Abstract
This review covers recent advances in gibberellin (GA) signaling. GA signaling is now understood to hinge on DELLA proteins. DELLAs negatively regulate GA response by activating the promoters of several genes including Xerico, which upregulates the abscisic acid pathway which is antagonistic to GA. DELLAs also promote transcription of the GA receptor, GIBBERELLIN INSENSITIVE DWARF 1 (GID1) and indirectly regulate GA biosynthesis genes enhancing GA responsiveness and feedback control. A structural analysis of GID1 provides a model for understanding GA signaling. GA binds within a pocket of GID1, changes GID1 conformation and increases the affinity of GID1 for DELLA proteins. GA/GID1/DELLA has increased affinity for an F-Box protein and DELLAs are subsequently degraded via the proteasome. Therefore, GA induces growth through degradation of the DELLAs. The binding of DELLA proteins to three of the PHYTOCHROME INTERACTING FACTOR (PIF) proteins integrates light and GA signaling pathways. This binding prevents PIFs 3, 4, and 5 from functioning as positive transcriptional regulators of growth in the dark. Since PIFs are degraded in light, these PIFs can only function in the combined absence of light and presence of GA. New analyses suggest that GA signaling evolved at the same time or just after the plant vascular system and before plants acquired the capacity for seed reproduction. An analysis of sequences cloned from Physcomitrella suggests that GID1 and DELLAs were the first to evolve but did not initially interact. The more recently diverging spike moss Selaginella has all the genes required for GA biosynthesis and signaling, but the role of GA response in Selaginella physiology remains a mystery.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gibberellins (GAs) are cyclic diterpenoid molecules that were first identified as compounds that stimulate the growth and elongation of rice seedlings (Kurosawa 1926). We now know there is a large family of different GAs produced in plants from the precursor geranylgeranyl diphosphate, but typically only a few are active (Takahashi et al. 1991). Furthermore, the active GAs can be inactivated by the addition of methyl, hydroxyl, or other functional groups and localized inactivation is an important regulatory mechanism (Yamaguchi 2008). GAs affect nearly all aspects of higher plant growth and development, including germination, hypocotyl elongation, stem growth, reproductive, organ and seed development, and circadian and light regulation (Olszewski et al. 2002). However, it can be argued that GA signaling is not absolutely required for leaf, stem and root development, since Arabidopsis plants that lack GA can form small stature non-self-reproducing plants if seed coats are removed to promote germination. In addition, GA signaling is not found in the moss Physcomitrella (Hirano et al. 2007; Vandenbussche et al. 2007; Yasumura et al. 2007); so, some ancient plant cellular processes that have been conserved though evolutionary history do not require GA. On the other hand, if reproduction is considered a required trait, then for many plants GA signaling is necessary. In some plants, GA is required to induce flowering. GA is often necessary for petal, stamen and pollen development and can be required for seed development and germination. Perhaps GA signaling first arose in the fine-tuning of required functions and its role expanded as plants acquired new traits such as seed production.
Metabolism of gibberellin
One of the first committed steps in the GA biosynthesis pathway is the formation of ent kaurene by copalyl synthase and ent kaurene synthase (Koornneef and van der Veen 1980; Wilson et al. 1992). Arabidopsis plants that have a knockout mutation in the gene encoding copalyl synthase, ga1, are small, dark green, lack petal and stamen development, and do not flower under short day lengths (Koornneef and van der Veen 1980; Wilson et al. 1992). Application of GA precursors to these plants restores normal growth and development, confirming the role of copalyl synthase as the first committed step in this pathway. The production of active GAs involves GA 20-oxidases (GA20ox) and GA 3-oxidases (GA3ox) (reviewed in Yamaguchi 2008). In contrast, GA 2-oxidases (GA2ox) can inactivate most active GAs. The pools of active GAs are maintained through a number of feed-back and feed-forward mechanisms regulating expression of GA20ox and GA3ox and oppositely regulating GA2ox.
Light, temperature, GA, cytokinin, ethylene signaling, and spaciotemporal regulation all affect the transcriptional regulation of genes for GA biosynthesis, catabolism, and deactivation (reviewed in Yamaguchi 2008). This regulation affects the levels and types of GA species that are present, but variability also exists in the sensitivity of tissues to GA. The complex nature of GA metabolism and signaling is exemplified by a recent study of flowering in Lolium (King et al. 2008), and previous observations in rice (Sakamoto et al. 2001) and Arabidopsis (Jasinski et al. 2005). In Lolium, there are two GA2ox genes that are expressed in a band just below the stem apex (Fig. 1). These subapically localized GA2ox enzymes inactivate GA1 and GA4 as the GAs are translocated from leaves to the shoot apex. However, GA5 is resistant to inactivation by GA2ox and can promote flowering. Until spaciotemporal regulation of GA2ox deactivation was understood, the florigenic role of GA5 was surprising because GA1 and GA4 are more active GAs having higher binding affinity for the GA receptor. In fact, further down the stem, where GA2ox is not expressed, it is mainly GA1 and GA4 that promote stem elongation. Therefore, the localized expression of GA2ox results in growth promotion by GA1 and GA4 and floral induction by GA5. In addition, the two Lolium GA2ox genes are differentially expressed in short day versus long day growing conditions.
Model for how GA deactivating enzymes affect GA distribution in Lolium. GA1, GA4 and GA5 produced in the leaves are transported to the apex. In a band between 1 and 4 mm from the tip, the expression of GA2ox and 16, 17 epoxidase deactivates GA1 and GA4. GA5 is resistant to deactivation, and presence of GA5 at the tip is correlated with flowering
Another recent discovery in GA signaling is the identification of additional proteins that inactivate GAs by addition to, or modification of, the GA molecule. The rice ELONGATED UPPERMOST INTERNODE (EUI) protein epoxidates GAs and is expressed in seeds, seedlings, and just below the stem apex (Zhang et al. 2008; Zhu et al. 2006). It also affects starch granule development and gravitropism in roots. GA5, which is resistant to GA2ox, is also resistant to epoxidation by EUI. A similar type of epoxidation activity has been identified below the shoot apex in Lolium (King et al. 2008). Other forms of gibberellin modification have been reported, but further research is needed to understand how these modifications affect activity (Schneider and Schliemann 1994; Varbanova et al. 2007).
DELLA proteins
DELLA proteins are negative regulators of GA-induced growth. The short stature of plants that are deficient for GA is relieved by recessive mutations in genes coding for members of the DELLA protein family (Koornneef et al. 1985; Wilson and Somerville 1995; Peng et al.1997; Silverstone et al. 1997; Dill and Sun 2001; King et al. 2001; Tyler et al. 2004). There are five DELLA proteins in Arabidopsis (GAI, RGA, RGL1, RGL2, and RGL3) with some overlapping and some unique functions (Peng et al. 1997; Silverstone et al. 2001; Lee et al. 2002; Jiang and Fu 2007). Plants where three or more genes are nonfunctional appear to exert little if any restraint on GA-induced growth, and their phenotype is tall and spindly (Achard et al. 2006). Rice and barley each have one DELLA gene, SLENDER RICE 1 (SLR1) and SLENDER1 (SLR1), respectively. Mutations in either SLR1 result in a slender plant that is mostly resistant to the effects of GA application (Ikeda et al. 2001; Chandler et al. 2002).
GA receptor
The gene for the GA receptor, gibberellin insensitive dwarf 1 (gid1), was identified among a group of rice mutants that were insensitive to GA, but its function was initially unknown (Ueguchi-Tanaka et al. 2005). The gid1 mutant phenotype is also similar to that of GA deficient plants, as might be expected if a sole GA receptor were knocked out. Homology-based searches of the rice genome suggest that there is only one GID1 gene in rice (Ueguchi-Tanaka et al. 2005). In Arabidopsis there are three GID1-like genes, GID1a, b, and c (Griffiths et al. 2006). The Arabidopsis genes have significant functional overlap with some differences in expression level between tissues. GID1b was shown to have a higher binding affinity for GA using in vitro binding assays (Nakajima et al. 2006), but this binding affinity may not be relevant in vivo, since gid1b single mutant plants were not noticeably different than other single gid1 mutants (Griffiths et al. 2006). The triple mutant gid1a,b,c was very small and unable to reproduce. This plant is smaller than the ga1-3 biosynthesis mutant, suggesting that GID1 may contribute to plant development through a GA-independent pathway. Another explanation is that, the more extensive growth in ga1-3 plants is due to a small amount of GA that is produced in ga1-3 plants (Talón et al. 1990; Silverstone et al. 2001). The observation that the global transcript expression profile of GA deficient plants is nearly identical to that of gid1a,b,c provides additional support for the hypothesis that all GA signaling works through GID1 (Willige et al. 2007).
The GID1 gene was cloned and predicted to encode a protein of unknown function containing a domain with similarity to hormone sensitive lipase, yet lacking a critical residue for lipase function. The protein was expressed in E. coli and did not have lipase activity. However, GID1 bound active GA with greater affinity than it bound inactive GAs.
Mechanism of GID1/DELLA signaling
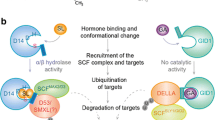
Ueguchi-Tanaka and colleagues have recently uncovered how the GID1 and DELLA proteins function in GA perception (Ueguchi-Tanaka et al. 2005, 2007a, b). Based on the structural homology between GID1 and hormone sensitive lipases (HSL), they propose that GID1 forms a receptor pocket that binds to GA. Upon binding GA, the GID1 protein undergoes a conformational change that allows interaction between the HSL-like “lid” of the GID1 receptor pocket and two of the DELLA protein domains, the signature “DELLA” domain and the THVYNP domain (Fig. 2). GA becomes enclosed within the pocket as the GID1 “lid” interacts with SLR1 (Fig. 3). Once the GA/GID1/DELLA complex is formed, binding to the F-box protein, SLEEPY1 (SLY1) is enhanced (Griffiths et al. 2006). This binding then results in the degradation of DELLAs through the 26S proteasome. The degradation of DELLAs relieves the growth suppression caused by DELLAs and results in GA-induced growth and other GA responses.
Domain structure of a typical DELLA protein based on RGA. Functional domains in the protein structure are in color with the name of each domain above or below the protein structure. The functional role of each domain is listed in italics above or below the domain name. DELLA, TVHYNP, poly serine/threonine/valine (pS/T/V), Leucine repeat (LR), and VHIID repeats were named for the consensus amino acids. See text for description of how domain functions were assigned and for SH2-like and SAW descriptions
Model for DELLA function in GA signaling. In the absence of GA, the DELLA protein is stable and acts as a transcriptional activator at promoters to turn on transcription of genes such as GID1 and Xerico that act as negative regulators of GA signaling (a). Many of the genes regulated by DELLAs contain promoter motifs known to be affected by ABA regulation, but this observation needs to be experimentally validated. DELLAs affect GID1 transcription directly, but their effects on GA biosynthesis and catabolism are indirect. The binding of GA to GID1, leads to further interaction with DELLAs, F-Box protein (FBP) and finally proteasomal degradation of DELLAs (b). Since DELLAs are required for transcription of DELLA regulated genes, these genes are not expressed in the absence of DELLAs. How DELLA dimerization affects function is unknown, but because dimerization domains are required for activity, DELLA dimers are depicted in the model. Hypothetical model for how posttranslational modification affects DELLA activity (c). DELLAs appear in multiple phosphorylated and non-phosphorylated forms, but it is not known whether phosphorylation is regulated. It is likely, but not necessary, that SPY modifies a dephosphorylated form of DELLA. O-GlcNAc modified DELLAs function as negative regulators of growth, possibly enhancing the interaction of DELLAs with DNA-binding proteins. DELLAs modified with O-GlcNAc interact with GA/GID1, F-Box protein and become degraded. It is possible that unmodified DELLAs have some activity (dotted line) in negatively regulating GA signaling since spy rga double mutants are more spindly than single mutants (Silverstone et al. 1997)
The GA/GID1/DELLA model helps to explain previous results, which were used to define the functional motifs within DELLA proteins. The DELLA proteins have several conserved domains that provide clues about how DELLAs function (Fig. 2). The first domain contains a 27-amino acid DELLA motif that has a role in GA response. Deletions within the DELLA domain cause a dwarf phenotype; the plants have a short stature that is not reversed by GA treatment (Koornneef et al. 1985; Peng et al. 1997, 1999; Dill et al. 2001; Boss and Thomas 2002; Chandler et al. 2002; Ikeda et al. 2002; Itoh et al. 2005b). Naturally occurring mutations resulted in DELLA domain deletions that were chosen by plant breeders when they created the short stature, high yielding, wheat, rice and maize cultivars that were part of the “green revolution” in twentieth century agriculture (Peng et al. 1999; Ogawa et al. 2000; Ikeda et al. 2001; Hedden 2003). Proteins with DELLA domain deletions cannot bind GID1, and are stable because they are not degraded as part of the DELLA/GID1 complex through the proteasome. These stable proteins still function as negative regulators of GA signaling restraining growth; however, in agricultural production, this partial growth restraint is advantageous, because it results in shorter plants that are high yielding and more resistant to lodging then previous varieties.
Additional studies examining natural DELLA domain deletions (Willige et al. 2007) or slr1 rice plants that were transformed with truncated SLR1 genes have helped to delineate the other functional domains of the DELLA proteins (Itoh et al. 2002, 2005b). As described above, the DELLA and TVHYNP domains function in GA signal recognition (Fig. 2; Itoh et al. 2002; Willige et al. 2007) by interacting with the lid of the GID1 receptor. The length of the “space” region between DELLA and TVHYNP is also important in GA signal recognition, but the amino acid sequence is not conserved (Itoh et al. 2002). Deletion of the pS/T/V domain creates a more active growth suppressor, suggesting that this is a regulatory domain that does not affect protein stability, and instead acts as a domain that regulates GA signal (Itoh et al. 2002). The leucine repeat (LR) and VHIID domains function in dimerization of the protein which is important for both GA perception and protein degradation (Itoh et al. 2002), but it is not known whether dimerization is required for interaction with GID1. The C-terminal domains of DELLAs interact with the F-box proteins, SLY1 of Arabidopsis, and the rice SLY1 homolog, GIBBERELLIN INSENSITIVE DWARF2 (GID2), leading to degradation of the DELLAs (Dill et al. 2004; Gomi et al. 2004). The binding of DELLAs to GID1 and GA must be essential to degradation, while the binding of DELLAs to F-box proteins is necessary but not sufficient for degradation. Proteins with DELLA domain mutations have an increased affinity for interaction with SLY1 but are stable and not degraded (Dill et al. 2004). Because DELLA deletion mutations inhibit protein degradation but continue to repress growth, it is inferred that other regions of the protein inhibit growth when they are activated by GA signaling.
The C-terminal domain of DELLA proteins is conserved among a larger group of proteins which all contain this domain, which is named GRAS [GAI, RGA and SCARECROW (SCR)]. There are approximately 33 GRAS domain-containing proteins in Arabidopsis that function in various aspects of growth and development and there is considerable variation in the presence of other motifs among these proteins (Pysh et al. 1999; Bolle 2004; Tian et al. 2004). The GRAS domain has two subdomains, SH2-like and SAW. Although neither were tested by deletion analysis, both are conserved among DELLA proteins, and mutations in these regions can affect the repressor function of the protein. Although the animal SH2-like domain binds phosphotyrosine-modified proteins, there is no evidence that any SH2-like domains are functional in plants (Richards et al. 2000). The function of the SAW domain is unknown. It is not clear whether the C-terminal domain that interacts with F-Box proteins is synonymous with the defined GRAS domain, whether GRAS domains always interact with F-box proteins, or whether GRAS domains have additional specialized functions. There is a separate rice GRAS protein, SLR1-like1 that is composed primarily of the C-terminal domain with a shortened pS/T/V and intact LR and VHIID domains (Itoh et al. 2005b). GA regulates the transcriptional expression of SLR1-like1, but not degradation of the protein. Itoh et al. (2005b) propose that this protein may act as a buffer against the effects of a sudden increase in GA level. This may be an important function as the Arabidopsis RGL1 is also resistant to GA-regulated degradation (Wen and Chang 2002).
DELLA activation
Understanding how DELLAs become activated is one of the most important issues for understanding GA signaling. It is unclear how GA activates DELLA while at the same time causing DELLA degradation through binding of GA/GID1/DELLA (Fig. 3). It has been proposed that these two separate functions are the result of GA acting on two different domains of the protein. Binding of GA/GID1/DELLA is dependent on the DELLA domain, while activation by GA is dependent on the pS/T/V domain. Transgenic plants that overexpress a DELLA lacking the pS/T/V domain remained responsive to GA and their protein stability was not affected suggesting that this domain is activated by GA but not affected in GA/GID1 binding and degradation (Itoh et al. 2002). However, transgenic plants containing the DELLAs lacking pS/T/V were shorter in stature than plants that overexpress the wild-type protein. Since these plants were shorter rather than taller, it was hypothesized that the pS/T/V site is subject to both positive and negative regulation via alternative posttranslational modification of serine and/or threonine amino acids with phosphorylation or the addition of N-acetyl glucosamine modification (Itoh et al. 2002). Such modification could determine DELLA protein interaction partners or localization of the protein. A further complication to the story is that degradation by the 26S proteasome typically involves phosphorylation of the protein that is to be degraded prior to interaction with F-Box proteins. Therefore, it is important to examine the posttranslational modification of specific amino acids within each domain of DELLA proteins.
To examine whether phosphorylation has any role in DELLA function, Hussain et al. (2005) compared the effects of serine/threonine kinase and phosphatase inhibitor treatments on stabilization of the Arabidopsis RGL2 DELLA protein in cultured tobacco cells. Surprisingly, their results suggested that RGL2 dephosphorylation, and not phosphorylation, is a necessary step in degradation. Next, they tested the effects of single serine/threonine to alanine substitutions on RGL2 stability. Six of these substitutions had greater stability than RGL2, but their GA repressor function was still active. Since GA signal repression was still functional, this result does not support the hypothesis that phosphorylation is key to the GA activation of DELLAs. The role of tyrosine phosphorylation in RGL2 stability was also tested, but the results were inconclusive (Hussain et al. 2007). The role of phosphorylation in rice SLR1 DELLA protein function is also unclear. Both phosphorylated and unphosphorylated forms had similar half-lives after GA application, and showed equal affinity toward the rice GID2 F-box protein (Itoh et al. 2005a). One possible explanation for these conflicting results is that these two SLR1 forms have differential protein interactions or that GA signaling differentially affects their subcellular localization. Another possible explanation for these puzzling results is that signaling may involve a different type of serine/threonine posttranslational modification. Modification with N-acetyl glucosamine (O-GlcNAc) to serine/threonine amino acids of substrate proteins can function alone or in competition with phosphorylation to affect protein function in animals (Hart et al. 2007). In some cases, the presence of either modification at a single serine or threonine can affect function, while sometimes the status of several sites affects function. The SPINDLY (SPY) protein was identified in a screen for negative regulators of GA signaling and functions as an O-GlcNAc transferase (Jacobsen et al. 1996; Thornton et al. 1999; Olszewski et al. 2002). Although the role of this modification in DELLA signaling has not been established, rice plants with reduced levels of SPY have increased levels of GA signaling and a change in the phosphorylation status of SLR1 protein (Shimada et al. 2006). SECRET AGENT is also an O-GlcNAc transferase and functions redundantly with SPY in some pathways, but has a minor, if any, role in GA signaling (Hartweck et al. 2006).
Additional experiments are needed to clarify whether posttranslational modification has a role in DELLA function, but a hypothetical model suggests a mechanism for DELLA modification and function (Fig. 3c). Several labs have shown that wild-type and mutant plants contain non-phosphorylated DELLAs as well as DELLAs with multiple phosphorylations (Itoh et al. 2002; Shimada et al. 2006; Silverstone et al. 2006; Hussain et al. 2007). The ratio does not change with GA application, but there are more phosphorylated or alternative mass forms when the levels of SPY protein are reduced (Shimada et al. 2006; Silverstone et al. 2007). It is possible that non-phosphorylated DELLAs have some suppressive activity without O-GlcNAc modification, or that it is a cumulative level of phosphorylation and O-GlcNAc modification that gives DELLAs suppressive activity; however, this model is speculative, as O-GlcNAc modification of DELLAs has not been shown.
Two other proteins of the GRAS family, SHORT ROOT (SHR) and SCR interact with each other to affect the subcellular localization of SHR and the transcription of SCR (Cui et al. 2007). The proteins interact through the LR and VHIID motifs that are also present in the DELLAs. It is known that the dimerization of DELLA proteins is important for their activity in GA signaling (Itoh et al. 2002). But it is an open question as to whether DELLA posttranslational modification affects dimerization or whether the formation of heterodimers between DELLAs and/or other GRAS proteins affect the GA pathway.
GA-regulated transcriptional networks
To learn more about how GA affects growth and development, global transcript profiling has been used to identify genes that are regulated by GA. To discriminate between genes that are directly regulated by GA and those that change due to secondary effects, Ogawa et al. (2003) performed a time course with wild-type or gibberellin deficient germinating Arabidopsis seeds that had been treated with GA or a control. The transcriptional levels of only a small number of genes (19) appeared to be affected by GA application at either 1 or 3 h. This observation that only a few genes are affected by GA application has been independently verified by researchers performing experiments involving Arabidopsis seedlings (Nemhauser et al. 2006; Zentella et al. 2007) and rice aleurone cells (Bethke et al. 2006). Many of the identified genes encode GA biosynthesis enzymes and GID receptors, suggesting that GA homeostasis is of major importance. The small number of identified genes suggests that GA works mainly through changes in only a few genes with large effects, through many genes with changes too small or localized to measure, and/or through changes that could not be observed in transcript profiling, such as posttranscriptional changes to existing proteins.
Since DELLAs appear to regulate most GA responses, another way to identify GA-regulated genes is to identify genes regulated by DELLAs. In an examination of GA deficient plants lacking full DELLA protein expression Cao et al. (2006) found that 40–60% of the 1,208 GA seed- and flower-regulated genes are also controlled by DELLAs. This larger number of genes reflects both primary and secondary effect transcripts. Most of the co-regulated genes were unique to either seeds or flowers. In an independent analysis, Zentella et al. (2007) used an inducible DELLA with a deletion in the DELLA domain (rga-Δ17) which stabilized the protein and resulted in dwarf GA-resistant plants. This inducible construct was used to identify genes that are primary targets of DELLAs and to avoid secondary effects of DELLAs on transcription. After induction and harvesting of 8-day-old seedlings, a small number of genes (14) were identified that are regulated by both GA and DELLA. At least 12 of these genes were not tissue specific because they were found in seedlings (Zentella et al. 2007) and also in seed and flower data sets (Cao et al. 2006). In support of the model that DELLAs are negative regulators of GA signaling, all were either GA induced and DELLA repressed (Cao et al. 2006) or GA repressed and DELLA induced (Cao et al. 2006; Zentella et al. 2007). The functional categories of the 14 co-regulated genes include GA biosynthesis/signaling, transcriptional regulation, protein binding, or protein degradation (Cao et al. 2006; Zentella et al. 2007). The remaining co-regulated genes identified by Cao et al. (2006) function in metabolism; have DNA/RNA, ion, or protein binding motifs; or have functions in biotic or abiotic stress. In another experiment to identify immediate DELLA transcriptional targets within flowers, DELLA transcription was induced at the same time as cyclohexamide was applied (Hou et al. 2008). Although most of the >700 identified genes were unique, the functional category profiles were similar with previous experiments (Hou et al. 2008). Two of the identified DELLA targets were also identified by Cao et al. (2006), but none were identified by Zentella et al. (2007), again suggesting that DELLAs regulate different genes at different times and places. One direct DELLA target encodes a glutaredoxin-like protein that when over-expressed had floral phenotypes similar to rga-Δ17 plants (Hou et al. 2008). Similar to the results of Zentella et al. (2007), only a portion of the genes affected by DELLAs were previously identified as being affected by GA (120 of 806; Hou et al. 2008). Whether all GA signaling functions through DELLAs as suggested by mutant studies is not clear. As mentioned above, Cao et al. (2006) found that only 40–60% of GA-regulated genes are also regulated by DELLAs. However, Zentella et al. (2007) found experimental difficulties assessing the relative changes in transcription levels compared to controls, and they propose that additional experiments will be needed to answer this question.
It has been postulated that DELLAs function as transcriptional repressors of genes involved in GA signaling (Olszewski et al. 2002). To test this hypothesis, Zentella et al. (2007) performed a chromatin immunoprecipitation (ChIP) experiment with a RGA tandem affinity tagged fusion protein and performed PCR to determine whether RGA was associated with the promoters of any of the DELLA and GA co-regulated genes identified by microarray analysis. Eight appeared to be transcriptional targets of RGA, but additional experiments such as electrophoretic mobility shift assays will be needed to test whether RGA binds DNA directly, or through interactions with other proteins. RGA activates transcription of these targets which are expected to be negative regulators of GA signaling. Interestingly, the putative promoter regions of GID1 genes, but not GA biosynthesis genes, were enriched by ChIP, demonstrating a complexity in feedback regulation that needs to be understood in greater detail. One of the targets of RGA is Xerico, a gene whose overexpression leads to upregulation of ABA levels. This exciting result suggests that one of the ways that DELLAs act as negative regulators of GA is by activating the ABA pathway that antagonizes GA signaling.
Interaction of GA and light signaling pathways
Light affects gibberellin biosynthesis, catabolism and GA response in numerous ways (Olszewski et al. 2002; Yamaguchi 2008). Recent experiments describe some of the ways light and gibberellin signaling converge to affect seedling growth (Alabadi et al. 2008; de Lucas et al. 2008; Feng et al. 2008). During Arabidopsis germination, the seed imbibes water and the root radical begins to grow, rupturing first the endosperm and then the testa. In darkness, seedlings demonstrate etiolated growth: the hypocotyl elongates, the cotyledons remain closed and form a hook until the seedling senses light. PHYTOCHROME INTERACTING FACTOR (PIF) proteins bind to promoter elements and activate the transcription of genes needed for etiolated growth. In light, hypocotyl elongation slows, the cotyledons open, and photosynthesis begins. One of the seedling light sensors is PHYTOCHROME B (PHYB). Upon illumination, PHYB translocates from the cytoplasm to the nucleus. In the nucleus PHYB and PIFs interact and are degraded. Without PIF, the genes needed for etiolated growth in the dark are turned off. PIFs typically have three conserved domains: a phytochrome interacting domain, a domain that binds DNA and a separate domain that contains a basic helix–loop–helix domain that can function in protein–protein interactions. Recent results show that DELLA proteins bind with PIF3, 4, and 5 and interfere with PIF binding at the promoters of genes for growth in the dark (de Lucas et al. 2008; Feng et al. 2008). GA promotes growth by causing the destruction of DELLAs though the GA/GID1/DELLA interaction, leaving PIF proteins to promote growth through transcriptional regulation (Fig. 4). Therefore, PIF-regulated etiolated growth requires both the absence of light and the presence of GA. In addition, light signaling through CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1), an E3 ubiquitin ligase, which is part of a light regulated proteasome complex, and GA signaling through DELLAs effect the accumulation of LONG HYPOCOTYL 5 (HY5), a basic leucine zipper transcription factor that regulates photomorphogenesis (Alabadi et al. 2008).
Model of the interaction between GA and light signaling pathways. In the absence of GA and light, DELLA and PIF proteins are stable and bind together (a). This prevents PIF protein binding to promoters of genes that promote elongation growth. In the presence of GA, binding of the GA receptor, GID1, leads to interaction with DELLAs, F-Box protein (FBP) and proteasomal degradation of DELLAs (b). In the absence of DELLAs, PIF proteins bind to promoters of genes that promote elongation growth. With combined light and GA signaling, PIFs and DELLAs are degraded, and the PIF-mediated promotion of growth is abolished (c). Note that these processes are not all-or-nothing. The levels of GA and light regulate the amount of elongation growth through PIF and DELLA proteins
PIF genes may have additional functions in GA signaling as well. The PIF1/PIL5 protein binds to G-box elements within RGA and GAI promoters and induces their transcription (Oh et al. 2007). An interesting question is whether PIF1 protein binds RGA or GAI protein. If so, it would create a negative feedback loop, whereby increased transcription/translation of RGA/GAI would provide increased RGA/GAI protein to bind PIF1 in the dark. It is known that PIF1 indirectly decreases the levels of active GAs, and increases the levels of ABA, a plant hormone that can function antagonistically to GA (Oh et al. 2007). It is not known how this indirect action is achieved. There are a number of other related PIF genes in the Arabidopsis genome that may act through DELLA proteins in diverse processes throughout the plant (Castillon et al. 2007). It will be important to discover whether the PIF/DELLA example can serve as a general model to explain how DELLA proteins function indirectly as transcription factors.
Interaction of GA and other hormone signaling pathways
A number of physiological studies have indicated significant crosstalk between GA and other plant hormones such as abscisic acid (ABA), auxin, cytokinin, and ethylene. However, some microarray experiments suggest that there is very little crosstalk between hormones (Nemhauser et al. 2006). Some explanations for this inconsistency are discussed below as the interactions between GA and other hormones are reviewed.
The relationship of ABA and GA highlights some of the difficulties in determining how hormones interact in plants. ABA and GA can have directly antagonistic effects, where each negatively regulates the transcription of the biosynthetic genes of the other (Gomez-Cadenas et al. 2001; Oh et al. 2007; Zentella et al. 2007; Gubler et al. 2008; Sawada et al. 2008; Toh et al. 2008). This direct effect on hormone levels can make it difficult to distinguish whether a signaling pathway influences only the biosynthesis of the other or whether the signaling pathway itself interacts with the signaling pathway of the other. Support that the two signaling pathways interact with each other comes from evidence that GA and ABA have antagonistic effects on the promoters of specific genes (Gomez-Cadenas et al. 2001; Xie et al. 2006; Oh et al. 2007; Zentella et al. 2007; Johnson et al. 2008; Sawada et al. 2008). Furthermore, global transcript profiling in both Arabidopsis and cereals confirms that many GA-regulated genes contain both GA- and ABA-responsive promoter elements (Ogawa et al. 2003; Cao et al. 2006; Xie et al. 2006; Sreenivasulu et al. 2008). Additionally, the evidence that RGA affects transcription of Xerico which affects ABA levels demonstrates crosstalk between the signaling pathways (Zentella et al. 2007). However, when hormone treatments were applied to embyroless rice seeds (Bethke et al. 2006) or when global changes to applied hormones were analyzed in Arabidopsis (Nemhauser et al. 2006), very few genes were regulated by both GA and ABA. One explanation for these conflicting results is that, microarray analysis fails to identify the many subtle transcript changes that are involved in crosstalk between hormones. An alternative explanation is that, crosstalk is rare and that DNA-based searching for regulatory promoter elements leads to a high number of false positive results.
Auxin can positively affect GA biosynthesis (Fu and Harberd 2003; Wolbang et al. 2007) and application of GA can positively affect auxin biosynthesis (Nemhauser et al. 2006; Bjorklund et al. 2007; Desgagne-Penix and Sponsel 2008). Conversely, removal of auxin through decapitation of the apex, or interfering with the auxin signal pathway results in lower levels of GA in Populus (Bjorklund et al. 2007), Pisum sativum (Ross et al. 2000) and Nicotiana tabacum (Wolbang and Ross 2001). Just as with ABA, the effects of GA and auxin on both biosynthesis and signaling can be confounding. GA signaling through DELLA proteins is mediated by auxin (Fu and Harberd 2003). Fu and Harberd (2003) showed that the GA-induced degradation of DELLA proteins is inhibited in plants with reduced polar auxin transport; therefore, auxin affects both GA biosynthesis and GA signal responsiveness. Global transcript profiling in Populus stems suggests that there is a great deal of overlap between GA and auxin signaling (Bjorklund et al. 2007). Over 95% of the transcriptional changes associated with GA application were included among the changes induced by IAA application. If this is generally true in other plants and organs, then it is important to note that auxin response appears to be under circadian regulation (Covington and Harmer 2007). Given the fact that much of the transcriptome is affected by the clock (Michael et al. 2008), as well as that many hormone-specific pathways demonstrate some circadian patterning (Mizuno and Yamashino 2008), future studies should view the interaction between auxin and GA within a circadian context.
GAs and cytokinins also influence each other through both biosynthesis and response pathways (Weiss and Ori 2007). The interaction of these hormones together with auxin is particularly important for understanding apical meristem differentiation (Shani et al. 2006). KNOTTED1-like homeodomain protein family members (KNOX) are needed to maintain a central nondifferentiating stem cell zone below the apex and to create peripheral regions where incipient leaves emerge by cell differentiation. Within the central zone, KNOX genes repress GA20ox, induce GA2ox (Sakamoto et al. 2001; Hay et al. 2002; Chen et al. 2004) and induce cytokinin biosynthesis gene expression (Rupp et al. 1999; Hamant et al. 2002; Jasinski et al. 2005; Yanai et al. 2005), leading to decreased GA and increased cytokinin levels. Polar auxin transport results in the creation of a site of high auxin concentration in the peripheral zone of the meristem where the incipient leaf will emerge (Heisler et al. 2005). When polar auxin transport is elevated, KNOX gene expression is down-regulated and GA levels increase and cytokinin levels decrease (Hay et al. 2002). This example highlights the need to understand hormone crosstalk in a specific time and place. In this case, global transcript analysis would be insufficient for understanding how GAs and cytokinins interact within the meristem.
Application of an ethylene precursor or increased ethylene signaling reduces endogenous levels of GAs through changes in transcription levels of biosynthesis genes and changes in DELLA protein stability (Achard et al. 2007; Vandenbussche et al. 2007). The formation of an apical hook during etiolated growth is regulated by ethylene and auxin, and is dependant on GA signaling through DELLA proteins (Achard et al. 2003). Both hormones promote hypocotyl growth in the light (Peng and Harberd 1997; Smalle et al. 1997; De Grauwe et al. 2007). The influence of ethylene on flowering is also dependent on DELLA proteins (Vriezen et al. 2004; Achard et al. 2007; De Grauwe et al. 2008;). A global transcript profiling analysis of the genes induced by GA application to either wild-type or ethylene response mutant plants suggests that ethylene and GA can have opposite, similar, or differential effects on the regulation of specific genes (De Grauwe et al. 2007). A recent tissue-specific transcript meta-analysis suggests that ethylene and GA have different zones of influence within root meristems (Dugardeyn et al. 2008). GA affects the apical tip of the root which contains the meristemic and elongation zones, and ethylene and auxin influence the elongation and differentiation zones where root hairs form and elongate. These observations underscore our need to have a more detailed picture of hormone biosynthesis and signaling in space, circadian time, and developmental time. This is particularly true given the recent observations that localized expression of GA inactivation genes can influence flowering as described above.
Evolution of the GA receptor/response system
Three recent papers have explored the evolution of the GA receptor/response system in plants (Hirano et al. 2007; Vandenbussche et al. 2007; Yasumura et al. 2007). The presence of homologs of GID1 and DELLAs in both Physcomitrella and Selaginella suggests an early evolutionary origin for the proteins (Hirano et al. 2007; Yasumura et al. 2007). Physcomitrella is a Bryophyte (moss) and is less related to modern flowering plants than Selaginella, a Lycophyte (spike moss) which is one of the first lineages of vascular plants to diverge from the flowering plant lineage (Kenrick and Crane 1997; Rensing et al. 2008).
Physcomitrella patens has homologs of DELLA and GID1 genes, but the proteins do not interact or functionally substitute for their flowering plants homologs (Fig. 5; Hirano et al. 2007; Yasumura et al. 2007). However, the DELLA and GID1 proteins of Selaginella moellendorffii were able to functionally substitute for SLR1 and GID1 in rice (Hirano et al. 2007) and the Selaginella kraussiana DELLA repressed growth in Arabidopsis (Yasumura et al. 2007). S. moellendorffii also contained functional homologs of GID2/SLY1 F-box proteins and GA20ox and GA3ox (Hirano et al. 2007). It seems that S. moellendorffii has a functional GA biosynthesis pathway. Some of the S. moellendorffii GA biosynthesis enzymes were cloned and expressed in E. coli and shown to be able to catalyze the synthesis of GA4 from precursors. An active species of GA, GA4, was detected in S. moellendorffii tissues. Application of active GA to S. moellendorffii caused down regulation of the GA receptor and biosynthesis genes and degradation of DELLA proteins suggesting that S. moellendorffii also has a functional GA response pathway providing feedback response (Hirano et al. 2007). However, GA application did not appear to have a measurable effect on the physiological responses in either S. moellendorffii or S. kraussiana (Hirano et al. 2007; Yasumura et al. 2007). Uniconizole, a GA biosynthesis inhibitor, reduced the growth of S. moellendorffii, but application of GA4 did not restore growth. These observations lead to the conclusion that DELLA and GID1 proteins were the first elements of the GA signaling system to evolve. The presence of functional components of the GA signaling system in Lycophytes suggests that GA signaling was acquired during or after divergence of vascular plants from the moss lineage. It will be important to determine whether GA signaling has any role in the Selaginella life cycle, and to understand how the GA signaling system evolved to affect the growth, development and reproduction of plants.
Proposed evolution of GA signaling and response pathway in plants. Before the divergence of Physcomitrella, GID1 and DELLA proteins were present, but were incapable of binding to each other. Sometime after Physcomitrella divergence but before Selaginella divergence, GA/GID1/DELLA complexes could form and species such as Selaginella could produce GA4. Although GA-regulated transcriptional feedback control of GA20ox, GA3ox and GID1 suggests that GA response is functional in Selaginella, application of GA had no effect on Selaginella physiology. Whether GA has any physiological effect in Selaginella or when GA began to have a role in plant growth and development is unknown
Conclusion
Recent advances in our understanding of the gibberellin receptor have suggested a relatively short signaling system composed of the receptor and the DELLA negative regulators. DELLA proteins function as partners in protein–protein interactions to activate or repress transcription. Upon receipt of the GA signal, GID1 binds to DELLAs and then DELLAs undergo degradation through the proteasome. GA signaling also activates DELLAs, but it is not known how activation is achieved. Many new questions are emerging from our improved understanding of GA signaling in plants:
-
Does the localized deactivation of GAs occur in tissues other than the band just below the flowering apex, and do other types of GA deactivation have a role in development?
-
Do GID1, DELLAs, and F-Box proteins have any regulated subcellular localization patterns?
-
Is DELLA dimerization important for interaction with other proteins and do DELLAs heterodimerize?
-
Why is RGL1 not destabilized by GA?
-
How are DELLAs activated?
-
Does all GA signaling occur through DELLAs?
-
Do DELLAs bind to all members of the PIF family? Do DELLAs bind other proteins containing basic helix–loop–helix domains? Does PIF1 bind RGA or GAI?
-
Is there circadian regulation of DELLAs or GID1?
-
How extensive is hormone crosstalk?
-
Are there specific spaciotemporal expression patterns in GA signaling that are undiscovered?
-
Does GA have any role in the life cycle of Selaginella? How did GA response evolve?
Abbreviations
- GA:
-
Gibberellin
- ABA:
-
Abscisic acid
- ChIP:
-
Chromatin immunoprecipitation
References
Achard P, Vriezen WH, Van Der Straeten D, Harberd NP (2003) Ethylene regulates Arabidopsis development via the modulation of DELLA protein growth repressor function. Plant Cell 15:2816–2825
Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, Van Der Straeten D, Peng J, Harberd NP (2006) Integration of plant responses to environmentally activated phytohormonal signals. Science 311:91–94
Achard P, Baghour M, Chapple A, Hedden P, Van Der Straeten D, Genschik P, Moritz T, Harberd NP (2007) The plant stress hormone ethylene controls floral transition via DELLA-dependent regulation of floral meristem-identity genes. Proc Natl Acad Sci USA 104:6484–6489
Alabadi D, Gallego-Bartolome J, Orlando L, Garcia-Carcel L, Rubio V, Martinez C, Frigerio M, Iglesias-Pedraz JM, Espinosa A, Deng XW, Blazquez MA (2008) Gibberellins modulate light signaling pathways to prevent Arabidopsis seedling de-etiolation in darkness. Plant J 53:324–335
Bethke P, Hwang Y, Zhu T, Jones R (2006) Global patterns of gene expression in the aleurone of wild-type and dwarf1 mutant rice. Plant Physiol 140:484–498
Bjorklund S, Antti H, Uddestrand I, Moritz T, Sundberg B (2007) Cross-talk between gibberellin and auxin in development of Populus wood: gibberellin stimulates polar auxin transport and has a common transcriptome with auxin. Plant J 52:499–511
Bolle C (2004) The role of GRAS proteins in plant signal transduction and development. Planta 218:683–692
Boss PK, Thomas MR (2002) Association of dwarfism and floral induction with a grape ‘green revolution’ mutation. Nature 416:847–850
Cao D, Cheng H, Wu W, Soo HM, Peng J (2006) Gibberellin mobilizes distinct DELLA-dependent transcriptomes to regulate seed germination and floral development in Arabidopsis. Plant Physiol 142:509–525
Castillon A, Shen H, Huq E (2007) Phytochrome interacting factors: central players in phytochrome-mediated light signaling networks. Trends Plant Sci 12:514–521
Chandler PM, Marion-Poll A, Ellis M, Gubler F (2002) Mutants at the Slender1 locus of ‘Himalaya’ barley: molecular and physiological characterization. Plant Physiol 129:181–190
Chen H, Banerjee AK, Hannapel DJ (2004) The tandem complex of BEL and KNOX partners is required for transcriptional repression of ga20ox1. Plant J 38:276–284
Covington MF, Harmer SL (2007) The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS Biol 5:e222
Cui HLM, Vernoux T, Jung JW, Paquette AJ, Gallagher KL, Wang JY, Blilou I, Scheres B, Benfey PN (2007) An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science 316:421–425
De Grauwe L, Vriezen WH, Bertrand S, Phillips A, Vidal AM, Hedden P, Van Der Straeten D (2007) Reciprocal influence of ethylene and gibberellins on response-gene expression in Arabidopsis thaliana. Planta 226:485–498
De Grauwe L, Chaerle L, Dugardeyn J, Decat J, Rieu I, Vriezen WH, Baghour M, Moritz T, Beemster GT, Phillips AL, Harberd NP, Hedden P, Van Der Straeten D (2008) Reduced gibberellin response affects ethylene biosynthesis and responsiveness in the Arabidopsis gai eto2-1 double mutant. New Phytol 177:128–141
de Lucas M, Daviere JM, Rodriguez-Falcon M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blazquez MA, Titarenko E, Prat S (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451:480–484
Desgagne-Penix I, Sponsel VM (2008) Expression of gibberellin 20-oxidase1 (AtGA20ox1) in Arabidopsis seedlings with altered auxin status is regulated at multiple levels. J Exp Bot 59:2057–2070
Dill A, Sun TP (2001) Synergistic de-repression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159:777–785
Dill A, Jung H-S, Sun TP (2001) The DELLA motif is essential for gibberellin-induced degradation of RGA. Proc Natl Acad Sci USA 98:14162–14167
Dill A, Thomas SG, Hu J, Steber CM, Sun TP (2004) The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16:1392–1405
Dugardeyn J, Vandenbussche F, Van Der Straeten D (2008) To grow or not to grow: what can we learn on ethylene-gibberellin cross-talk by in silico gene expression analysis? J Exp Bot 59:1–16
Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, Schafer E, Fu X, Fan LM, Deng XW (2008) Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451:475–479
Fu X, Harberd NP (2003) Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature 421:740–743
Gomez-Cadenas A, Zentalla R, Walker-Simmons M, Ho TH (2001) Gibberellin/abscisic acid antagonism in barley aleurone cells: site of action of the protein kinase PKABA1 in relation to gibberellin signaling molecules. Plant Cell 13:667–679
Gomi K, Sasaki A, Itoh H, Ueguchi-Tanaka M, Ashikari M, Kitano H, Matsuoka M (2004) GID2, an F-box subunit of the SCF E3 complex, specifically interacts with phosphorylated SLR1 protein and regulates the gibberellin-dependent degradation of SLR1 in rice. Plant J 37:626–634
Griffiths J, Murase K, Rieu I, Zentella R, Zhang ZL, Powers SJ, Gong F, Phillips AL, Hedden P, Sun TP, Thomas SG (2006) Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18:3399–3414
Gubler F, Hughes T, Waterhouse P, Jacobsen J (2008) Regulation of dormancy in barley by blue light and after-ripening: effects on abscisic acid and gibberellin metabolism. Plant Physiol 147:886–896
Hamant O, Nogue F, Belles-Boix E, Jublot D, Grandjean O, Traas J, Pautot V (2002) The KNAT2 homeodomain protein interacts with ethylene and cytokinin signaling. Plant Physiol 130:657–665
Hart GW, Housley MP, Slawson C (2007) Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature 446:1017–1022
Hartweck LM, Genger RK, Grey WM, Olszewski NE (2006) SECRET AGENT and SPINDLY have overlapping roles in the development of Arabidopsis thaliana L. Heyn. J Exp Bot 57:865–875
Hay A, Kaur H, Phillips A, Hedden P, Hake S, Tsiantis M (2002) The gibberellin pathway mediates KNOTTED1-type homeobox function in plants with different body plans. Curr Biol 12:1557–1565
Hedden P (2003) The genes of the Green Revolution. Trends Genet 19:5–9
Heisler MG, Ohno C, Das P, Sieber P, Reddy GV, Long JA, Meyerowitz EM (2005) Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr Biol 15:1899–1911
Hirano K, Nakajima M, Asano K, Nishiyama T, Sakakibara H, Kojima M, Katoh E, Xiang H, Tanahashi T, Hasebe M, Banks JA, Ashikari M, Kitano H, Ueguchi-Tanaka M, Matsuoka M (2007) The GID1-mediated gibberellin perception mechanism is conserved in the Lycophyte Selaginella moellendorffii but not in the Bryophyte Physcomitrella patens. Plant Cell 19:3058–3079
Hou X, Hu W-W, Shen L, Lee L, Tao Z, Han J-H, Yu H (2008) DELLA target genes in flower development. Plant Physiol (in press)
Hussain A, Cao D, Cheng H, Wen Z, Peng J (2005) Identification of the conserved serine/threonine residues important for gibberellin-sensitivity of Arabidopsis RGL2 protein. Plant J 44:88–99
Hussain A, Cao D, Peng J (2007) Identification of conserved tyrosine residues important for gibberellin sensitivity of Arabidopsis RGL2 protein. Planta 226:475–483
Ikeda A, Ueguchi-Tanaka M, Sonoda Y, Kitano H, Koshioka M, Futsuhara Y, Matsuoka M, Yamaguchi J (2001) slender rice, a constitutive gibberellin response mutant is caused by a null mutation of the SLR1 gene, an orthologue of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13:999–1010
Ikeda A, Sonoda Y, Vernieri P, Perata P, Hirochika H, Yamaguchi J (2002) The slender rice mutant, with constitutively activated gibberellin signal transduction, has enhanced capacity for abscisic acid level. Plant Cell Physiol 43:974–979
Itoh H, Ueguchi-Tanaka M, Sato Y, Ashikari M, Matsuoka M (2002) The gibberellin signaling pathway is regulated by appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14:57–70
Itoh H, Sasaki A, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Hasegawa Y, Minami E, Ashikari M, Matsuoka M (2005a) Dissection of the phosphorylation of rice DELLA protein, SLENDER RICE1. Plant Cell Physiol 46:1392–1399
Itoh H, Shimada A, Ueguchi-Tanaka M, Kamiya N, Hasegawa Y, Ashikari M, Matsuoka M (2005b) Overexpression of a GRAS protein lacking the DELLA domain confers altered gibberellin responses in rice. Plant J 44:669–679
Jacobsen SE, Binkowski KA, Olszewski NE (1996) SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc Nat Acad Sci USA 93:9292–9296
Jasinski S, Piazza P, Craft J, Hay A, Woolley L, Rieu I, Phillips A, Hedden P, Tsiantis M (2005) KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr Biol 15:1560–1565
Jiang C, Fu X (2007) GA action: turning on de-DELLA repressing signaling. Curr Opin Plant Biol 10:461–465
Johnson RR, Shin M, Shen JQ (2008) The wheat PKABA1-interacting factor TaABF1 mediates both abscisic acid-suppressed and abscisic acid-induced gene expression in bombarded aleurone cells. Plant Mol Biol 68:93–103
Kenrick P, Crane PR (1997) The origin and early evolution of plants on land. Nature 389:33–39
King K, Moritz T, Harberd N (2001) Gibberellins are not required for normal stem growth in Arabidopsis thaliana in the absence of GAI and RGA. Genetics 159:767–776
King RW, Mander LN, Asp T, MacMillan CP, Blundell CA, Evans LT (2008) Selective deactivation of gibberellins below the shoot apex is critical to flowering but not to stem elongation of Lolium. Mol Plant 1:295–307
Koornneef M, van der Veen JH (1980) Induction and analysis of gibberellin-sensitive mutants in Arabidopsis thaliana (L.) Heynh. Theor Appl Genet 58:257–263
Koornneef M, Elgersma A, Hanhart CJ, van Loenen MEP, van Rijn L, Zeevaart JAD (1985) A gibberellin insensitive mutant of Arabidopsis thaliana. Physiol Plant 65:33–39
Kurosawa E (1926) Experimental studies on the nature of the substance secreted by the “bakanae” fungus. Nat Hist Soc Formos 16:213–227
Lee S, Cheng H, King KE, Wang W, He Y, Hussain A, Lo J, Harberd NP, Peng J (2002) Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev 16:646–658
Michael TP, Mockler TC, Breton G, McEntee C, Byer A, Trout JD, Hazen SP, Shen R, Priest HD, Sullivan CM, Givan SA, Yanovsky M, Hong F, Kay SA, Chory J (2008) Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet 4:e14
Mizuno T, Yamashino T (2008) Comparative transcriptome of diurnally oscillating genes and hormone-responsive genes in Arabidopsis thaliana: insight into circadian clock-controlled daily responses to common ambient stresses in plants. Plant Cell Physiol 49:481–487
Nakajima M, Shimada A, Takashi Y, Kim YC, Park SH, Ueguchi-Tanaka M, Suzuki H, Katoh E, Iuchi S, Kobayashi M, Maeda T, Matsuoka M, Yamaguchi I (2006) Identification and characterization of Arabidopsis gibberellin receptors. Plant J 46:880–889
Nemhauser JL, Hong F, Chory J (2006) Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126:467–477
Ogawa M, Kusano T, Katsumi M, Sano H (2000) Rice gibberellin-insensitive gene homolog, OsGAI encodes a nuclear-localized protein capable of gene activation at transcriptional level. Gene 245:21–29
Ogawa M, Hanada A, Yamauchi Y, Kuwahara A, Kamiya Y, Yamaguchi S (2003) Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15:1591–1604
Oh E, Yamaguchi S, Hu J, Yusuke J, Jung B, Paik I, Lee HS, Sun TP, Kamiya Y, Choi G (2007) PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19:1192–1208
Olszewski N, TP Sun, Gubler F (2002) Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell 14:S61–S80
Peng J, Harberd NP (1997) Gibberellin deficiency and response mutations suppress the stem elongation phenotype of phytochrome-deficient mutants of Arabidopsis. Plant Physiol 113:1051–1058
Peng J, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP (1997) The Arabidopsis GAI gene defines a signalling pathway that negatively regulates gibberellin responses. Genes Dev 11:3194–3205
Peng J, Richards DE, Hartley NM, Murphy GP, Devos KM, Flintham JE, Beales J, Fish LJ, Worland AJ, Pelica F, Sudhakar D, Christou P, Snape JW, Gale MD, NPet Harberd (1999) ‘Green Revolution’ genes encode mutant gibberellin response modulators. Nature 400:256–261
Pysh LD, Wysocka-Diller JW, Camilleri C, Bouchez D, Benfey PN (1999) The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J 18:111–119
Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T et al (2008) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319:64–69
Richards DE, Peng J, Harberd NP (2000) Plant GRAS and metazoan STATs: one family? Bioessays 22:573–577
Ross JJ, O’Neill DP, Smith JJ, Kerckhoffs LHJ, Elliott RC (2000) Evidence that auxin promotes gibberellin A1 biosynthesis in pea. Plant J 21:547–552
Rupp HM, Frank M, Werner T, Strnad M, Schmulling T (1999) Increased steady state mRNA levels of the STM and KNAT1 homeobox genes in cytokinin overproducing Arabidopsis thaliana indicate a role for cytokinins in the shoot apical meristem. Plant J 18:557–563
Sakamoto T, Kamiya N, Ueguchi-Tanaka M, Iwahori S, Matsuoka M (2001) KNOX homeodomain protein directly suppresses the expression of a gibberellin biosynthetic gene in the tobacco shoot apical meristem. Genes Dev 15:581–590
Sawada Y, Aoki M, Nakaminami K, Mitsuhashi W, Tatematsu K, Kushiro T, Koshiba T, Kamiya Y, Inoue Y, Nambara E, Toyomasu T (2008) Phytochrome- and gibberellin-mediated regulation of abscisic acid metabolism during germination of photoblastic lettuce seeds. Plant Physiol 146:1386–1396
Schneider G, Schliemann W (1994) Gibberellin conjugates: an overview. Plant Growth Regul 15:247–260
Shani E, Yanai O, Ori N (2006) The role of hormones in shoot apical meristem function. Curr Opin Plant Biol 9:484–489
Shimada A, Ueguchi-Tanaka M, Sakamoto T, Fujioka S, Takatsuto S, Yoshida S, Sazuka T, Ashikari M, Matsuoka M (2006) The rice SPINDLY gene functions as a negative regulator of gibberellin signaling by controlling the suppressive function of the DELLA protein, SLR1, and modulating brassinosteroid synthesis. Plant J 48:390–402
Silverstone AL, C-w Chang, Krol E, T-p Sun (1997) Developmental regulation of the gibberellin biosynthetic gene GA1 in Arabidopsis thaliana. Plant J 12:9–19
Silverstone AL, Jung H-S, Dill A, Kawaide H, Kamiya Y, T-p Sun (2001) Repressing a repressor: gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13:1555–1566
Silverstone AL, Tseng TS, Swain SM, Dill A, Jeong SY, Olszewski NE, Sun TP (2007) Functional analysis of SPINDLY in gibberellin signaling in Arabidopsis. Plant Physiol 143:987–1000
Smalle J, Haegman M, Kurepa J, Van Montagu M, Straeten D (1997) Ethylene can stimulate Arabidopsis hypocotyl elongation in the light. Proc Natl Acad Sci USA 94:2756–2761
Sreenivasulu N, Usadel B, Winter A, Radchuk V, Scholz U, Stein N, Weschke W, Strickert M, Close TJ, Stitt M, Graner A, Wobus U (2008) Barley grain maturation and germination: metabolic pathway and regulatory network commonalities and differences highlighted by new MapMan/PageMan profiling tools. Plant Physiol 146:1738–1758
Takahashi N, Phinney BO, MacMillan J (1991) Gibberellins. Springer-Verlag, New York, p 426
Talón M, Koornneef M, Zeevaart JA (1990) Endogenous gibberellins in Arabidopsis thaliana and possible steps blocked in the biosynthetic pathways of the semidwarf ga4 and ga5 mutants. Proc Natl Acad Sci USA 87:7983–7987
Thornton T, Kreppel L, Hart G, Olszewski N (1999) Genetic and biochemical analysis of Arabidopsis SPY. In: Altman A, Ziv M, Izhar S (eds) Plant biotechnology and in vitro biology in the 21st century. Kluwer Academic Publishers, Dordrecht, pp 445–448
Tian C, Wan P, Sun S, Li J, Chen M (2004) Genome-wide analysis of the GRAS gene family in rice and Arabidopsis. Plant Mol Biol 54:519–532
Toh S, Imamura A, Watanabe A, Nakabayashi K, Okamoto M, Jikumaru Y, Hanada A, Aso Y, Ishiyama K, Tamura N, Iuchi S, Kobayashi M, Yamaguchi S, Kamiya Y, Nambara E, Kawakami N (2008) High temperature-induced abscisic acid biosynthesis and its role in the inhibition of gibberellin action in Arabidopsis seeds. Plant Physiol 146:1368–1385
Tyler L, Thomas SG, Hu J, Dill A, Alonso JM, Ecker JR, Sun TP (2004) Della proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol 135:1008–1019
Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, Chow TY, Hsing YI, Kitano H, Yamaguchi I, Matsuoka M (2005) GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437:693–698
Ueguchi-Tanaka M, Nakajima M, Katoh E, Ohmiya H, Asano K, Saji S, Hongyu X, Ashikari M, Kitano H, Yamaguchi I, Matsuoka M (2007a) Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1, and gibberellin. Plant Cell 19:2140–2155
Ueguchi-Tanaka M, Nakajima M, Motoyuki A, Matsuoka M (2007b) Gibberellin receptor and its role in gibberellin signaling in plants. Annu Rev Plant Biol 58:183–198
Vandenbussche F, Fierro AC, Wiedemann G, Reski R, Van Der Straeten D (2007) Evolutionary conservation of plant gibberellin signalling pathway components. BMC Plant Biol 7:65
Varbanova M, Yamaguchi S, Yang Y, McKelvey K, Hanada A, Borochov R, Yu F, Jikumaru Y, Ross J, Cortes D, Ma CJ, Noel JP, Mander L, Shulaev V, Kamiya Y, Rodermel S, Weiss D, Pichersky E (2007) Methylation of gibberellins by Arabidopsis GAMT1 and GAMT2. Plant Cell 19:32–45
Vriezen WH, Achard P, Harberd NP, Van Der Straeten D (2004) Ethylene-mediated enhancement of apical hook formation in etiolated Arabidopsis thaliana seedlings is gibberellin dependent. Plant J 37:505–516
Weiss D, Ori N (2007) Mechanisms of cross talk between gibberellin and other hormones. Plant Physiol 144:1240–1246
Wen CK, Chang C (2002) Arabidopsis RGL1 encodes a negative regulator of gibberellin responses. Plant Cell 14:87–100
Willige BC, Ghosh S, Nill C, Zourelidou M, Dohmann EM, Maier A, Schwechheimer C (2007) The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 19:1209–1220
Wilson RN, Somerville CR (1995) Phenotypic suppression of the gibberellin-insensitive mutant (gai) of Arabidopsis. Plant Physiol 108:495–502
Wilson RN, Heckman JW, Somerville CR (1992) Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiol 100:403–408
Wolbang CM, Ross JJ (2001) Auxin promotes gibberellin biosynthesis in decapitated tobacco plants. Planta 214:153–157
Wolbang CM, Davies NW, Taylor SA, Ross JJ (2007) Gravistimulation leads to asymmetry of both auxin and gibberellin levels in barley pulvini. Physiol Plant 131:140–148
Xie Z, Zhang Z, Zou X, Yang G, Komatsu S, Shen Q (2006) Interactions of two abscisic-acid induced WRKY genes in repressing gibberellin signaling in aleurone cells. Plant J 46:232–242
Yamaguchi S (2008) Gibberellin metabolism and its regulation. Annu Rev Plant Biol 59:225–259
Yanai O, Shani E, Dolezal K, Tarkowski P, Sablowski R, Sandberg G, Samach A, Ori N (2005) Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr Biol 15:1566–1571
Yasumura Y, Crumpton-Taylor M, Fuentes S, Harberd NP (2007) Step-by-step acquisition of the gibberellin-DELLA growth-regulatory mechanism during land-plant evolution. Curr Biol 17:1225–1230
Zentella R, Zhang ZL, Park M, Thomas SG, Endo A, Murase K, Fleet CM, Jikumaru Y, Nambara E, Kamiya Y, Sun TP (2007) Global analysis of della direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 19:3037–3057
Zhang Y, Zhu Y, Peng Y, Yan D, Li Q, Wang J, Wang L, He Z (2008) Gibberellin homeostasis and plant height control by EUI and a role for gibberellin in root gravity responses in rice. Cell Res 18:412–421
Zhu Y, Nomura T, Xu Y, Zhang Y, Peng Y, Mao B, Hanada A, Zhou H, Wang R, Li P, Zhu X, Mander LN, Kamiya Y, Yamaguchi S, He Z (2006) ELONGATED UPPERMOST INTERNODE encodes a cytochrome P450 monooxygenase that epoxidizes gibberellins in a novel deactivation reaction in rice. Plant Cell 18:442–456
Acknowledgments
The author has been supported by the US Department of Energy Grant DE-FG01-04ER04 and thanks to Dr. Neil E. Olszewski for support and NEO and anonymous reviewers for suggestions to improve the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hartweck, L.M. Gibberellin signaling. Planta 229, 1–13 (2008). https://doi.org/10.1007/s00425-008-0830-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-008-0830-1