Abstract
Plant root-associated bacteria (rhizobacteria) elicit plant basal immunity referred to as induced systemic resistance (ISR) against multiple pathogens. Among multi-bacterial determinants involving such ISR, the induction of ISR and promotion of growth by bacterial volatile compounds was previously reported. To exploit global de novo expression of plant proteins by bacterial volatiles, proteomic analysis was performed after exposure of Arabidopsis plants to the rhizobacterium Bacillus subtilis GB03. Ethylene biosynthesis enzymes were significantly up-regulated. Analysis by quantitative reverse transcriptase polymerase chain reaction confirmed that ethylene biosynthesis-related genes SAM-2, ACS4, ACS12, and ACO2 as well as ethylene response genes, ERF1, GST2, and CHIB were up-regulated by the exposure to bacterial volatiles. More interestingly, the emission of bacterial volatiles significantly up-regulated both key defense mechanisms mediated by jasmonic acid and salicylic acid signaling pathways. In addition, high accumulation of antioxidant proteins also provided evidence of decreased sensitivity to reactive oxygen species during the elicitation of ISR by bacterial volatiles. The present results suggest that the proteomic analysis of plant defense responses in bacterial volatile-mediated ISR can reveal the mechanisms of plant basal defenses orchestrated by endogenous ethylene production pathways and the generation of reactive oxygen species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The rhizosphere surrounding plant roots is a biologically active area where rich root exudates serve as food supply for root-associated bacteria, and where pathogenic and beneficial microorganisms compete (Kloepper et al. 1999). Beneficial bacteria include specialized microbes named plant growth-promoting rhizobacteria (PGPR) that generate an array of biologically active compounds. During the last decades, several groups have explored the agricultural utilization of PGPR-elicited plant growth promotion and suppression of plant diseases in modern agricultural systems (Vallad and Goodman 2004; Kloepper et al. 1999; Glick 2005; MacDonald et al. 1986; Ryu et al. 2005a, 2006; Timmusk et al. 1999). Recent studies on molecular signaling mechanisms between plants and PGPR explain the diversity of microbial signaling molecules and their perception by plants (Vallad and Goodman 2004; Kloepper et al. 2004; Ryu et al. 2003, 2004a). A well-understood strategy for PGPR-mediated plant growth promotion is the biosynthesis and secretion of molecules mimicking plant growth regulators such as indole-3-acetic acid, cytokinins, and gibberellins by PGPR, and the reduction of endogenous ethylene through the breakdown of the ethylene precursor 1-aminocyclopropane-1-carboxylate by 1-aminocyclopropane-1-carboxylate deaminase produced by PGPR (Loper and Schroth 1986; Glick 2005; MacDonald et al. 1986; Timmusk et al. 1999). Among plant hormones, there has recently been much interest in the role of ethylene in eliciting both plant defenses and plant growth. The ethylene insensitive 2 (ein2) mutant impaired the bacteria capacity on induced systemic resistance (ISR) and plant growth promotion when grown in soils with PGPR strains (Ryu et al. 2004a, 2005b). Similarly, the growth of the same mutant plants was not promoted by a plant growth-promoting fungus, Piriformospora indica, indicating that ethylene can be a general signaling component of plant growth promotion pathways and ISR by PGPR and PGPF (Camehl et al. 2010).
New evidence was recently found that the emission of bacterial volatiles from specific strains of PGPR without physical contact with plant roots also resulted in growth promotion and ISR in Arabidopsis seedlings (Ryu et al. 2003, 2004a). Two strains out of several PGPR strains were tested, Bacillus subtilis GB03 and B. amyloliquefaciens IN937a, promoted plant growth and elicited an ISR brought about by volatile emissions. To elucidate the signaling networks involved in growth promotion and ISR via PGPR VOCs, a series of mutant and transgenic plant lines was tested by Ryu et al. (2003, 2004a). Among several mutant lines tested for regulatory control of ISR, only the ethylene-insensitive line (ein2) did not show an improvement of disease symptoms when Arabidopsis plants were pre-treated with GB03 volatiles. To examine if ISR is mediated at the level of transcription, three transgenic β-glucuronidase (GUS) fusion lines involved in plant-defense signaling were tested. Of these lines assayed, the combined JA/ethylene-activated PDF1/2-GUS line alone exhibited elevated GUS activity (>1,000-fold) over untreated control plants (Ryu et al. 2004a).
Although the molecular and genetic basis of plant responses to PGPR-elicited ISR has been studied extensively, little is known about the molecular and biochemical mechanisms of bacterial VOC-elicited ISR. More significantly, genetic changes in plants are sometimes not followed by differential expression of the corresponding proteins. Therefore, proteome analysis has recently become a critical method to understand the direct relationship between proteins and their biological functions (Anderson and Anderson 1998). Although this method has had broad applications, its use in the plant–microbe interaction field has been limited mostly to biological control. Proteome analysis was conducted on cucumber plants after the application of the beneficial Trichoderma asperellum strain T34, which is known to elicit ISR when applied at 105 spores/ml (Segarra et al. 2007). However, proteome analysis of the plant responses to PGPR treatment during elicitation of ISR has not been assessed. In the current study, proteomic analysis was used to understand the changes of expression of plant proteins in response to bacterial volatiles, and changes in the proteome of Arabidopsis leaf tissue induced by bacteria VOCs, with special emphasis on the systemic changes in bacterial VOC-elicited plant proteins, were investigated. The analysis revealed that bacterial volatile compound-elicited ethylene biosynthesis and antioxidant production can play a critical role in the ISR against bacterial pathogens. In addition to proteomics analysis, we also evaluated transcriptional expression of SA- JA-, and ET-response genes following Arabidopsis seedling exposure to bacterial volatile using quantitative reverse transcriptase polymerase chain reaction (qRT-PCR). The information obtained should allow a better understanding of communication between plant and bacteria via volatiles.
Materials and methods
Plant growth and bacterial inoculation
Plant and bacterial preparations were prepared as described previously (Ryu et al. 2003, 2004a). Briefly, Bacillus subtilis strain GB03 was grown on TSA media for 24 h before inoculation into one compartment of an I-plate containing 1/2 Murashige and Skoog medium 1.5% (w/v) agar and 1.5% (w/v) sucrose 0.4% (w/v). Arabidopsis thaliana ecotype Columbia (Col-0) seedlings were transferred to the other compartment of the I-plate and cultivated in a growth chamber for 14 days at 21°C before collection of plant samples for proteome analysis. For the ISR assay, 2 μl of 106 colony forming units per 1 ml Erwinia carotovora subsp. carotovora was drop-inoculated on leaves 2 weeks after bacterial volatile emission as previously described (Ryu et al. 2004a). The disease severity (0–5) was assessed 24 h after pathogen challenge. 1 mM salicylic acid and sterile distilled water used as positive control and negative control, respectively.
Chlorophyll content
Total chlorophyll content was determined after its extraction from leaf tissue with dimethyl sulfoxide (DMSO) according to Hiscox and Israelstam (1979). Leaf slices weighing 0.5 g were placed in 10 ml of DMSO in 15-ml culture tubes protected from light. The vials were incubated at 65°C overnight in a water bath. Absorbance of the extract was read at 645 and 663 nm. Total chlorophyll (mg/g fresh weight) = [(20.2 × OD645) + (8.02 × OD663)] × V/(1,000 × W), where V is the volume of the extract and W is the weight of tissue in grams.
Concentration of inorganic ions
Leaf and root P, K, Mg, Ca, Na, Fe, Cu, Zn, and S were determined as dry-ash pulverized material at 400°C for 8 h after dissolving the leaves in 1 N HCl (Kalra 1998). The samples were filtered through Whatman No. 2 filters and diluted to 10 ml, followed by analysis using ICP-MS (Optima 5300 DV, PerkinElmer Inc., USA).
Assay of antioxidant enzyme activities
Plant leaves treated with water and GB03 VOCs were ground in liquid N2 and the resulting powder was used to measure antioxidant enzyme activities as previously described. The prepared powder was mixed in 50 mM potassium phosphate buffer (pH 7.0) and centrifuged at 12,000 rpm for 20 min. The supernatants were used for the enzyme activity assays. A standard protocol was used for the measurement of SOD activity based on the generation of superoxide radicals by xanthine and xanthine oxidase, which then react with INT to form a red formazan dye product (McCord and Fridovich 1969). Ascorbate peroxidase (APX) (EC 1.11.1.7) was determined according to Nakano and Asada (1981). The decrease in ascorbate concentration was followed as the decline in absorbance at 290 nm, and ascorbate peroxidase activity was calculated using the extinction coefficient (2.8 mM−1 cm−1 at 290 nm) for ascorbate. Glutathione reductase activity (GR) was determined by the method of Halliwell and Foyer (1976). Catalase (CAT) activity was assayed by measuring the initial rate of disappearance of H2O2 (Kato and Shimizu 1987). The decrease in H2O2 was followed as the decline in absorbance at 240 nm, and CAT activity was calculated using the extinction coefficient (40 mM−1 cm−1 for at 240 nm) for H2O2 (Kato and Shimizu 1987). Dehydroascorbic acid reductase (DHAR) (EC 1.8.5.4) activity was assayed according to Hossain and Asada (1984) by following the increase of A 290 as dehydroascorbate was reduced to ascorbate.
Protein sample preparation
Arabidopsis leaf proteins were isolated according to a modified TCA/acetone/phenol extract method (Wang et al. 2008). Two grams of leaf material were ground into a fine powder under liquid nitrogen using a pre-cooled mortar and pestle. The powder was mixed with cold TCA/acetone buffer (10% TCA, 0.07% 2-ME in acetone) and the mixture was shaken for 1 h and centrifuged at 8,000 rpm for 15 min at 4°C. The supernatant was washed twice with a cold acetone solution followed by air-drying. SDS extraction buffer [30% sucrose, 1–2% SDS, 0.1 M Tris–HCL (pH 8.8)] was added, followed by equal volumes of saturated phenol, and mixed by vortexing for 1 h (room temp.). The phenol phase was separated by centrifugation (8,000 rpm, 10 min, 4°C) and the phenol phase was transferred to a new tube. Five volumes of 0.1 M ammonium acetate in methanol were added and incubated at −20°C for at least 2 h. Proteins were pelleted by centrifugation, and washed once with cold methanol and twice with acetone. The pellet was air-dried and dissolved in protein lysis buffer (7 M urea, 2 M thiourea, 4% CHAPS, 1 mM PMSF, 50 mM DTT, 0.5% IPG buffer; Amersham Biosciences, San Francisco, CA, USA). The protein concentration was determined using the 2D-Quant kit (Amersham Biosciences).
2-DE and image analysis
Proteins (150 μg) were separated using an 18-cm pH 5-8L IPG strip in a Protean IEF cell (Bio-Rad, Hercules, CA, USA) and in-gel rehydrated for 12 h. Isoelectric focusing was performed at 20°C for 15 min at 250 V, 3 h at 10,000 V, and 9 h at 90,000 V. The strips were equilibrated with equilibrating solution [50 mM Tris–HCL (pH 8.8), 6 M urea, 30% glycerol, 2% SDS] containing 1% DTT, and then treated with the same buffer containing 2.5% iodoacetamide. In the second dimension, the strips were transferred onto 11% SDS-polyacrylamide gels and visualized by silver staining (Blum et al. 1987). The 2-DE of each treatment was repeated three times. Gel images were scanned using a GS-800 Imaging Densitometer (Bio-Rad) and analyzed with the software PDQuest version 7.2.0 (Bio-Rad). The volume of each spot was normalized to a relative volume, and mean values calculated from triplicate data were compared. Quantitative analysis sets were created between each control and bacteria-treated leaves. Proteins showing a statistical level of p < 0.05 by Student’s t test were selected for identification. The cut-off for differentially regulated proteins was a 1.5-fold change in expression.
Protein identification using MALDI-TOF MS and MALDI-TOF/TOF MS
Selected protein spots were excised from 2-DE gels and subjected to in-gel digestion with trypsin as described previously (Lee et al. 2004). MALDI-MS was carried out using a Voyager-DE-STR Mass spectrometer (Applied Biosystems, Framingham, MA, USA), and spectra were obtained in the reflection/delayed extraction mode. Data were subjected to SequazymeTM Peptide Mass Standard Kit (Calibration 1, Applied Biosystems, Framingham, MA, USA). Monoisotopic peptide masses were analyzed with MoverZ (http://www.proteomics.com), and the NCBInr database was searched using Mascot software (http://www.matrixscience.com). The following parameters were used for database searches: mass tolerance of 50 ppm, one missed cleavage, oxidation of methionine, and cysteine modified by iodoacetamide.
MS and MS/MS analyses were performed on the ABI 4800 Plus TOF-TOF Mass Spectrometer (Applied Biosystems, Framingham, MA, USA), which uses a 200 Hz ND:YAG laser operating at 355 nm. The ten most and least intense ions per MALDI spot, with signal/noise ratios >25, were selected for subsequent MS/MS analysis in 1 kV mode and 800–1,000 consecutive laser shots. During MS/MS analysis, air was used as the collision gas. Data were subjected to a Mass Standard Kit for the 4700 Proteomics Analyzer (calibration Mixture 1). MS/MS spectra were searched against the NCBInr database by ProteinPilot v.3.0 (with MASCOT as the database search engine) with peptide and fragment ion mass tolerance of 50 ppm. Carbamidomethylation of cysteines and oxidation of methionines were allowed during the search of the peptides. One missing trypsin cleavage was allowed. Peptide mass tolerance and fragment mass tolerance of the selected 95 proteins were set to 50 ppm. High confidence identifications had statistically significant search scores (greater than 95% confidence, equivalent to MASCOT expect value p < 0.05), were consistent with the protein’s experimental pI and MW, and accounted for the majority of ions present in the mass spectra.
Reverse transcriptase (RT)-PCR, and quantitative (Q)-RT-PCR
Total RNA was isolated from inoculated 14-day-old leaf tissues according to the protocol of Yang et al. (2009a). Total RNA was treated with 1 U of RNase-free DNase (Promega, USA) for 10 min (min) at 37°C, and then subjected to a second round of purification using the TRI reagent. First-strand cDNA synthesis was carried out in 20 l of AccuPower RT PreMix (Bioneer, Korea) containing 1 g of DNase-treated total RNA, oligo(dT) primers, and Moloney murine leukemia virus reverse transcriptase (MMLV-RT; Invitrogen, USA). PCR reactions were carried out according to the manufacturer’s instructions. The candidate gene was analyzed using the following primers: 5′-AGACCCAACCAAAGTCGACAGAAG-3′ (SAM-2_F) and 5′-CTGGAATCAACCCTGTTCCGTAAG-3′ (SAM-2_R), and 5′-GCTCTCAGTGTTTGTTGACACCTA-3′ (MAT3_F) and 5′-AACCTGAAGTTACCTCCCCTCTTA-3′ (MAT3_R). Additional genes and the primer sets used to detect them were as follows: 5′-GGGTCGACATGAGACCTCTCCTTA-3′ (ACS4_F), 5′-CGCGAAACAAACTCTAAACCAACC-3′ (ACS4_R); 5′-ATGTCAGAGAATTTGATGGAGTCA-3′ (ACS12_F), 5′-CAAATGACACATTACCTCCCATTA-3′ (ACS12_R); 5′-CAGATGTGTCTGATGAATACAGGA-3′ (ACO2_F), 5′-CCCTAGATTCTCACACAGTAGATCC-3′ (ACO2_R); 5′-TCAGAAGACCCCAAAAGCTCCTCA-3′ (ERF1_F), 5′-TTGATCACCGCTCCGTGAAGTTAG-3′ (ERF1_R); 5′-AATATGGTTTTGCTTCAGTCA-3′ (GST2_F), 5′-TGCCAAAGATACTCTCAAGAG-3′ (GST2_R); 5′-GCTTCAGACTACTGTGAACC-3′ (ChiB_F), 5′-TCCACCGTTAATGATGTTCG-3′ (ChiB_R); 5′-AATGAGCTCTCATGGCTAAGTTTGCTTCC-3′ (PDF1.2_F), 5′-AATCCATGGAATACACACGATTTAGCACC-3′ (PDF1.2_R); 5′-GGTTAGGAATAGCCTTGTGAAGAA-3′ (VSP1_F), 5′-GTAGAGTGGATTTGGGAGCTTAAA-3′ (VSP1_R); 5′-TTCCACAACCAGGCACGAGGAG-3′ (PR1_F), 5′-CCAGACAAGTCACCGCTACCC-3′ (PR1_R); and 5′-CCAAACCATTCTTTCGAGGA-3′ (FMO1_F) and 5′-CTCAAGCCAAGTTCGGAAAG-3′ (FMO1_R). AGI codes were MAT3 (At2g36880), ACS4 (AT2G22810), ACS12 (AT5G51690), ACO2 (AT1G62380), ERF1 (AT3G23240), GST2 (AT4G02520), CHIB (AT3G12500), PDF1.2 (AT5G44420), VSP1 (AT5G24780), VSP1 (AT5G24780), PR1 (AT2G14610), FMO1 (AT1G19250), and AtACT2 (AT3G18780).
As a control to ensure that equal amounts of RNA were analyzed in each experiment, AtActin was also assayed using the primers 5′-GTTAGCAACTGGGATGATATGG-3′ and 5′-CAGCACCAATCGTGATGACTTGCCC-3′. Candidate priming genes were amplified from 100 ng of cDNA by PCR using an annealing temperature of 60°C. Amplified PCR products were separated by 2% agarose gel electrophoresis. qRT-PCR was carried out using a Chromo4 real-time PCR system (Bio-Rad). Reaction mixtures (10 μl) contained 5 μl of 2× Brilliant SYBR Green QPCR master mix (Bio-Rad), cDNA, and 10 pM of each primer. The thermocycle parameters were as follows: initial polymerase activation for 10 min at 95°C, and then 40 cycles of 30 s at 95°C, 30 s at 60°C, and 42 s at 72°C. Conditions were determined by comparing the threshold values in a series of dilutions of the RT product, followed by a non-RT template control and a non-template control for each primer pair. Relative RNA levels were calibrated and normalized to the level of AtAct2 mRNA.
Data analysis
Data was subjected to analysis of variance using JMP software (SAS Institute Inc., Cary, NC). Significance of effects of strain GB03 VOC treatment was determined by the magnitude of the F value at P = 0.05. When a significant F value was obtained for treatments, separation of means was accomplished using Fisher’s protected least significant difference (LSD) at P = 0.05. Results of repeated trials of each experiment outlined above were similar. Hence, one representative trial of each experiment is reported in the result section.
Results and discussion
Plant growth promotion and induced systemic resistance by bacterial volatiles
The PGPR strain Bacillus subtilis GB03 was previously found to promote seedling growth of Arabidopsis through the production of volatile organic compounds including 2,3-butanediol and acetoin (Ryu et al. 2003). The same strain triggered a plant systemic resistance response referred to as “induced systemic resistance (ISR)” against a soft-rot bacterial pathogen, Erwinia carotovora subsp. carotovora (Ryu et al. 2004a). Further study revealed that the ethylene signaling pathway was involved in the activation of ISR by the bacterial volatile emission of strain GB03. A transcriptome analysis showed the up-regulation of ethylene biosynthesis and ethylene response genes among hormone-related genes in a functional classification of genes that were differentially expressed after 72 h of exposure to GB03 VOCs (Zhang et al. 2007). To investigate the involvement of ethylene in the ISR elicited by strain GB03 VOCs, a proteomics technique was employed. Before the proteomics analysis, the capability of strain GB03 to elicit plant growth promotion and ISR was validated using the I-plate system previously described (Ryu et al. 2003, 2004a). Six days after bacterial inoculation on one side of an I-plate, the foliar shoot growth (shoot fresh weight) and root growth (root fresh weight) were significantly increased in strain GB03 VOC-treated Arabidopsis plants (Fig. 1a, b-inset). Disease severity of Arabidopsis seedlings treated by strain GB03 VOC was twofold less than water control (Fig. 1c). ISR capacity of 1 mM salicylic acid treatment was not significantly different with strain GB03 treatment (Fig. 1c). The results were consistent with previously reported data (Ryu et al. 2003, 2004a).For studying mode of action on growth promotion by bacterial volatiles, the possible mechanisms mediating the increase in plant growth were assessed by measuring the mineral content of the leaves and root systems. Leaf samples showed a significant decrease in the concentration of all minerals tested, including K, S, Fe, P, Na, Zn, Ca, and Mg, with the exception of Cu, in the bacterial VOC-treated plants compared to water-treated plants, while the S, Fe, and Cu contents in the root systems of plants treated with strain GB03 VOC were greater compared to water controls (Fig. 1d). Under conditions of spatial separation between plants and bacteria, diverse soil bacterial siderophores have been proposed to facilitate iron acquisition in plants (Kloepper et al. 1980). Zhang et al. (2009a) demonstrated that strain GB03 VOC indirectly acidified the plant root system and directly induced plant iron uptake. Strain GB03 VOCs were also shown to increase photosynthesis rates through FIT1-dependent iron acquisition (Zhang et al. 2009a). Under experimental conditions using half strength MS medium carrying 50 μM Fe-EDTA, GB03 VOC exposure increased iron content by boosting the accessibility of iron to the roots, resulting in a significant increase in the photosynthetic effectiveness of Arabidopsis seedlings. The photosynthetic apparatus contains 2 or 3 iron atoms in each photosystem II (PSII), 12 iron atoms in each photosystem I; 5 iron atoms per cytochrome b6-f complex, and two iron atoms per ferredoxin molecule (Varotto et al. 2002). The inability of photosynthesis negatively affects the electron transport system by light intensity-dependent damage of PSII.
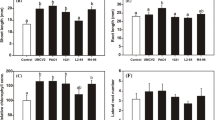
Plant growth promotion in Arabidopsis by bacterial volatiles. a Sample plates from 3, 6, 9, 12, and 15 days after inoculation of Bacillus subtilis strain GB03 on I plates with airborne exposure to bacterium and water treatment. b Enhancement of shoot fresh weight and (inset) root fresh weight by bacterial volatile emission at 3, 6, 9, 12, and 15 days after bacterial inoculation. c Induced systemic resistance against Erwinia carotovra subsp. carotovora by strain GB03 volatiles at 14 days after bacteria treatment. 1 mM salicylic acid used as a positive control. Concentration of inorganic ions in leaves (d) and roots (e) of A. thaliana grown in the absence (Control) or presence (GB03) of B. subtilis GB03. The concentration of inorganic ions was measured 14 days after B. subtilis GB03 treatment. Data represent the means ± SEM of three replicates. Asterisks indicate significant difference (P ≤ 0.05) between GB03 and control treatments
Overview of 2-DE and characterization of the differentially expressed proteins
Strain GB03 VOC-specific proteins were analyzed 14 days after inoculating plants with strain GB03. The proteome profiles of bacterial VOC-treated Arabidopsis leaf samples were obtained using 2-DE. Three replicates of the 2-DE gel images of water control and strain GB03 treatments are shown in Fig. 2a. A total of 95 spots were detected based on the differential expression of the proteins between the two treatments. The spots that reproducibly changed by more than the 1.5-fold were subjected to peptide mass fingerprinting. The comparative intensities of all spots were analyzed regardless of the statistical significance of the differences (P = 0.05) between water controls and GB03-treated samples. Besides many examples of proteome analysis on plant–pathogen interactions, to understand the nature of plant response by beneficial bacteria, most results were reported previously on rhizobia–legume interactions (for recent review Chen et al. 2010). Few studies were conducted on PGPR–plant interactions (Kandasamy et al. 2009; Miché et al. 2006). The differential regulation of the compatible and incompatible rice plants by the bacterial endophyte Azoarcus sp. strain BH72 that is capable of fixing nitrogen was attributed to the distinct protein profiles (Miché et al. 2006). In the other study, 23 proteins were differentially expressed by rice root application of Pseudomonas fluorescens strain KH-1 that exhibited plant growth promotion of rice in the field (Kandasamy et al. 2009). Similar to our results, proteome analysis of leaf sheath from rice seedlings treated by the strain KH-1 showed the up-regulation of RuBisCo that act in photosynthesis and accumulation of chlorophyll (Agrios 2005). In this study, 17 spots involved in photosynthesis and development (mostly ATP synthase subunits) were selected (Fig. 2a). Among the selected 17 spots, spots 7, 8, 43, 73, 76, 77, and 89 showed down-regulation of expression in parallel with the up-regulation of the expression of the other spots (Fig. 2b). To investigate the increase in photosynthetic rates caused by bacterial volatiles, a biochemical assessment of the levels of photosynthesis by measurement of chlorophyll content was carried out, which revealed significant increases in photosynthesis rates of up to 0.15 mg/g FW in bacterial VOC-treated plants compared to water control-treated plants (Fig. 3f).
2-DE analysis of Arabidopsis plants treated with bacterial volatiles. a Representative 2-DE gel images of water control (Control) and Arabidopsis plants treated with bacterial volatile emissions (GB03). b Graphical representation of differentially expressed proteins identified from bacterial volatile compound-treated Arabidopsis plants. The relative intensities of three replicates (±SD) are presented. All identified spots show statistically significant differences (P < 0.05) according to Student’s t test. The spot numbers are the same as those specified in a. Functional distribution of identified proteins: identified proteins were categorized based on their subcellular location (c); molecular function (d); and biological processes (e). Pie-chart values represent the percentage of the identified proteins
The activity of antioxidant enzymes in A. thaliana grown in the absence or presence of B. subtilis GB03. Enzyme activities were measured 14 days after B. subtilis GB03 treatment. Data represent means ± SE of three replicates. The activities of ascorbate peroxidase (a), catalase (b), glutathione reductase (c), dehydroascorbate reductase (d), and superoxide dismutase (e) are shown. f The chloroplast activity of 14-day-old A. thaliana seedlings grown on I plates with airborne exposure to bacterial strains and water treatment. The extraction method was described in the “Materials and methods” section. Asterisks indicate significant difference (P ≤ 0.05) between GB03 and control treatments
GB03-VOCs specific response proteins are listed in Table 1. The normalized volumes of spots were analyzed using the ANOVA test (P = 0.05) with three replications of water control and GB03 VOC-treated seedlings. The expression level of a total of 95 proteins derived from 2-D gel spots were altered following VOC treatment (61 spots up- and 34 spots down-regulated) (Table 1). The 95 spots were identified by MALDI-TOF/MS and peptide mass fingerprinting (PMF), and the sequenced proteins were identified using the dbEST database. A Blast search identified homologous proteins. The number of peptide matches and the ratio of peptide coverage are presented in Table 1. To increase sensitivity for identification, the sequenced peptides were analyzed using the ABI 4800 Plus MALDI-TOF/TOF analyzer (supplemental Table 1). The identified proteins were classified into three main groups according to subcellular location (8 types), molecular function (9 types), and biological process (8 types). Additionally, many proteins could not be identified. Within the groups of differentially expressed proteins, the subcellular location group was divided into 8 sub-groups, namely chloroplast (28%), apoplast (18%), chloroplast/plasma membrane (15%), mitochondrion (12%), cell wall (10%), cytosol (7%), peroxisome (6%), and others (4%) (Fig. 2c). The molecular function group was divided into nine sub-groups, namely binding proteins (34%), transferases (18%), lyases (15%), oxidoreductases (14%), isomerases (6%), transporters (4%), antioxidants (3%), structural molecules (2%), and others (4%). The spots corresponding to the biological processes group were divided into eight sub-groups, namely response to stimulus (32%), carbohydrate metabolism (18%), biosynthesis of plant hormones (17%), photosynthesis (10%), amino acid metabolism (10%), nitrogen metabolism (4%), oxidative stress (3%), and others (6%) (Fig. 2d, e). Within the listed analysis, focus was placed on the antioxidant protein/oxidative stresses and ethylene biosynthesis proteins on the basis of previous findings showing that the two most abundant bacterial VOCs in strain GB03, 2,3-butanediol and 3-hydroxy-2-butanone, played a major role in the elicitation of ISR against Erwinia carotovora subsp. carotovora. In the assessment of the regulatory control of ISR, several hormonal mutants, including ein2, npr1, NahG, and jar1, were tested, but only the ethylene-insensitive mutant (ein2) failed to elicit ISR, indicating that the induction of systemic resistance by bacterial volatile compounds occurs through the ethylene-dependent signaling pathway (Ryu et al. 2004a). It is noteworthy that the greatest one among the biological processes groups was “response to stimulus” as much as 32%, indicating that plant can recognize bacterial volatiles as an exogenous stimulus resulting in modulating protein expression (Fig. 2e).
Enzymatic activities of antioxidant proteins
Among the differentially expressed proteins responding to bacterial VOCs, antioxidants and oxidative stress-related proteins, including thioredoxin-dependent peroxidase 1 (spot 1), dehydroascorbate reductase (spot 11), Fe superoxide dismutase 1 (spot 16), glutathione S-transferase (spot 17), and structure of glutathione S-transferase (spot 19) were the most highly expressed. The changes in the intensity of spots were confirmed by measurement of antioxidants including superoxide dismutase (SOD), ascorbate peroxidase (APX), glutathione reductase (GR), catalase (CAT), dehydroascorbic acid reductase (DHAR), ascorbate, and glutathione (GSH) (Foyer et al. 1994). The expression level of two antioxidants, APX and CAT, which detoxify H2O2 to H2O, was significantly increased by 1.6- and 1.4-fold, respectively (Fig. 3a, b). GR expression in Arabidopsis leaves exposed to GB03-VOCs was up-regulated as much as 1.3-fold compared to the water control (Fig. 3c). DHAR, which converts dehydroascorbate to ascorbate using glutathione as a substrate, was increased by 2.4-fold (Fig. 3d). Intriguingly, the most active antioxidant, SOD, which converts ·O2 − to H2O2, showed the highest induction (eightfold) triggered by the emission of the strain GB03 VOCs compared to control (Fig. 3e). A previous microarray analysis revealed the up-regulation of expression of several antioxidant genes, including glutathione S-transferase (At3g02885, At1g78370, At3g03190, At2g30860, and At1g02930), peroxidase (At3g49110, At2g37130, At2g35380, and At5g15180), APX (At2g25080), SOD (At5g51100 and At5g23310), and M and H type thioredoxin (At1g52990, At1g04980, At1g08570, At1g11530, and At1g52990) at 72 h after volatile emission (Zhang et al. 2007). Although our previous results provided that bacterial volatiles play a major role in plant defense against pathogens (Ryu et al. 2004a), the involvement of reactive oxygen species (ROS) in ISR had not been reported. In this study, our proteome data suggest the potential role of ROS scavenging enzymes induced by bacterial volatile emission indicating that bacterial volatile emissions increased the levels of antioxidant enzymes. Besides induction of resistance against pathogens, ROS play an important role in abiotic stresses such as drought and high salt (Mittler 2002). Recently accumulated studies presented that PGPR also augmented plant resistance to abiotic stresses referred to as “induced systemic tolerance” (Yang et al. 2009b; Raudales et al. 2009). More interestingly, among bacterial determinants, two recent studies demonstrated that bacterial volatiles conferred resistance to salt and drought (Zhang et al. 2009b; Cho et al. 2008). Collectively, under severe stress condition, ROS production in plants can exceed the descavenging capacity and accumulate to level that can damage cell components (Mittler 2002). It seems that bacterial volatile could modulate the damaging level of ROS, thus symptoms of biotic and abiotic stress can be limited.
Role of ethylene in plant responses to bacterial volatiles
The gaseous hormone ethylene regulates a diversity of plant physiological functions and also serves as a defense mechanism against pathogen attacks (Bleecker and Kende 2000). Many contradictory reports exist on the role of ethylene in plant defense systems, including studies showing induction of plant defense or increases in susceptibility using diverse ethylene mutants and pathosystems (Van Loon et al. 2006). In the present study, bacterial VOCs induced key enzymes involved in ethylene biosynthesis as shown in Fig. 4a. The last step in the biosynthesis of ethylene from oxaloacetate requires four enzymes in Arabidopsis (Fig. 4a). Four enzymes that play a critical role in the production of ethylene are aspartate aminotransferase (NCBI ID: 531555, spot 57); aspartate-semialdehyde dehydrogenase precursor (NCBI ID: 11228579, spot 82); MAT3, methionine adenosyltransferase 3 (NCBI ID: 15228048, spot 52); and SAM-2, S-adenosylmethionine synthetase 2 (NCBI ID: 15234354, spot 90). These enzymes, which catalyze the conversion of oxaloacetate to l-aspartate, l-methionine, and S-adenosyl-l-methionine, were identified due to their differential expression induced by bacterial VOC emission. The expression of three enzymes, aspartate aminotransferase (NCBI ID: 531555); MAT3, methionine adenosyltransferase 3; and SAM-2, S-adenosylmethionine synthetase 2, was significantly up-regulated by 2.23, 2.51, and 2.11 fold, respectively, compared to water control treatment. Interestingly, the expression of aspartate-semialdehyde dehydrogenase precursor (NCBI ID: 11228579) was suppressed by 2.65-fold in Arabidopsis plants exposed to bacterial VOCs. Further experiments are needed to examine the possible epigenetic regulation of the aspartate-semialdehyde dehydrogenase precursor and to measure ethylene level after bacterial VOC treatment. S-adenosylmethionine synthetase (MAT, EC:2.5.1.6) is the enzyme that catalyzes the formation of S-adenosylmethionine (AdoMet) from methionine and ATP (Horikawa et al. 1990). AdoMet is an important methyl donor for transmethylation and is also the propylamino donor in polyamine biosynthesis that is strongly related to the induction of resistance to biotic and abiotic stresses (Horikawa et al. 1990) In bacteria, there is a single isoform of AdoMet synthetase (gene metK), while there are two in budding yeast and in mammals (genes SAM1 and SAM2), and in plants this enzyme is part of a multigene family. In addition to the effect on ethylene biosynthesis, bacterial VOC-mediated overexpression of four enzymes that process alpha-d-glucose-6-phosphate into acetyl Co-A led to the recruitment of high levels of the substrate acetyl Co-A for the production of ATP through the TCA cycle. This activity could be due to a requirement for increased plant growth triggered by bacterial VOCs (Ryu et al. 2003). The present work did not reveal the involvement of cytokinin-related proteins, as was reported in prior studies.
Ethylene biosynthesis and signaling in bacterial volatile compound-treated Arabidopsis plants. a Overview of ethylene biosynthesis and possible roles of up- or down-regulated proteins in bacterial volatile compound-exposed Arabidopsis plants. b Gene expression levels as determined by quantitative reverse transcriptase (qRT)-PCR. The expression ratio, a ratio of the expression in the B. subtilis GB03-inoculated treatment relative to the water-treated control, is shown as the mean ± SEM
To verify ethylene signaling, RT-PCR and qRT-PCR experiments were conducted to examine the transcriptional level of ET biosynthesis (SAM-2, ACS4, ACS12, and ACO2), ET response (ERF1, GST2, and CHIB), JA response (PDF1.2 and VSP1), and SA response genes (PR1 and FMO1). The gene expression of the early ET biosynthesis enzymes, SAM-2 and MAT1, was relatively higher than the other two downstream enzymes, ACS2 and ACO2. This result was in agreement with the observed protein expression pattern. The intensity of SAM-2 (spot no. 90) was significantly different between bacterial volatile and water control treatments. However, ACS2 and ACO2 proteins could not be detected by 2-DE analysis. Interestingly, the transcriptional level of ET-response genes was strongly up-regulated by bacterial volatiles. Furthermore, the up-regulation of ERF1, GST1, and CHIB expression strongly indicated the activation of ET signaling in bacterial volatile-emitted Arabidopsis plants. The induction of ethylene response genes occurred despite the undetectable levels of the last two enzymes in the ET production pathway. There are two possible explanations for this finding: (1) although no reports exist showing the induction of ET by a precursor of ET such as SAM, it is possible that SAM plays a critical role in eliciting ET signaling. This possibility should be tested in future studies. (2) The data obtained by RT-PCR revealed that the expression of the remaining genes, ACS2, ACS12, and ACO2, was slightly increased in the bacterial volatile-treated plants, suggesting that lower levels of ET can be produced. The low level of ET production could be enough to initiate ET signaling. To answer these questions, endogenous ET production should be measured. However, the current technology is not capable of detecting low levels of ET production. More interestingly, the JA response marker genes PDF1.2 and VSP2, which have been reported to cross-talk with the components of ET signaling pathways, were significantly up-regulated by as much as 11- and 8-fold compared with water controls. The induction of ET signaling is synchronized to JA signaling (Smith et al. 2009; Bostock 2005). Unexpectedly, the PR1 gene, which is an SA marker, was also increased by as much as 11-fold in bacterial volatile compound-treated samples. These results suggest that bacterial volatiles induce ET, JA, and SA signaling. However, SA–ET/JA signaling was antagonized, as reported in the many previous studies (Robert-Seilaniantz et al. 2007). More recently, the PGPR-elicited, ISR-inducing ET/JA signaling was reported to have a synergistic effect on SA-mediated induced resistance, referred to as systemic acquired resistance (SAR) (Van Wees et al. 2000).
Data obtained by microarray analysis indicated that ten auxin transport and response genes, including nitrilase 1 (At3g44310), auxin efflux carrier family protein (At2g17500), IAA-amino acid hydrolase 3 (At1g51760), auxin-responsive protein, which is putatively similar to auxin-inducible SAUR (Small Auxin Up RNAs) (At5g18060), and anthranilate phosphoribosyltransferase (At5g17990) were highly expressed and associated with an increase in the number of leaf epidermis cells (Zhang et al. 2007). Intriguingly, Arabidopsis plants treated with the same bacterial VOC showed an enhanced ISR against a necrotrophic bacterial pathogen, Erwinia carotovora subsp. carotovora (Ryu et al. 2004a). Recent reports on the contribution of auxin to plant defense mechanisms suggest that treatment with the biotrophic pathogen P. syringae results in negative cross-talk between the auxin signaling pathway and the SA signaling network (Navarro et al. 2006; Chen et al. 2007; Zhang et al. 2007). In contrast, another report showed that a null mutant for auxin signaling and biosynthesis had significantly reduced plant resistance to the necrotrophic fungi Botrytis cinerea and Plectosphaerella cucumerina (Llorente et al. 2008). SAR induced in an auxin biosynthesis mutant of Arabidopsis by pre-challenge with incompatible P. syringae pv. tomato DC3000 carrying avrRpm1 resulted in plants compromised in the establishment and maintenance of SAR, indicating the significant role of IAA or IAA-derivative compounds in the establishment of SAR (Truman et al. 2010). Of the nine up-regulated genes that were classified as other hormone genes at 72 h after bacterial VOC treatment, five genes were ethylene-inducible and ethylene-synthesis genes (At2g38230, At3g16050, At2g25450, At5g43450, and At4g16690) (Zhang et al. 2007). These results indicate that ethylene biosynthesis and inducible genes were activated by constitutive emission of bacterial VOCs, although the plant sample collection time points were different.
A study using Nicotiana attenuata as a model system and its isogenic transformants deficient in ET biosynthesis and reception demonstrated a regulation of the bacterial endophyte community, which resulted in the enhancement of bacterial diversity in wild-type and ethylene-signaling impaired plants (Long et al. 2010). The present results showed that the induction of proteins related to ethylene synthesis by VOC from the endophyte GB03 can be recruited by many species that are capable of eliciting ISR.
Conclusion
Bacterial VOCs have been reported to increase plant growth and elicit plant defenses. These results were obtained from assessment at the phenotypic and molecular level, rather than the enzymatic or protein level. To obtain solid evidence of bacterial VOC-mediated plant responses at the protein level, the present study used 2-DE and detected 95 differentially expressed protein spots. To assess the effect of bacterial VOCs on plant growth, the uptake of iron was assessed because of its critical role in plant metabolism and photosynthesis. Emission of B. subtilis GB03 VOC significantly increased shoot and root growth. This was attributed to the uptake of minerals such as S, Cu, and Fe in the root system, and to increased photosynthetic capacity, which was related to the up-regulation of plastid-specific proteins. Thus, bacterial VOCs induce an increase in mineral uptake for plant energy metabolism and photosynthesis. In the study of the induction of resistance, 7 antioxidant proteins and 20 ethylene biosynthesis-related proteins were selected for further characterization. The functional analysis of the proteins differentially expressed in response to bacterial VOCs revealed the up-regulation of proteins associated with gluconeogenesis and the TCA cycle, which would be necessary to generate the necessary energy to support the increased rate of plant growth. Furthermore, the overexpression of ethylene biosynthesis proteins belonging to the TCA cycle indicates an increase in the endogenous level of ethylene in the plant tissues. Additionally, antioxidant protein detection and enzymatic validation showed that bacterial VOCs conferred tolerance through the generation of ROS. Overall, the present results provide strong evidence of the orchestrated regulation of many plant traits related to both plant growth promotion and induced resistance at the protein level.
Abbreviations
- ET:
-
Ethylene
- ISR:
-
Induced systemic resistance
- JA:
-
Jasmonic acid
- MS:
-
Murashige and Skoog
- PGPR:
-
Plant growth-promoting rhizobacteria
- qRT-PCR:
-
Quantitative RT-PCR
- VOC:
-
Volatile organic compound
- SA:
-
Salicylic acid
References
Agrios GN (2005) Plant pathology. Elsevier Academic Press, pp 635
Anderson NL, Anderson NG (1998) Proteome and proteomics: new technologies, new concepts, and new words. Electrophoresis 19:1853–1861
Bleecker AB, Kende H (2000) Ethylene: a gaseous signal molecule in plants. Annu Rev Cell Dev Biol 16:1–18
Blum H, Beier H, Gross HJ (1987) Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 8:93–99
Bostock RM (2005) Signal crosstalk and induced resistance: straddling the line between cost and benefit. Annu Rev Phytopathol 43:545–580
Camehl I, Sherameti I, Venus Y, Bethke G, Varma A, Lee J, Oelmüller R (2010) Ethylene signalling and ethylene-targeted transcription factors are required to balance beneficial and nonbeneficial traits in the symbiosis between the endophytic fungus Piriformospora indica and Arabidopsis thaliana. New Phytol 185:1062–1073
Chen Z, Agnew JL, Cohen JD, He P, Shan L, Sheen J, Kunkel BN (2007) Pseudomonas syringae type III effector AvrRpt2 alters Arabidopsis thaliana auxin physiology. Proc Natl Acad Sci USA 104:20131–20136
Chen Z, McConkey BJ, Glick BR (2010) Proteomic studies of plant–bacteria interactions. Soil Biol Biochem. doi:10.1016/j.soilbio.2010.05.033 (in press)
Cho SM, Kang BR, Han SH, Anderson AJ, Park J-Y, Lee Y-H, Cho BH, Yang K-Y, Ryu C-M, Kim YC (2008) 2R, 3R-Butanediol, a bacterial volatile produced by Pseudomonas chlororaphis O6, is involved in induction of systemic tolerance to drought in Arabidopsis thaliana. Mol Plant Microbe Interact 21:1067–1075
Foyer CH, Lescure JC, Lefebvre C, Morot-Gaudry JF, Vincentz M, Vaucheret H (1994) Adaptations of photosynthetic electron transport, carbon Assimilation, and carbon partitioning in transgenic Nicotiana plumbaginifolia plants to changes in nitrate reductase activity. Plant Physiol 104:171–178
Glick BR (2005) Modulation of plant ethylene levels by the bacterial enzyme ACC deaminase. FEMS Microbiol Lett 251:1–7
Halliwell B, Foyer CH (1976) Ascorbic acid, metal ions and the superoxide radical. Biochem J 155:697–700
Hiscox JD, Israelstam GF (1979) A method for the extraction of chlorophyll from leaf tissue without maceration. Can J Bot 57:1332–1334
Horikawa S, Sasuga J, Shimizu K, Ozasa H, Tsukada K (1990) Molecular cloning and nucleotide sequence of cDNA encoding the rat kidney S-adenosylmethionine synthetase. J Biol Chem 265:13683–13686
Hossain MA, Asada K (1984) Purification of dehydroascorbate reductase from spinach and its characterization as a thiol enzyme. Plant Cell Physiol 25:85–92
Kalra YP (1998) Handbook of standard methods of plant analysis. CRC Press, Boca Raton, FL
Kandasamy S, Loganathan K, Muthuraj R, Duraisamy S, Seetharaman S, Thiruvengadam R, Ponnusamy B, Ramasamy S (2009) Understanding the molecular basis of plant growth promotional effect of Pseudomonas fluorescens on rice through protein profiling. Proteome Sci 7:47
Kato M, Shimizu S (1987) Chlorophyll metabolism in higher plants. VII. Chlorophyll degradation in senescing tobacco leaves: phenolic-dependent peroxidative degradation. Can J Bot 65:729–735
Kloepper JW, Leong J, Teintze M, Schroth MN (1980) Enhanced plant growth by siderophores produced by plant growth promoting rhizobacteria. Nature 286:885–886
Kloepper JW, Rodriguez-Kabana R, Zehnder GW, Murphy J, Sikora E, Fernandez C (1999) Plant root–bacterial interactions in biological control of soilborne diseases and potential extension to systemic and foliar diseases. Aust J Plant Pathol 28:27–33
Kloepper JW, Ryu CM, Zhang S (2004) Induced systemic resistance and promotion of plant growth by Bacillus spp. Phytopathology 94:1259–1266
Lee K, Kye M, Jang JS, Lee OJ, Kim T, Lim D (2004) Proteomic analysis revealed a strong association of a high level of alpha1-antitrypsin in gastric juice with gastric cancer. Proteomics 4:3343–3352
Llorente F, Muskett P, Sánchez-Vallet A, López G, Ramos B, Sánchez-Rodríguez C, Jordá L, Parker J, Molina A (2008) Repression of the auxin response pathway increases Arabidopsis susceptibility to necrotrophic fungi. Mol Plant 1:496–509
Long HH, Sonntag DG, Schmidt DD, Baldwin IT (2010) The structure of the culturable root bacterial endophyte community of Nicotiana attenuata is organized by soil composition and host plant ethylene production and perception. New Phytol 185:554–567
Loper JE, Schroth MN (1986) Influence of bacterial sources of indole-3-acetic acid on root elongation of sugar beet. Phytopathology 76:386–389
MacDonald EMS, Powell GK, Regier DA, Glass NL, Roberto F, Kosuge T, Morris RO (1986) Secretion of zwatin, ribosylzeatin and ribosyl-1″-methylzeatin by Pseudomonas savastanoi plasmid coded cytokinin biosynthesis. Plant Physiol 82:742–747
McCord JM, Fridovich I (1969) The utility of superoxide dismutase in studying free radical reactions. I. Radicals generated by the interaction of sulfite, dimethyl sulfoxide, and oxygen. J. Biol Chem 244:6056–6063
Miché L, Battistoni F, Gemmer S, Belghazi M, Reinhold-Hurek B (2006) Upregulation of jasmonate-inducible defense proteins and differential colonization of roots of Oryza sativa cultivars with the endophyte Azoarcus sp. Mol Plant Microbe Interact 19:502–511
Mittler R (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7:405–410
Nakano Y, Asada K (1981) Hydrogen peroxide is scavenged by ascorbate-specific peroxidase in spinach chloroplasts. Plant Cell Physiol 22:867–880
Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JD (2006) A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312:436–439
Raudales RE, Stone E, McSpadden Gardener BB (2009) Seed treatment with 2,4-diacetylphloroglucinol-producing pseudomonads improves crop health in low pH soils by altering patterns of nutrient uptake. Phytopathology 99:506–511
Robert-Seilaniantz A, Navarro L, Bari R, Jones JD (2007) Pathological hormone imbalances. Curr Opin Plant Biol 10:372–379
Ryu C-M, Farag MA, Hu CH, Reddy MS, Wei HX, Pare PW, Kloepper JW (2003) Bacterial volatiles promote growth in Arabidopsis. Proc Natl Acad Sci USA 100:4927–4932
Ryu C-M, Farag MA, Hu CH, Reddy MS, Kloepper JW, Paré PW (2004a) Bacterial volatiles induce systemic resistance in Arabidopsis. Plant Physiol 134:1017–1026
Ryu C-M, Murphy JF, Mysore KS, Kloepper JW (2004b) Plant growth-promoting rhizobacteria systemically protect Arabidopsis thaliana against Cucumber mosaic virus by a salicylic acid and NPR1-independent and jasmonic acid-dependent signaling pathway. Plant J 39:381–392
Ryu C-M, Kim J-W, Choi O-H, Park SY, Park SH, Park C-S (2005a) Nature of a root-associated Paenibacillus polymyxa from field-grown winter barley in Korea. J Microbiol Biotechnol 15:984–991
Ryu C-M, Hu C-H, Locy RD, Kloepper JW (2005b) Study of mechanisms for plant growth promotion elicited by rhizobacteria in Arabidopsis thaliana. Plant Soil 286:285–292
Ryu C-M, Kim J, Choi O, Kim SH, Park CS (2006) Improvement of biological control capacity of Paenibacillus polymyxa E681 by seed pelleting on sesame. Biol Cont 39:282–289
Segarra G, Casanova E, Bellido D, Odena MA, Oliveira E, Trillas I (2007) Proteome, salicylic acid, and jasmonic acid changes in cucumber plants inoculated with Trichoderma asperellum strain T34. Proteomics 7:3943–3952
Smith JL, De Moraes CM, Mescher MC (2009) Jasmonate- and salicylate-mediated plant defense responses to insect herbivores, pathogens and parasitic plants. Pest Manag Sci 65:497–503
Timmusk S, Nicander B, Granhall U, Tillberg E (1999) Cytokinin production by Paenibacillus polymyxa. Soil Biol Biochem 31:1847–1852
Truman WM, Bennett MH, Turnbull CG, Grant MR (2010) Arabidopsis auxin mutants are compromised in systemic acquired resistance and exhibit aberrant accumulation of various indolic compounds. Plant Physiol 152:1562–1573
Vallad GE, Goodman RM (2004) Systemic acquired resistance and induced systemic resistance in conventional agriculture. Crop Sci 44:1920–1934
Van Loon LC, Geraats BP, Linthorst HJ (2006) Ethylene as a modulator of disease resistance in plants. Trends Plant Sci 11:184–191
Van Wees SC, de Swart EA, van Pelt JA, van Loon LC, Pieterse CM (2000) Enhancement of induced disease resistance by simultaneous activation of salicylate- and jasmonate-dependent defense pathways in Arabidopsis thaliana. Proc Natl Acad Sci USA 97:8711–8716
Varotto C, Maiwald D, Pesaresi P, Jahns P, Salamini F, Leister D (2002) The metal ion transporter IRT1 is necessary for iron homeostasis and efficient photosynthesis in Arabidopsis thaliana. Plant J 31:589–599
Wang W, Tai F, Chen S (2008) Optimizing protein extraction from plant tissues for enhanced proteomics analysis. J Sep Sci 31:2032–2039
Yang JW, Yu SH, Ryu C-M (2009a) Priming of defense-related genes confers root-colonizing bacilli-elicited induced systemic resistance in pepper. Plant Pathol J 25:389–399
Yang J, Kloepper JW, Ryu C-M (2009b) Rhizosphere bacteria help plants tolerate abiotic stress. Trends Plant Sci 14:1–4
Zhang H, Kim MS, Krishnamachari V, Payton P, Sun Y, Grimson M, Farag MA, Ryu CM, Allen R, Melo IS, Paré PW (2007) Rhizobacterial volatile emissions regulate auxin homeostasis and cell expansion in Arabidopsis. Planta 226:839–851
Zhang H, Sun Y, Xie X, Kim M-S, Dowd SE, Paré PW (2009a) A soil bacterium regulates plant acquisition of iron via deficiency-inducible mechanisms. Plant J 58:568–577
Zhang H, Xie X, Kim MS, Kornyeyev DA, Holaday S, Paré PW (2009b) Soil bacteria augment Arabidopsis photosynthesis by decreasing glucose sensing and abscisic acid levels in planta. Plant J 56:264–273
Acknowledgments
This work was supported by an EB-NCRC grant (#R15-2003-012-02003-0) and the World Class University Program (#R32-10148) funded by MOEST, the 21C Frontier Microbial Genomics and Application Center Program, KRIBB initiative program, and partly by the Biogreen 21 program (20080401034023/20070401034005) funded by the Rural Development Administration, the Industrial Source Technology Development Program of the Ministry of Knowledge Economy (MKE) of Korea, and the National Academy of Agricultural Science, Rural Development Administration. Y.S.K. was supported by scholarships from the BK21 program funded by MOEST in Korea.
Author information
Authors and Affiliations
Corresponding author
Additional information
Y. S. Kwon and C.-M. Ryu contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kwon, Y.S., Ryu, CM., Lee, S. et al. Proteome analysis of Arabidopsis seedlings exposed to bacterial volatiles. Planta 232, 1355–1370 (2010). https://doi.org/10.1007/s00425-010-1259-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-010-1259-x