and specific symbiotic pairs of crop plants and nitrogen-fixing bacteria. The knowledge obtained from these studies and the translation of this knowledge to crop plants will provide groundbreaking tools for agriculture.
Future direction
The increased expansion of our knowledge about the molecular, biochemical, and structural biology of NFs perception in legumes for nodule formation has raised a number of new questions. Firstly, the study of molecular players involved in NFs perception and decoding has been largely limited to the model legume such as M. trancatula and L. japonicus, so exploring the role of these molecular players and their associated mechanism in economically important crops like Cicer aeritinum, and Phaseolus vulgaris would be very exciting. Secondly, it is well known that components of the exocyst complex, and the cell cycle components, play an important role in infection thread formation (Singh et al. 2023), so it will be interesting to see if there is any interplay among the components of the exocyst complex, cell cycle components and regulators involved in NFs signaling and its associated partners, if yes then how they regulate infection thread and nodule symbiosis together. Thirdly, exploration of NFs perception and associated downstream signaling have majorly focused on the model legume such as M. trancatula and L. japonicus, where rhizobia enter into legume through infection threads, and detailed analysis of each component of NFs signaling in allotetraploid legumes such as Arachis hypogaea (peanut) remains to be done. Finally, with the diversity in the number of NFs and the rhizobial modes of entry, our knowledge regarding their recognition and downstream signaling mechanisms in the host legumes is still incomplete and warrants further research.
Author contribution statement
JS and PKV conceived the idea, designed the study, wrote the review, and prepared the figures.
Data availability
The datasets generated or analyzed during the current study are presented in the article.
References
Ané J-MM, Kiss GB, Riely BK et al (2004) Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science (80-) 303:1364–1367. https://doi.org/10.1126/science.1092986
Antolín-Llovera M, Ried MK, Parniske M (2014) Cleavage of the symbiosis receptor-like kinase ectodomain promotes complex formation with nod factor receptor 5. Curr Biol 24:422–427. https://doi.org/10.1016/j.cub.2013.12.053
Arrighi J, Barre A, Amor B et al (2006) The Medicago truncatula lysine motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol 142(1):265–279. https://doi.org/10.1104/pp.106.084657
Bensmihen S, de Billy F, Gough C (2011) Contribution of NFP LysM domains to the recognition of Nod factors during the Medicago truncatula/Sinorhizobium meliloti symbiosis. PLoS ONE. https://doi.org/10.1371/JOURNAL.PONE.0026114
Bozsoki Z, Gysel K, Hansen SB et al (2020) Ligand-recognizing motifs in plant LysM receptors are major determinants of specificity. Science 369:663–670. https://doi.org/10.1126/science.abb3377
Brewin NJ (1991) Development of the legume root nodule. Annu Rev Cell Biol 7:191–226. https://doi.org/10.1146/annurev.cb.07.110191.001203
Broghammer A, Krusell L, Blaise M et al (2012) Legume receptors perceive the rhizobial lipochitin oligosaccharide signal molecules by direct binding. Proc Natl Acad Sci USA 109:13859–13864. https://doi.org/10.1073/pnas.1205171109
Capoen W, Goormachtig S, De Rycke R et al (2005) SrSymRK, a plant receptor essential for symbiosome formation. Proc Natl Acad Sci USA 102:10369–10374. https://doi.org/10.1073/pnas.0504250102
Cerri MR, Wang Q, Stolz P et al (2017) The ERN1 transcription factor gene is a target of the CCaMK/CYCLOPS complex and controls rhizobial infection in Lotus japonicus. New Phytol 215:323–337. https://doi.org/10.1111/nph.14547
Chakraborty S, Nguyen B, Wasti SD, Xu G (2019) Plant leucine-rich repeat receptor kinase (LRR-RK): structure, ligand perception, and activation mechanism. Molecules 24:3081. https://doi.org/10.3390/MOLECULES24173081
Charpentier M, Bredemeier R, Wanner G et al (2008) Lotus japonicus Castor and Pollux are ion channels essential for perinuclear calcium spiking in legume root endosymbiosis. Plant Cell 20:3467–3479. https://doi.org/10.1105/tpc.108.063255
Charpentier M, Sun J, Martins TV et al (2016) Nuclear-localized cyclic nucleotide-gated channels mediate symbiotic calcium oscillations. Science 352(6289):1102–1105. https://doi.org/10.1126/science.aae0109
Chen T, Zhu H, Ke D et al (2012) A MAP kinase kinase interacts with SymRK and regulates nodule organogenesis in Lotus japonicus. Plant Cell 24:823–838. https://doi.org/10.1105/tpc.112.095984
den Herder G, Yoshida S, Antolín-Llovera M et al (2012) Lotus japonicus E3 ligase SEVEN IN ABSENTIA4 destabilizes the symbiosis receptor-like Kinase SYMRK and negatively regulates rhizobial infection. Plant Cell 24:1691–1707. https://doi.org/10.1105/tpc.110.082248
Endre G, Kereszt A, Kevei Z et al (2002) A receptor kinase gene regulating symbiotic nodule development. Nature 417:962–966. https://doi.org/10.1038/nature00842
Ferguson BJ, Indrasumunar A, Hayashi S et al (2010) Molecular analysis of legume nodule development and autoregulation. J Integr Plant Biol 52:61–76. https://doi.org/10.1111/j.1744-7909.2010.00899.x
Fonouni-Farde C, Tan S, Baudin M et al (2016) DELLA-mediated gibberellin signalling regulates Nod factor signalling and rhizobial infection. Nat Commun 7:1–13. https://doi.org/10.1038/ncomms12636
Frugier F, Kosuta S, Murray JD et al (2008) Cytokinin: secret agent of symbiosis. Trends Plant Sci 13:115–120. https://doi.org/10.1016/j.tplants.2008.01.003
Gage DJ (2004) Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol Mol Biol Rev 68:280–300. https://doi.org/10.1128/mmbr.68.2.280-300.2004
Gamas P, Brault M, Jardinaud MF, Frugier F (2017) Cytokinins in symbiotic nodulation: when, where, what for? Trends Plant Sci 22:792–802. https://doi.org/10.1016/j.tplants.2017.06.012
Gherbi H, Markmann K, Svistoonoff S et al (2008) SymRK defines a common genetic basis for plant root endosymbioses with arbuscular mycorrhiza fungi, rhizobia, and Frankia bacteria. Proc Natl Acad Sci USA 105:4928–4932. https://doi.org/10.1073/pnas.0710618105
Gonzalez-Rizzo S, Crespi M, Frugier F (2006) The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti. Plant Cell 18:2680–2693. https://doi.org/10.1105/tpc.106.043778
Göttfert M (1993) Regulation and function of rhizobial nodulation genes. FEMS Microbiol Rev 10:39–63. https://doi.org/10.1111/J.1574-6968.1993.TB05863.X
Haney CH, Riely BK, Tricoli DM et al (2011) Symbiotic rhizobia bacteria trigger a change in localization and dynamics of the Medicago truncatula receptor kinase LYK3. Plant Cell 23:2774–2787. https://doi.org/10.1105/tpc.111.086389
Hayashi T, Shimoda Y, Sato S et al (2014) Rhizobial infection does not require cortical expression of upstream common symbiosis genes responsible for the induction of Ca 2+ spiking. Plant J 77:146–159. https://doi.org/10.1111/tpj.12374
He J, Zhang C, Dai H et al (2019) A LysM receptor heteromer mediates perception of arbuscular mycorrhizal symbiotic signal in rice. Mol Plant 12:1561–1576. https://doi.org/10.1016/j.molp.2019.10.015
He C, Gao H, Wang H et al (2021) GSK3-mediated stress signaling inhibits legume–rhizobium symbiosis by phosphorylating GmNSP1 in soybean. Mol Plant 14:488–502. https://doi.org/10.1016/j.molp.2020.12.015
Indrasumunar A, Wilde J, Hayashi S et al (2015) Functional analysis of duplicated Symbiosis Receptor Kinase (SymRK) genes during nodulation and mycorrhizal infection in soybean (Glycine max). J Plant Physiol 176:157–168. https://doi.org/10.1016/j.jplph.2015.01.002
Isidra-Arellano MC, Pozas-Rodríguez EA, Rocío Reyero-Saavedra M et al (2020) Inhibition of legume nodulation by Pi deficiency is dependent on the autoregulation of nodulation (AON) pathway. Plant J 103:1125–1139. https://doi.org/10.1111/tpj.14789
Jin Y, Liu H, Luo D et al (2016) DELLA proteins are common components of symbiotic rhizobial and mycorrhizal signalling pathways. Nat Commun 7:1–14. https://doi.org/10.1038/ncomms12433
Kalo P, Kaló P, Gleason C et al (2005) Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science (80-) 308:1786–1789. https://doi.org/10.1126/science.1110951
Kawaharada Y, Kelly S, Nielsen MW et al (2015) Receptor-mediated exopolysaccharide perception controls bacterial infection. Nature 523:308–312. https://doi.org/10.1038/nature14611
Kawaharada Y, James EK, Kelly S et al (2017a) The ethylene responsive factor required for nodulation 1 (ERN1) transcription factor is required for infection-thread formation in Lotus japonicus. Mol Plant-Microbe Interact 30:194–204. https://doi.org/10.1094/MPMI-11-16-0237-R
Kawaharada Y, Nielsen MW, Kelly S et al (2017b) Differential regulation of the Epr3 receptor coordinates membrane-restricted rhizobial colonization of root nodule primordia. Nat Commun 8:1–11. https://doi.org/10.1038/ncomms14534
Ke D, Fang Q, Chen C et al (2012) The small GTPase ROP6 interacts with NFR5 and is involved in nodule formation in Lotus japonicus. Plant Physiol 159:131–143. https://doi.org/10.1104/pp.112.197269
Kelly S, Sullivan JT, Kawaharada Y et al (2018) Regulation of Nod factor biosynthesis by alternative NodD proteins at distinct stages of symbiosis provides additional compatibility scrutiny. Environ Microbiol 20:97–110. https://doi.org/10.1111/1462-2920.14006
Kevei Z, Lougnon G, Mergaert P et al (2007) 3-Hydroxy-3-methylglutaryl coenzyme A reductase1 interacts with NORK and is crucial for nodulation in Medicago truncatula. Plant Cell 19:3974–3989. https://doi.org/10.1105/tpc.107.053975
Kobe B, Deisenhofer J (1994) The leucine-rich repeat: a versatile binding motif. Trends Biochem Sci 19:415–421. https://doi.org/10.1016/0968-0004(94)90090-6
Kosuta S, Held M, Hossain MS et al (2011) Lotus japonicus symRK-14 uncouples the cortical and epidermal symbiotic program. Plant J 67:929–940. https://doi.org/10.1111/j.1365-313X.2011.04645.x
Krusell L, Sato N, Fukuhara I et al (2011) The Clavata2 genes of pea and Lotus japonicus affect autoregulation of nodulation. Plant J 65:861–871. https://doi.org/10.1111/j.1365-313X.2010.04474.x
Lévy J, Bres C, Geurts R et al (2004) A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science (80-) 303:1361–1364. https://doi.org/10.1126/science.1093038
Li H, Chen M, Duan L et al (2018) Domain swap approach reveals the critical roles of different domains of SYMRK in root nodule symbiosis in Lotus japonicus. Front Plant Sci 9:697. https://doi.org/10.3389/fpls.2018.00697
Li X, Zheng Z, Kong X et al (2019) Atypical receptor kinase RINRK1 required for rhizobial infection but not nodule development in Lotus japonicus. Plant Physiol 181:804–816. https://doi.org/10.1104/pp.19.00509
Liang P, Stratil TF, Popp C et al (2018) Symbiotic root infections in Medicago truncatula require remorin-mediated receptor stabilization in membrane nanodomains. Proc Natl Acad Sci USA 115:5289–5294. https://doi.org/10.1073/pnas.1721868115
Limpens E, Franken C, Smit P et al (2003) LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science (80-) 302:630–633. https://doi.org/10.1126/science.1090074
Limpens E, Mirabella R, Fedorova E et al (2005) Formation of organelle-like N2-fixing symbiosomes in legume root nodules is controlled by DMI2. Proc Natl Acad Sci USA 102:10375–10380. https://doi.org/10.1073/pnas.0504284102
Madsen EB, Madsen LH, Radutoiu S et al (2003) A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425:637–640. https://doi.org/10.1038/nature02045
Madsen EB, Antolín-Llovera M, Grossmann C et al (2011) Autophosphorylation is essential for the in vivo function of the Lotus japonicus Nod factor receptor 1 and receptor-mediated signalling in cooperation with Nod factor receptor 5. Plant J 65:404–417. https://doi.org/10.1111/j.1365-313X.2010.04431.x
Markmann K, Giczey G, Parniske M (2008) Functional adaptation of a plant receptor- kinase paved the way for the evolution of intracellular root symbioses with bacteria. PLoS Biol 6:e68. https://doi.org/10.1371/journal.pbio.0060068
Mbengue M, Camut S, de Carvalho-Niebel F et al (2010) The medicago truncatula E3 ubiquitin ligase PUB1 interacts with the LYK3 symbiotic receptor and negatively regulates infection and nodulation. Plant Cell 22:3474–3488. https://doi.org/10.1105/tpc.110.075861
Mergaert P, Uchiumi T, Alunni B et al (2006) Eukaryotic control on bacterial cell cycle and differentiation in the Rhizobium-legume symbiosis. Proc Natl Acad Sci USA 103:5230–5235. https://doi.org/10.1073/pnas.0600912103
Moling S, Pietraszewska-Bogiel A, Postma M et al (2014) Nod factor receptors form heteromeric complexes and are essential for intracellular infection in Medicago nodules. Plant Cell 26:4188–4199. https://doi.org/10.1105/tpc.114.129502
Monahan-Giovanelli H, Pinedo CA, Gage DJ (2006) Architecture of infection thread networks in developing root nodules induced by the symbiotic bacterium Sinorhizobium meliloti on Medicago truncatula. Plant Physiol 140:661–670. https://doi.org/10.1104/pp.105.072876
Murakami E, Cheng J, Gysel K et al (2018) Epidermal LysM receptor ensures robust symbiotic signalling in Lotus japonicus. Elife 7:e33506. https://doi.org/10.7554/eLife.33506
Nishida H, Suzaki T (2018) Nitrate-mediated control of root nodule symbiosis. Curr Opin Plant Biol 44:129–136. https://doi.org/10.1016/j.pbi.2018.04.006
Oldroyd GED, Murray JD, Poole PS, Downie JA (2011) The rules of engagement in the legume–rhizobial symbiosis. Annu Rev Genet 45:119–144. https://doi.org/10.1146/annurev-genet-110410-132549
Ott T (2017) Membrane nanodomains and microdomains in plant–microbe interactions. Curr Opin Plant Biol 40:82–88. https://doi.org/10.1016/j.pbi.2017.08.008
Ouma EW, Asango AM, Maingi J, Njeru EM (2016) Elucidating the potential of native rhizobial isolates to improve biological nitrogen fixation and growth of common bean and soybean in smallholder farming systems of Kenya. Int J Agronomy 2016:1–7. https://doi.org/10.1155/2016/4569241
Pan H, Stonoha-Arther C, Wang D (2018) Medicago plants control nodulation by regulating proteolysis of the receptor-like kinase DMI2. Plant Physiol 177:792–802. https://doi.org/10.1104/pp.17.01542
Peiter E, Sun J, Heckmann AB et al (2007) The Medicago truncatula DMI1 protein modulates cytosolic calcium signaling. Plant Physiol 145:192–203. https://doi.org/10.1104/pp.107.097261
Radutoiu S, Madsen L, Madsen E et al (2003) Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425(6958):585–592. https://doi.org/10.1038/nature02039
Ried MK, Antolín-Llovera M, Parniske M (2014) Spontaneous symbiotic reprogramming of plant roots triggered by receptor-like kinases. Elife 3:1–17. https://doi.org/10.7554/eLife.03891
Rutten L, Miyata K, Roswanjaya YP et al (2020) Duplication of symbiotic Lysin Motif-receptors predates the evolution of nitrogen-fixing nodule symbiosis. Plant Physiol. https://doi.org/10.1104/pp.19.01420
Saha S, Paul A, Herring L et al (2016) Gatekeeper tyrosine phosphorylation of SYMRK is essential for synchronizing the epidermal and cortical responses in root nodule symbiosis. Plant Physiol 171:71–81. https://doi.org/10.1104/pp.15.01962
Sánchez-López R, Jáuregui D, Nava N et al (2011) Down-regulation of SymRK correlates with a deficiency in vascular bundle development in Phaseolus vulgaris nodules. Plant Cell Environ 34:2109–2121. https://doi.org/10.1111/j.1365-3040.2011.02408.x
Schallus T, Jaeckh C, Fehér K et al (2008) Malectin: a novel carbohydrate-binding protein of the endoplasmic reticulum and a candidate player in the early steps of protein N-glycosylation. Mol Biol Cell 19:3404. https://doi.org/10.1091/MBC.E08-04-0354
Schauser L, Roussis A, Stiller J, Stougaard J (1999) A plant regulator controlling development of symbiotic root nodules. Nature 402:191–195. https://doi.org/10.1038/46058
Schroeder JI, Delhaize E, Frommer WB et al (2013) Using membrane transporters to improve crops for sustainable food production. Nature 497:60–66. https://doi.org/10.1038/nature11909
Singh J, Valdés-López O (2022a) A nodule peptide confiscates haem to promote iron uptake in rhizobia. Trends Plant Sci 28:125–127. https://doi.org/10.1016/j.tplants.2022.11.005
Singh J, Valdés-López O (2022b) Discovering the genetic modules controlling root nodule symbiosis under abiotic stresses: salinity as a case study. New Phytol 237(4):1082–1085. https://doi.org/10.1111/nph.18627
Singh J, Verma PK (2021) NSP1 allies with GSK3 to inhibit nodule symbiosis. Trends Plant Sci 26:999–1001. https://doi.org/10.1016/j.tplants.2021.07.001
Singh J, Verma PK (2023) Plant transcription factors and nodule development. In: Plant transcription factors. Elsevier, pp 175–196. https://doi.org/10.1016/B978-0-323-90613-5.00020-0
Singh S, Katzer K, Lambert J et al (2014) CYCLOPS, A DNA-binding transcriptional activator, orchestrates symbiotic root nodule development. Cell Host Microbe 15:139–152. https://doi.org/10.1016/j.chom.2014.01.011
Singh J, Varshney V, Mishra V (2023) AUR1 and its pals: orchestration of intracellular rhizobia infection in legume for nitrogen fixation. Plant Cell Rep 1:3. https://doi.org/10.1007/s00299-023-02979-x
Smit P, Raedts J, Portyanko V et al (2005) NSP1 of the GRAS protein family is essential for rhizobial nod factor-induced transcription. Science (80-) 308:1789–1791. https://doi.org/10.1126/science.1111025
Smit P, Limpens E, Geurts R et al (2007) Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol 145(1):183–191. https://doi.org/10.1104/pp.107.100495
Soyano T, Kouchi H, Hirota A, Hayashi M (2013) NODULE INCEPTION directly targets NF-Y subunit genes to regulate essential processes of root nodule development in Lotus japonicus. PLoS Genet 9:e1003352. https://doi.org/10.1371/journal.pgen.1003352
Stracke S, Kistner C, Yoshida S et al (2002) A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417:959–962. https://doi.org/10.1038/nature00841
Takeda N, Maekawa T, Hayashi M (2012) Nuclear-localized and deregulated calcium- and calmodulin-dependent protein kinase activates rhizobial and mycorrhizal responses in Lotus japonicus. Plant Cell 24:810–822. https://doi.org/10.1105/tpc.111.091827
Tirichine L, Sandal N, Madsen LH et al (2007) A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science (80-) 315:104–107. https://doi.org/10.1126/science.1132397
Tóth K, Stratil TF, Madsen EB et al (2012) Functional domain analysis of the remorin protein LjSYMREM1 in Lotus japonicus. PLoS ONE 7:e30817. https://doi.org/10.1371/journal.pone.0030817
Van Brussel AAN, Bakhuizen R, Van Spronsen PC et al (1992) Induction of pre-infection thread structures in the leguminous host plant by mitogenic lipo-oligosaccharides of rhizobium. Science (80-) 257:70–72. https://doi.org/10.1126/science.257.5066.70
van Velzen R, Holmer R, Bu F et al (2018) Comparative genomics of the nonlegume Parasponia reveals insights into evolution of nitrogen-fixing rhizobium symbioses. Proc Natl Acad Sci USA 115:E4700–E4709. https://doi.org/10.1073/pnas.1721395115
Vernié T, Kim J, Frances L et al (2015) The NIN transcription factor coordinates diverse nodulation programs in different tissues of the medicago truncatula root. Plant Cell 27:3410–3424. https://doi.org/10.1105/tpc.15.00461
Vernié T, Camut S, Camps C et al (2016) PUB1 interacts with the receptor kinase DMI2 and negatively regulates rhizobial and arbuscular mycorrhizal symbioses through its ubiquitination activity in medicago truncatula. Plant Physiol 170:2312–2324. https://doi.org/10.1104/pp.15.01694
Wais RJ, Galera C, Oldroyd G et al (2000) Genetic analysis of calcium spiking responses in nodulation mutants of Medicago truncatula. Proc Natl Acad Sci USA 97:13407–13412. https://doi.org/10.1073/pnas.230439797
Wang D, Griffitts J, Starker C et al (2010) A nodule-specific protein secretory pathway required for nitrogen-fixing symbiosis. Science (80-) 327:1126–1129. https://doi.org/10.1126/science.1184096
Wong JEMM, Nadzieja M, Madsen LH et al (2019) A Lotus japonicus cytoplasmic kinase connects Nod factor perception by the NFR5 LysM receptor to nodulation. Proc Natl Acad Sci USA 116:14339–14348. https://doi.org/10.1073/pnas.1815425116
Wong JEMM, Gysel K, Birkefeldt TG et al (2020) Structural signatures in EPR3 define a unique class of plant carbohydrate receptors. Nat Commun. https://doi.org/10.1038/s41467-020-17568-9
Xie F, Murray JD, Kim J et al (2012) Legume pectate lyase required for root infection by rhizobia. Proc Natl Acad Sci USA 109:633–638. https://doi.org/10.1073/pnas.1113992109
Yoshida S, Parniske M (2005) Regulation of plant symbiosis receptor kinase through serine and threonine phosphorylation. J Biol Chem 280:9203–9209. https://doi.org/10.1074/jbc.M411665200
Yuan S, Zhu H, Gou H et al (2012) A ubiquitin ligase of symbiosis receptor kinase involved in nodule organogenesis. Plant Physiol 160:106–117. https://doi.org/10.1104/pp.112.199000
Zhu H, Chen T, Zhu M et al (2008) A Novel ARID DNA-binding protein interacts with SymRK and is expressed during early nodule development in Lotus japonicus. Plant Physiol 148:337–347. https://doi.org/10.1104/pp.108.119164
Acknowledgements
JS thanks the Council of Scientific & Industrial Research, Government of India, for the fellowship.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The author declare that there is no conflict of interest.
Additional information
Communicated by Gerhard Leubner.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Singh, J., Verma, P.K. Role of Nod factor receptors and its allies involved in nitrogen fixation. Planta 257, 54 (2023). https://doi.org/10.1007/s00425-023-04090-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-023-04090-7