Abstract
Evolved resistance to the herbicide glyphosate has been reported in eleven weed species, including Lolium multiflorum. Two glyphosate-resistant L. multiflorum populations were collected, one from Chile (SF) and one from Oregon, USA (OR), and the mechanisms conferring glyphosate resistance were studied. Based on a Petri dish dose–response bioassay, the OR and the SF populations were two and fivefold more resistant to glyphosate when compared to the susceptible (S) population, respectively; however, based on a whole-plant dose–response bioassay, both OR and SF populations were fivefold more resistant to glyphosate than the S population, implying that different resistance mechanisms might be involved. The S population accumulated two and three times more shikimic acid in leaf tissue 96 h after glyphosate application than the resistant OR and SF populations, respectively. There were no differences between the S and the glyphosate-resistant OR and SF populations in 14C-glyphosate leaf uptake; however, the patterns of 14C-glyphosate translocation were significantly different. In the OR population, a greater percentage of 14C-glyphosate absorbed by the plant moved distal to the treated section and accumulated in the tip of the treated leaf. In contrast, in the S and in the SF populations, a greater percentage of 14C-glyphosate moved to non-treated leaves and the stem. cDNA sequence analysis of the EPSP synthase gene indicated that the glyphosate-resistant SF population has a proline 106 to serine amino acid substitution. Here, we report that glyphosate resistance in L. multiflorum is conferred by two different mechanisms, limited translocation (nontarget site-based) and mutation of the EPSP synthase gene (target site-based).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glyphosate was commercialized in 1974, and has become the leading postemergence, systemic, non-selective, broad-spectrum herbicide for the control of annual and perennial weeds (Baylis 2000). Although it was first used as a non-crop and plantation crop herbicide, now it is also used in non-tillage systems and in glyphosate resistant crops, such as soybean (Glycine max (L.) Merril.), cotton (Gossypium hirsutum L.), canola (Brassica napus L.), and maize (Zea mays L.), for selective weed control (Shaner 2000).
Glyphosate inhibits the enzyme 5-enolpyruvylshikimate-3-phosphate (EPSP) synthase (EC 2.5.1.19) (Steinrücken and Amrhein 1980). EPSP synthase is the sixth enzyme of the shikimic acid pathway, which is essential for the biosynthesis of aromatic amino acids in algae, higher plants, bacteria, and fungi (Kishore and Shah 1988). EPSP synthase catalyzes the conversion of shikimate-3-phosphate (S3P) and phosphoenolpyruvate (PEP) to yield EPSP and inorganic phosphate (Pi) (Geiger and Fuchs 2002). Glyphosate is a competitive inhibitor of PEP, as it occupies the binding site of PEP, mimicking an intermediate state of the ternary enzyme-substrates complex (Schönbrunn et al. 2001).
It is well established that glyphosate exerts its herbicidal effect through inhibition of EPSP synthase, which prevents the biosynthesis of the aromatic amino acids phenylalanine, tyrosine and tryptophan that are required for protein synthesis (Siehl 1997). However, a more rapid and dramatic effect than reduction in aromatic amino acid pools, is the increase in shikimic acid and, to a lesser extend, shikimate-derived benzoic acids. This increase in shikimic acid has been related to a decline in carbon fixation intermediates (e.g., ribulose bisphosphate) and a reduction of photosynthesis (Duke et al. 2003).
Glyphosate was used worldwide for more than 20 years with no reports of evolved resistance in weed species (Bradshaw et al. 1997). However, in 1996 glyphosate resistance was reported in Lolium rigidum L. in Australia (Pratley et al. 1996). Today, evolved resistance to glyphosate has been reported in 11 weed species in 8 different countries, including L. rigidum in Australia (Powles et al. 1998; Pratley et al. 1999) and in the USA (Simarmata et al. 2003), Eleusine indica L. Gaertn. in Malaysia (Tran et al. 1999; Lee and Ngim 2000), Conyza canadensis L. Cronq. in the USA (VanGessel 2001; Koger et al. 2004; Main et al. 2004), L. multiflorum Lam. in Chile (Perez and Kogan 2003), the USA (Perez-Jones et al. 2005) and Brazil (Heap 2006), C. bonariensis L. Cronq. in South Africa (Heap 2006) and Spain (Urbano et al. 2005), Plantago lanceolata L. in South Africa (Heap 2006), Euphorbia heterophylla L. in Brazil (Heap 2006), Sorghum halepense L. in Argentina (Heap 2006), and Ambrosia artemisiifolia L. (Sellers et al. 2005), Amaranthus rudis S. (Zelaya and Owen 2005), and A. palmeri S. Wats (Culpepper et al. 2006) in the USA.
In previous studies, two different mechanisms, limited translocation (nontarget site-based) and mutation of the EPSP synthase gene (target site-based), have been shown to confer glyphosate resistance in weed species. On the contrary, metabolism of glyphosate has not been found to be a mechanism of resistance (Feng et al. 1999, 2004; Tran et al. 1999; Lorraine-Colwill et al. 2003). Thus, in several L. rigidum populations from Australia, glyphosate resistance was directly correlated with limited translocation (nontarget site-based resistance) of the herbicide to meristematic tissues (Lorraine-Colwill et al. 2003; Wakelin et al. 2004). Likewise, impaired glyphosate translocation to other leaves and roots appeared to be the only mechanism of resistance in several C. canadensis populations from the USA (Feng et al. 2004; Koger and Reddy 2005; Dinelli et al. 2006). On the other hand, mutations of the EPSP synthase gene causing amino acid changes of the enzyme (target site-based resistance) have been shown to confer glyphosate resistance. In E. indica, two different mutations, a proline to serine and a proline to threonine substitution at amino acid 106, were found in glyphosate-resistant populations from Malaysia (Baerson et al. 2002a; Ng et al. 2003). In L. rigidum, two different mutations, a proline to threonine and a proline to alanine substitution at amino acid 106, were found in glyphosate-resistant populations from Australia and South Africa, respectively (Wakelin and Preston 2006; Yu et al. 2007).
Glyphosate resistance in L. multiflorum was first discovered in Chilean orchards (Perez and Kogan 2003), and later in a filbert orchard in Oregon, USA (Perez-Jones et al. 2005). However, it is still not clear what mechanisms of resistance are involved. Here, we investigate the mechanisms of glyphosate resistance present in two resistant L. multiflorum populations, exploring both target site- and nontarget site-based mechanisms.
Materials and methods
Plant material
Two glyphosate-resistant L. multiflorum populations were examined in this study. Seeds of the SF population were collected from an almond orchard in Region VI of Chile in 2001, and seeds of the OR population were collected from a filbert orchard in Oregon, USA, in 2003. Both sites had been intensively treated with glyphosate during the last 15 years, with two to three applications per year at 1.44–1.68 kg ae ha−1. Seeds were collected only from plants that had survived a recommended field application of glyphosate and were grown in the greenhouse. Subsequently, plants at the 3-leaf stage were treated with glyphosate (Roundup®, 0.36 kg ae l−1, Monsanto, Saint Louis, MO, USA) at 0.84 kg ae ha−1 to increase selection of resistant individuals. Seeds from surviving plants were collected and used in all the experiments. A known susceptible (S) L. multiflorum population collected in the Willamette Valley, OR, USA, was included as a control in all the experiments.
Petri dish dose–response bioassay
The Petri dish experiments were conducted using 100 × 15 mm polystyrene Petri dishes (VWR International Inc, Brisbane, CA, USA) containing one layer of blue blotter germination paper (Hoffman Manufacturing Inc, Albany, OR, USA) and 5-ml aliquots of different glyphosate (Roundup®, 0.36 kg ae l−1) concentrations (0, 12.5, 25, 50, 100, 200, and 400 mg ae l−1). Twenty-five seeds were placed per Petri dish and the dishes were transferred to a growth chamber set at 20°C and a 12-h photoperiod. Percent germination was recorded seven days later, and the LD50 (herbicide concentration required to inhibit germination by 50%) was determined for each L. multiflorum population. Two experiments were conducted with three replications per glyphosate concentration per population.
Whole-plant dose–response bioassay
Seeds of both susceptible and glyphosate-resistant L. multiflorum populations were planted in 267-ml plastic pots containing commercial potting mix (Sunshine Mix #1, Sun Gro Horticulture Inc, Bellevue, WA, USA). Plants were grown in the greenhouse under 25/20°C day/night temperature and a 16-h photoperiod. Plants at the 3-leaf stage were sprayed with glyphosate (Roundup®, 0.36 kg ae l−1) (0.01, 0.05, 0.11, 0.21, 0.42, 0.84, 1.68, and 3.37 kg ae ha−1) using an overhead compressed air sprayer calibrated to deliver 187 l ha−1. Shoot biomass was harvested 3 weeks after herbicide treatment, dried at 70°C for 48 h and weighed. The GR50 (herbicide rate required to reduce growth by 50%) was determined for each L. multiflorum population. Two experiments were conducted with four replications per glyphosate rate per population. Biomass data are reported as percent of the untreated control.
Whole-plant shikimic acid bioassay
Shikimic acid extraction was performed according to Singh and Shaner (1998) with some modifications. Plants of both susceptible and glyphosate-resistant L. multiflorum populations were grown in the greenhouse and treated at the 3-leaf stage with glyphosate at 0.42 kg ae ha−1 as described previously. Plant leaves (second and third leaf) were harvested for shikimic acid extraction 24, 48, 72, and 96 h after treatment. Leaf tissues were chopped and 0.05 g fresh weight samples were placed in 1.5-ml tubes containing 1 ml 0.25 N HCl. The samples were immediately mixed, placed at −20°C until frozen, thawed at room temperature, and incubated at 37°C for 45 min. Shikimic acid was measured spectrophotometrically using the method of Cromartie and Polge (2000). Three 25-μl aliquots per sample were mixed with 100 μl 0.25% periodic acid/0.25% sodium(meta)periodate solution in different wells in a 96-well plate . The plate was incubated at 37°C for 30 min to allow shikimic acid oxidation. After incubation, the samples were mixed with 100 μl 0.6 N NaOH/0.22 M Na2SO3 and optical density was measured spectrophotometrically at 380 nm in a VERSAmaxTM microtiter plate reader (Molecular Devices, Sunnyvale, CA, USA). Shikimic acid in μg g−1 fresh weight was determined based on a standard curve. The standard curve was determined using untreated plants and known concentrations of shikimic acid (Acros Organics, Geel, Belgium). One experiment was conducted with six replications per harvest time per population.
14C-glyphosate leaf uptake and translocation
Seeds of both susceptible and glyphosate-resistant L. multiflorum populations were planted in 169-ml plastic pots containing a 2:1 mixture by volume of turf sand and potting mix (Premier Pro-Mix BX, Premier Horticultural Inc, Red Hill, PA, USA). Each pot was fertilized by adding 1.7 g of controlled released fertilizer (17-6-12 plus minors, Scotts-Sierra Horticultural Products Co, Marysville, OH, USA) and then the pots were transferred to a greenhouse under 28/20°C day/night temperature. Plants at the 3- to 4-leaf stage were treated with glyphosate (Touchdown HiTechTM, 0.6 kg ae l−1, Syngenta Crop Protection Inc, Greensboro, NC, USA) at 1.2 kg ae ha−1 as described previously. The nonionic surfactant X-77 (Loveland Industries Inc, Greeley, Co, USA) at 0.2% v/v was added to the herbicide solution. A 2.5 cm section on the adaxial surface at the middle of the third leaf of each plant was covered with aluminum foil and did not receive the overall spray application. The plants were left to air dry for 30 min before radiolabel treatment. Five 0.2-μl drops of radiolabeled 14C-glyphosate (phosphonomethyl-14C; 0.0814 GBq mmol−1 specific activity, Sigma, Saint Louis, MO, USA) solution were applied to the nontreated section of the third leaf of each plant, using a 10-μl syringe with a repeating dispenser (Hamilton Company, Reno, NV, USA). Approximately 500 Bq of 14C-glyphosate was applied to each plant in 1 μl (5 × 0.2-μl) of treatment solution. Plants were harvested 24, 48, and 72 h after treatment and divided into five sections: treated section, above treated section (tip of the treated leaf), below treated section and rest of leaves (untreated leaves), stem, and roots. The treated leaf was washed with 5 ml of an acidified (pH 1.5) washing solution (0.1 M HCl plus methanol, 50:50 by volume) in a 20-ml glass vial for 15 s to remove unabsorbed herbicide. The acid washing regime was used to effectively remove poorly soluble salts of glyphosate that might form on the leaf surface as described by Hall et al. (2000). A 1-ml subsample of the washing solutions was mixed with 15-ml of Ready SafeTM (Beckman Coulter Inc, Fullerton, CA, USA) cocktail and radioactivity was quantified using an LS 6000 SC liquid scintillation counter (Beckman Coulter Inc) to determine 14C-glyphosate leaf uptake. Plant sections were oven dried at 70°C for 24 h, weighed, and combusted for 2 min in an OX-300 biological sample oxidizer (R.J. Harvey Instruments Corp, Hillsdale, NJ, USA). Evolved 14CO2 was trapped in 14C-cocktail solution purged with N2 (R.J. Harvey Instruments Corp) and radioactivity was measured by liquid scintillation as described previously. 14C-glyphosate present in the different sections of the plants is expressed as percentage of total absorbed radioactivity. One experiment was conducted with six replications per harvest time per population.
Phosphorimaging
Visualization of 14C-glyphosate translocation was performed using a Fujifilm BAS-2500 phosphorimager (Fujifilm Corporation, Tokyo, Japan). The plants used for phosphorimaging were treated with glyphosate and 14C-glyphosate, respectively, as described for the 14C-glyphosate leaf uptake and translocation experiment. Plants were harvested 72 h after treatment and the soil was gently washed from the roots. The treated leaf of each plant was washed with 10 ml of the acidified washing solution for 30 s to remove unabsorbed herbicide. Then, the plants were blotted dry, pressed, oven dried at 70°C for 24 h, and exposed to a phosphorimager plate for 24 h before scanning for radioactivity. A total of three plants were scanned for each population.
EPSP synthase gene sequencing
Total RNA was extracted from leaf tissue of both susceptible and glyphosate-resistant L. multiflorum populations using a RNeasy® isolation kit (Qiagen Inc, Valencia, CA, USA). First strand complementary DNA (cDNA) synthesis was performed from total RNA using a SuperscriptTM III first strand synthesis system (Invitrogen Corp, Carlsbad, CA, USA) and the oligo(dT)20 primer. A pair of primers (sense: 5′-AGCTGTAGTCGTTGGCTGTG-3′; antisense: 5′-GCCAAGAAATAGCTCGCACT-3′) was designed based on the EPSP synthase gene sequence of L. multiflorum (GeneBank Accession number DQ153168) to amplify a 564 kb fragment of the epsps gene containing codon 106. Polymerase chain reaction (PCR) was conducted in a 50-μl reaction using a Primus96 plus thermocycler (MWG Biotech Inc, High Point, NC, USA). The reaction mixture contained 1× PCR buffer, 0.2 μM of each primer, 0.2 mM of each deoxynucleotide, 1 unit of Taq DNA polymerase (Fermentas Inc, Hanover, MD, USA), and 50–100 ng of template cDNA. The cycling program consisted of one denaturation step of 3 min at 94°C, 35 cycles of 30 s at 94°C, 30 s at 55°C, and 1 min at 72°C, followed by a final extension step of 10 min at 72°C. The amplified cDNA fragments were cloned using a TOPO® TA cloning kit (Invitrogen Corp), purified using a QIAquick® PCR purification kit (Qiagen Inc), and sequenced using an automatic ABI PRISM® 3771 DNA sequencer (Perkin–Elmer Applied Biosystem, Foster City, CA, USA) with fluorescence dye-labeled dideoxynucleotides. RNA extraction and amplification of the EPSP synthase gene was performed on four plants from each L. multiflorum population. To exclude PCR errors, four clones per PCR product were sequenced and aligned.
Statistical analysis
Dose–response curves for the Petri dish and whole-plant bioassays were obtained by a non-linear regression using the log-logistic equation (Streibig 1988; Streibig et al. 1993; Seefeldt et al. 1995):
where y represents percentage germination or shoot biomass (percentage of control) at herbicide concentration or rate x, C is the mean response at very high herbicide concentration or rate (lower limit), D is the mean response when the herbicide concentration or rate is zero (upper limit), b is the slope of the line at LD50 or GR50, and LD50 and GR50 are the herbicide concentration required for 50% percentage germination inhibition, and the herbicide rate required for 50% growth reduction, respectively. The regression parameters for each L. multiflorum population were obtained using Sigma Plot® (version 9.0, SPSS Inc, Chicago, IL, USA) and compared to test significant differences using a sum of square reduction test. The level of resistance was determined by calculating the ratio of the LD50 or GR50 of the glyphosate-resistant populations to the ones of the susceptible population. Analysis of variance for the Petri dish and whole-plant dose–response bioassays showed no significant interaction between experiments and treatments; therefore, data from repeated experiments were combined. Statistical analyses among the three L. multiflorum populations in the 14C-glyphosate leaf uptake and translocation experiments were performed using PROC MIXED in SAS (version 9.1, SAS Institute Inc, Cary, NC, USA). The LSMEANS statement was used to generate treatment averages, standard errors, and 95% confidence intervals (CI).
Results
Petri dish dose–response bioassay
Percent germination in each L. multiflorum population decreased as glyphosate concentration increased (Fig. 1). However, the dose–responses from the glyphosate-resistant OR and SF populations were different from the S population. Thus, the LD50’s for the OR (LD50 = 73.81 ± 6.87 mg ae l−1) and the SF (LD50 = 160.32 ± 3.77 mg ae l−1) populations were two and fivefold greater than for the S (LD50 = 32.03 ± 1.72 mg ae l−1) population (Table 1).
Whole-plant dose–response bioassay
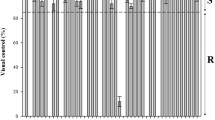
Shoot biomass in each L. multiflorum population decreased as glyphosate rate increased (Fig. 2). However, there was a different dose–response between the glyphosate-resistant OR and SF populations and the S population. Based on the whole-plant dose–response bioassay, both OR and SF (GR50 = 0.3 ± 0.31 kg ae ha−1) populations are fivefold more resistant to glyphosate than the S population (GR50 = 0.06 ± 0.01 kg ae ha−1) (Table 2).
Whole-plant shikimic acid bioassay
When plants were treated with glyphosate at 0.42 kg ae ha−1, more shikimic acid accumulated in leaf tissue of the S population compared to the glyphosate-resistant OR and SF populations (Fig. 3). At 96 h after glyphosate treatment, the S population accumulated approximately two and three times more shikimic acid than the OR and SF populations, respectively.
14C-glyphosate leaf uptake and translocation
On average, 94% of the radioactivity applied as 14C-glyphosate was recovered. Leaf uptake of 14C-glyphosate between the S and the glyphosate-resistant OR and SF populations was not different. At 72 h after treatment, the percentage of leaf uptake of 14C-glyphosate for the S, the OR, and the SF populations was 39, 35, and 37%, respectively (Fig. 4). Although leaf uptake was similar in all three L. multiflorum populations, the patterns of 14C-glyphosate translocation were different. There was a difference in the proportion of 14C-glyphosate translocated from the treated leaf section to the rest of the plant in the OR population compared to the S and SF populations. At 24 h after treatment, 51.5% of the 14C-glyphosate absorbed by the plant moved above the treated section (tip of the treated leaf) in the OR population, in contrast to 29.1 and 38.2% in the S and SF populations, respectively (Table 3). A greater percentage of 14C-glyphosate absorbed by the plant moved to non-treated leaves in the S and SF populations compared to the OR population. A similar pattern was observed 48 and 72 h after treatment. Translocation of 14C-glyphosate to roots among the three L. multiflorum populations was not different, while a greater percentage of 14C-glyphosate moved to the stem in the S and SF populations compared to the OR population (Table 3). The differences in 14C-glyphosate translocation among the L. multiflorum populations were confirmed with the phosphorimaging. As shown in Fig. 5, more 14C-glyphosate remained in the treated leaf and moved upwards to the tip of the leaf in the OR population compared to the S and SF populations.
EPSP synthase gene sequencing
cDNA sequence analysis of the EPSP synthase gene in both glyphosate-resistant OR and SF populations revealed several nucleotide substitutions resulting in silent mutations. However, in the SF population, two nucleotide changes of codon 106 in the first and third positions (from cytosine to thymine and from adenine to guanine) resulted in a proline to serine amino acid substitution (Fig. 6).
Partial and deduced amino acid sequence alignment of the EPSP synthase gene of glyphosate-susceptible (S) and glyphosate-resistant (OR and SF) L. multiflorum populations. The boxed codon shows a proline (P) to serine (S) substitution at amino acid 106 (amino acid number based on Arabidiopsis thaliana sequence)
Discussion
Both Petri dish and whole-plant dose response bioassays were successful in identifying the glyphosate-resistant L. multiflorum populations. Based on the Petri dish bioassay, the OR and the SF populations were two and fivefold more resistant to glyphosate when compared to the S population, respectively; however, based on the whole-plant bioassay, both OR and SF populations were fivefold more resistant to glyphosate than the S population. This difference between the Petri dish and the whole-plant dose–response bioassays might be due to different mechanisms of resistance that are involved in the L. multiflorum populations.
The Petri dish bioassay is a simple, quick and inexpensive method that has been used before to identify glyphosate-resistant populations in L. multiflorum (Perez and Kogan 2003) and L. rigidum (Neve et al. 2004). However, the whole-plant bioassay provides a more realistic level of herbicide resistance because the plant growth stage, time and rate of application are comparable with a field situation. In a situation when several weed populations are to be tested for glyphosate resistance, the Petri dish bioassay can be effectively used. However, the results obtained should be always confirmed with a whole-plant bioassay.
Shikimic acid accumulation in leaf tissue 96 h after glyphosate treatment was two and three times greater in the S population than in the OR and SF populations, respectively. The greater accumulation of shikimic acid in the S population further confirms that the OR and SF populations are glyphosate-resistant. However, the low levels of shikimic acid accumulation in the glyphosate-resistant populations indicates that either glyphosate is not totally excluded from its target site (EPSP synthase) in vivo, or that EPSP synthase can be partially inhibited by glyphosate. Shikimic acid also accumulated in leaf tissue after glyphosate treatment in other glyphosate-resistant weed species such as L. rigidum (Baerson et al. 2002b; Simarmata et al. 2003; Wakelin and Preston 2006), E. indica (Tran et al. 1999) and C. canadensis (Mueller et al. 2003). On the other hand, shikimic acid did not accumulate in leaf tissue after glyphosate treatment in engineered glyphosate-resistant crops such as soybean (Singh and Shaner 1998) and cotton (Pline et al. 2002), where glyphosate insensitive EPSP synthase is highly overexpressed (Padgette et al. 1996).
No differences were found between the S and the glyphosate-resistant OR and SF populations in 14C-glyphosate leaf uptake; however, the patterns of 14C-glyphosate translocation were significantly different. In the OR population, a greater percentage of 14C-glyphosate absorbed by the plant moved above the treated section and accumulated in the tip of the treated leaf. Similar results were found in several glyphosate-resistant L. rigidum populations from Australia, in which the resistant plants accumulated more glyphosate in the leaf tip compared with susceptible plants (Lorraine-Colwill et al. 2003; Wakelin et al. 2004). Likewise, in a glyphosate-resistant L. rigidum population from South Africa, more glyphosate remained in treated leaves and less glyphosate translocated to young leaves compared to susceptible plants (Yu et al. 2007). In contrast, in the S and in the SF populations, a greater percentage of 14C-glyphosate moved to non-treated leaves and the stem. Because glyphosate tends to be actively phloem transported and accumulates in meristematic tissue (Sprankle et al. 1975; McWhorter et al. 1980; Arnaud et al. 1994), the different translocation pattern in the OR population and other L. rigidum populations is associated with glyphosate resistance. 14C-glyphosate translocation patterns between the SF and the S populations were similar at 48 h but different at 24 and 72 h after treatment. In the S population, the percentage of 14C-glyphosate located in the treated section of the leaf increased from 24 to 72 h, while this percentage in the SF population decreased. This increase observed in the S population is due to a decrease in the translocation of the herbicide due to its phytotoxic effect, while it is still being absorbed passively through the cuticle of the leaf. Because glyphosate does not tend to accumulate in the tip of the treated leaf in the SF population, a different mechanism of glyphosate resistance must be involved.
cDNA sequence analysis of the EPSP synthase gene indicated that the glyphosate-resistant SF population has a proline 106 to serine amino acid substitution. This same amino acid substitution is present in the mutated glyphosate-resistant EPSP synthase encoded by the aroA locus in Salmonella typhimurium (Stalker et al. 1985) and is known to confer moderate levels of glyphosate resistance. In petunia, an EPSP synthase carrying the proline 106 to serine amino acid substitution was constructed by site-directed mutagenesis and expressed in Escherichia coli. The analysis of the purified enzyme showed an approximately 7.5-fold increase in K i(app)(glyphosate) (from 0.4 to 3.0 μM) resulting in decreased glyphosate binding (Padgette et al. 1991). In E. indica, EPSP synthase from a resistant population having the proline 106 to serine amino acid substitution was expressed in E. coli and compared with EPSP synthase from a susceptible population. The kinetic characterization of the E. coli-expressed EPSP synthase variants showed a 16-fold increase in K i(app)(glyphosate) (from 47.8 to 759 nM) indicating reduced sensitivity to glyphosate (Baerson et al. 2002a). Target site-based glyphosate resistance was confirmed in E. indica by determining the glyphosate concentration required to inhibit EPSP synthase by 50% (IC50). The IC50 values for the resistant and the susceptible populations were determined to be approximately 16.0 and 3.0 μM, respectively (Baerson et al. 2002a).
Here, we have found that glyphosate resistance in L. multiflorum is conferred by two different mechanisms, limited translocation (nontarget site-based) and mutation of the EPSP synthase gene (target site-based). The biochemical and/or physicochemical basis of the nontarget site-based mechanism (i.e., limited translocation) are still unclear. Glyphosate and its salts (e.g., isopropylamine and potassium) are highly polar, water-soluble molecules with low lipophilic character that probably penetrate the overall lipophilic cuticle via diffusion through a hydrophilic pathway (hydrated cutin and pectin strands) into the apoplast (Caseley and Coupland 1985; Hess 1985; Franz et al. 1997). Absorption of glyphosate by plant cells through the plasma membrane into the symplast is a slow process and involves a passive diffusion mechanism, and also an active transport mechanism (phosphate carrier) (Caseley and Coupland 1985; Sterling 1994; Franz et al. 1997). It seems that in the glyphosate-resistant OR population, and in several glyphosate-resistant L. rigidum populations, glyphosate is either trapped in the apoplast, or the mechanisms of absorption through the plasma membrane are malfunctioning, promoting movement of the herbicide through the xylem with the transpiration stream to the tip of the leaf. Lorraine-Colwill et al. (2003) suggested the existence of a cellular pump in the resistant plants that can pump glyphosate out of the cells, but this theory is yet to be proved. Wakelin and Preston (2006) suggested that perhaps the mechanism by which glyphosate is retained in the symplast is malfunctioning in the resistant plants. Thus, the biochemical and/or physicochemical basis of the nontarget site-based mechanism controlling limited glyphosate translocation to meristematic tissue and increased movement of the herbicide to the tip of the leaves are still to be determined. On the other hand, the molecular basis of the target site-based mechanism is well understood. There is adequate evidence in the literature that demonstrates that a proline 106 to serine amino acid substitution of EPSP synthase decreases glyphosate binding and confers moderate levels of glyphosate resistance.
Abbreviations
- EPSP:
-
5-Enolpyruvylshikimate-3-phosphate
References
Arnaud L, Nurit F, Ravanel P, Tissut M (1994) Distribution of glyphosate and of its target enzyme inside wheat plants. Pestic Sci 40:217–223
Baerson SR, Rodriguez D, Tran M, Feng Y, Biest NA, Dill GM (2002a) Glyphosate-resistant goosegrass. Identification of a mutation in the target enzyme 5-enolpyruvylshikimate-3-phosphate synthase. Plant Physiol 129:1265–1275
Baerson SR, Rodriguez DJ, Biest NA, Tran M, You J, Kreuger RW, Dill GM, Pratley JE, Gruys KJ (2002b) Investigating the mechanism of glyphosate resistance in rigid ryegrass (Lolium rigidum). Weed Sci 50:721–730
Baylis A (2000) Why glyphosate is a global herbicide: strengths, weaknesses and prospects. Pest Manag Sci 56:299–308
Bradshaw LD, Padgette SR, Kimball SL, Wells BH (1997) Perspectives on glyphosate resistance. Weed Technol 11:189–198
Caseley JC, Coupland D (1985) Environmental and plant factors affecting glyphosate uptake, movement and activity. In: Grossbard E, Atkinson D (eds) The herbicide glyphosate. Butterworths, London, pp 92–123
Cromartie TH, Polge ND (2000) An improved assay for shikimic acid and its use as a monitor for the activity of sulfosate. Proc Weed Sci Soc Am 40:291
Culpepper AS, Grey TL, Vencill WK, Kichler JM, Webster TM, Brown SM, York AC, Davis JW, Hanna WW (2006) Glyphosate-resistant palmer amaranth (Amaranthus palmeri) confirmed in Georgia. Weed Sci 54:620–626
Dinelli G, Marotti I, Bonetti A, Minelli M, Catizone P, Barnes J (2006) Physiological and molecular insight on the mechanisms of resistance to glyphosate in Conyza canadensis (L.) Cronq. biotypes. Pest Biochem Physiol 86:30–41
Duke SO, Baerson SR, Rimando AM (2003) Glyphosate. Encyclopedia of agrochemicals. Wiley, New York. Article on line. http://www.interscience.wiley.com
Feng PCC, Pratley JE, Bohn JA (1999) Resistance to glyphosate in Lolium rigidum. II. Uptake, translocation, and metabolism. Weed Sci 47:412–415
Feng PCC, Tran M, Chiu T, Sammons RD, Heck GR, Cajacob CA (2004) Investigations into glyphosate-resistant horseweed (Conyza canadensis): retention, uptake, translocation, and metabolism. Weed Sci 52:498–505
Franz JE, Mao MK, Sikorski JA (1997) Glyphosate: a unique global herbicide. ACS Monograph 189. American Chemical Society, Washington USA
Geiger DR, Fuchs MA (2002) Inhibitors of aromatic amino acid biosynthesis (glyphosate). In: Böger P, Wakabayashi K, Hirai K (eds) Herbicide classes in development. Springer, Heidelberg, pp 59–85
Heap IM (2006) International survey of herbicide-resistant weeds. Available at http://www.weedscience.org (accessed October, 2006)
Hall GJ, Hart CA, Jones CA (2000) Plants as sources of cations antagonistic to glyphosate. Pest Manag Sci 56:351–358
Hess FD (1985) Herbicide absorption and translocation and their relationship to plant tolerances and susceptibility. In: Duke SO (ed) Weed physiology, vol II. Herbicide physiology. CRC Press, Boca Raton, pp 191–214
Kishore GM, Shah DM (1988) Amino acid biosynthesis inhibitors as herbicides. Annu Rev Biochem 57:627–663
Koger CH, Poston DH, Hayes RM, Montgomery RF (2004) Glyphosate-resistant horseweed in Mississippi. Weed Technol 18:820–825
Koger CH, Reddy KN (2005) Role of absorption and translocation in the mechanism of glyphosate resistance in horseweed (Conyza canadensis). Weed Sci 53:84–89
Lee LJ, Ngim J (2000) A first report of glyphosate-resistant goosegrass (Eleusine indica (L) Gaertn) in Malaysia. Pest Manag Sci 56:336–339
Lorraine-Colwill DF, Powles SB, Hawkes TR, Hollinshead PH, Warner SAJ, Preston C (2003) Investigations into the mechanism of glyphosate resistance in Lolium rigidum. Pestic Biochem Physiol 74:62–72
Main CL, Mueller TC, Hayes RM, Wilkerson JB (2004) Response of selected horseweed (Conyza canadensis (L.) Cronq.) populations to glyphosate. J Agric Food Chem 52:879–883
McWhorter CG, Jordan TN, Will GD (1980) Translocation of 14C-glyphosate in soybean (Glycine max) and johnsongrass (Sorghum halepense). Weed Sci 28:113–118
Mueller TC, Massey JH, Hayes RM, Main CL, Stewart N Jr (2003) Shikimate accumulates in both glyphosate-sensitive and glyphosate-resistant horseweed (Conyza canadensis L. Cronq.) J Agric Food Chem 51:680–684
Neve P, Sadler J, Powles SB (2004) Multiple herbicide resistance in a glyphosate-resistant rigid ryegrass (Lolium rigidum) population. Weed Sci 52:920–928
Ng CH, Wickneswari R, Salmijah S, Teng YT, Ismail BS (2003) Gene polymorphisms in glyphosate-resistant and -susceptible biotypes of Eleusine indica from Malaysia. Weed Res 43:108–115
Padgette SR, Re DB, Gasser CS, Eichholtz DA, Frazier RB, Hironaka CM, Levine EB, Shah DM, Fraley RT, Kishore GM (1991) Site-directed mutagenesis of a conserved region of the 5-enolpyruvylshikimate-3-phosphate synthase active site. J Biol Chem 266:22364–22369
Padgette SR, Re DB, Barry GF, Eichholtz DE, Delannay X, Fuchs RL, Kishore GM, Fraley RT (1996) New weed control opportunities: development of soybeans with a Roundup ReadyTM gene. In: Duke SO (ed) Herbicide-resistant crops: agricultural, environmental, economic, regulatory, and technical aspects. CRC Press, Inc. Boca Raton, pp 53–84
Perez A, Kogan M (2003) Glyphosate-resistant Lolium multiflorum in Chilean orchards. Weed Res 43:12–19
Perez-Jones A, Park KW, Colquhoun J, Mallory-Smith C, Shaner D (2005) Identification of glyphosate resistant Italian ryegrass (Lolium multiflorum) in Oregon. Weed Sci 53:775–779
Pline WA, Wilcut JW, Duke SO, Edmisten KL, Wells R (2002) Tolerance and accumulation of shikimic acid in response to glyphosate applications in glyphosate-resistant and nonglyphosate-resistant cotton (Gossypium hirsutum L.) J Agric Food Chem 50:506–512
Powles SB, Lorraine-Colwill DF, Dellow JJ, Preston C (1998) Evolved resistance to glyphosate in rigid ryegrass (Lolium rigidum) in Australia. Weed Sci 46:604–607
Pratley J, Baines P, Eberbach P, Incerti M, Broster J (1996) Glyphosate resistance in annual ryegrass. In: Virgona J, Michalk D (eds) Proceedings of the 11th annual conference of the Grassland Society of New South Wales. Wagga Wagga, Australia, p 126
Pratley J, Urwin N, Stanton R, Baines P, Broster J, Cullis K, Schafer D, Bohn J, Krueger R (1999) Resistance to glyphosate in Lolium rigidum. I. Bioevaluation. Weed Sci 47:405–411
Seefeldt S, Jensen JE, Fuerst EP (1995) Log–logistic analysis of herbicide dose–response relationships. Weed Technol 9:218–225
Sellers BA, Pollard JM, Smeda RJ (2005) Two common ragweed (Ambrosia artemisiifolia) biotypes differ in biology and response to glyphosate. Proc Weed Sci Soc 45:156
Shaner DL (2000) The impact of glyphosate-tolerant crops on the use of other herbicides and on resistance management. Pest Manag Sci 56:320–326
Siehl DL (1997) Inhibitors of EPSPS synthase, glutamine synthetase and histidine synthesis. In: Roe RM, Burton JD, Kuhr RJ (eds) Herbicide activity: toxicology, biochemistry and molecular biology. IOS Press, Amsterdam, pp 37–67
Simarmata M, Kaufmann JE, Penner D (2003) Potential basis of glyphosate resistance in California rigid ryegrass (Lolium rigidum). Weed Sci 51:678–682
Singh BK, Shaner DL (1998) Rapid determination of glyphosate injury to plants and identification of glyphosate-resistant plants. Weed Tech 12:527–530
Schönbrunn E, Eschenburg S, Shuttleworth WA, Schloss JV, Amrhein N, Evans JNS, Kabsch W (2001) Interaction of the herbicide glyphosate with its target enzyme 5-enolpyruvylshikimate 3-phosphate synthase in atomic detail. PNAS 98:1376–1380
Sprankle P, Meggitt WF, Penner D (1975) Absorption, action, and translocation of glyphosate. Weed Sci 23:235–240
Stalker DM, Hiatt WR, Comai L (1985) A single amino acid substitution in the enzyme 5-enolpyruvylshikimate-3-phosphate synthase confers resistance to the herbicide glyphosate. J Biol Chem 260:4724–4728
Steinrücken H, Amrhein N (1980) The herbicide glyphosate is a potent inhibitor of 5-enolpyruvylshikimic acid-3-phosphate synthase. Biochem Biophys Res Commun 94:1207–1212
Sterling TM (1994) Mechanisms of herbicide absorption across plant membranes and accumulation in plant cells. Weed Sci 42:263–276
Streibig JC (1988) Herbicide bioassay. Weed Res 28:479–484
Streibig JC, Rudemo M, Jensen JE (1993) Dose–response curves and statistical models. In: Streibig JC, Kudsk P (eds) Herbicide bioassays. CRC Press, Boca Raton, pp 29–56
Tran M, Baerson S, Brinker R, Casagrande L, Faletti M, Feng Y, Nemeth M, Reynolds T, Rodriguez D, Shaffer D, Stalker D, Taylor N, Teng Y, Dill G (1999) Characterization of glyphosate resistant Eleusine indica biotypes from Malaysia. In: Proceedings 1 (B) of the 17th Asian-Pacific weed science society conference. The Asian-Pacific Weed Science Society, Bangkok, pp 527–536
Urbano JM, Borrego A, Torres V, Jimenez C, Leon JM, Barnes J (2005) Glyphosate-resistant hairy fleabane (Conyza bonariensis) in Spain. Proc Weed Sci Soc 45:394
VanGessel MJ (2001) Glyphosate-resistant horseweed from Delaware. Weed Sci 49:703–705
Wakelin AM, Preston C (2006) A target-site mutation is present in a glyphosate-resistant Lolium rigidum population. Weed Res 46:432–440
Wakelin AM, Lorraine-Colwill DF, Preston C (2004) Glyphosate resistance in four different population of Lolium rigidum is associated with reduced translocation of glyphosate to meristematic zones. Weed Res 44:453–459
Yu Q, Cairns A, Powles S (2007) Glyphosate, paraquat and ACCase multiple herbicide resistance evolved in a Lolium rigidum biotype. Planta 225:499–513
Zelaya IA, Owen MDK (2005) Differential response of Amaranthus tuberculatus (Moq ex DC) JD Sauer to glyphosate. Pest Manag Sci 61:936–950
Acknowledgments
The 14C-glyphosate leaf uptake and translocation experiments were carried out at Syngenta Vero Beach Research Center. We thank Syngenta Crop Protection for their economic and technical support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Perez-Jones, A., Park, KW., Polge, N. et al. Investigating the mechanisms of glyphosate resistance in Lolium multiflorum . Planta 226, 395–404 (2007). https://doi.org/10.1007/s00425-007-0490-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-007-0490-6