Abstract
Condensin complexes are thought to play essential roles in mitotic chromosome assembly and segregation in eukaryotes. To date, two condensin complexes (condensin I and II) have been identified. Both complexes contain two structural maintenance of chromosome (SMC) subunits and three non-SMC subunits. In plants, little is known about the localization and function of all the condensin subunits. Here, we report the analyses on the localization of a non-SMC subunit of Arabidopsis condensin I and II, AtCAP-H, and AtCAP-H2, respectively. Our study indicated that localization of AtCAP-H and AtCAP-H2 is dynamically changed through the mitotic cell cycle using GFP-tagged AtCAP-H and AtCAP-H2 in tobacco cultured cells. They are localized at mitotic chromosomes from prometaphase to telophase. However, their localization in interphase is quite different. AtCAP-H was mainly found in the cytoplasm whereas AtCAP-H2 was mainly found in a nucleolus. It is revealed using GFP-tagged deletion mutant s of AtCAP-H that the kleisin-γ middle domain (GM domain) is a unique domain only in AtCAP-H, responsible for chromosomal localization. We propose that the GM domain of CAP-H is essential for its chromosomal localization at mitosis and thus proper function of CAP-H. Differences in localization of AtCAP-H and AtCAP-H2 at interphase also suggest their functional differentiation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Proper chromatin condensation is essential for the normal segregation of chromatids during cell division. A condensin complex plays an essential role in mitotic chromosome assembly and segregation in eukaryotes (reviewed by Hearing and Nasmyth 2003). Condensin was identified first in Xenopus egg extract (Hirano et al. 1997). The condensin complex is composed of five subunits. These five proteins are conserved among eukaryotes (Sutani et al. 1999; Freeman et al. 2000; Schmiseing et al. 2000; Kimura et al. 2001). The two core subunits (CAP-E/SMC2 and CAP-C/SMC4) belong to SMC (structural maintenance of chromosomes) family protein. Two of the other three non-SMC subunits (CAP-D2 and CAP-G) share a highly degenerated repeating motif known as a HEAT repeat (Neuwald and Hirano 2000), and the rest of the three non-SMC proteins is CAP-H/kleisin-γ that belongs to a kleisin superfamily (Schleiffer et al. 2003). Immunodepletion or antibody blocking of condensin subunits for Xenopus egg extracts has shown that condensin subunits were essential for chromosome condensation and segregation in vitro (Hirano and Mitchison 1994; Hirano et al. 1997; Kimura and Hirano 2000; Wignall et al. 2003). Genetic studies in yeasts and metazoans have also revealed that condensin subunits are necessary for chromosome condensation and segregation in vivo (Bhat et al. 1996; Freeman et al. 2000; Sutani et al. 1999; Hagstrom et al. 2002).
Recently, a second condensin complex (condensin II) has been identified in vertebrate cells (Ono et al. 2003; Yeong et al. 2003). The condensin II has the same pair of SMC subunits with condensin I but contains a different set of non-SMC subunits (CAP-D3, CAP-G2, and CAP-H2). The CAP-D3 and CAP-G2 also contain the HEAT repeat and CAP-H2 belongs to the kleisin superfamily. The condensin II also localized on mitotic chromosomes. Depletion of the condensin I or II specific subunits produces a highly characteristic defect in the chromosome morphology of HeLa cells. The condensin I and II show alternative distributions along the axis of a chromosome (Ono et al. 2004) and condensin II and I associate with chromosomes in this order, so they might have different functions to mitotic chromosome assembly (Hirota et al. 2004).
Proteins of the kleisin superfamily are found in archaea, bacteria, and eukaryote (Schleiffer et al. 2003). An alignment of the N- and C-terminal domains of the kleisin superfamily shows almost complete identity of the hydrophobic pattern and some conservation of functional residues. In eukaryotes, these proteins are classified into three types (kleisin-α, -β, and -γ). Kleisin-α includes non-SMC subunits of cohesin, Scc1, and Rec8. CAP-H and CAP-H2 belong to kleisin-γ and kleisin-β, respectively.
Studies on yeast cohesin complex showed that the N-terminal domain of the kleisin-α subunit (Scc1) interacts with the head of the SMC3 subunit while the C-terminal domain interacts with the head of the SMC1 subunit (Arumugam et al. 2003; Gruber et al. 2003; Haering et al. 2004). The middle domain contains cleavage sites for a thiol protease, separase. The proteolytic cleavage of Scc1’s middle domain triggers the metaphase-to-anaphase transition (Uhlmann et al. 2000). From the finding that the N- and C-terminal sequences of the kleisin-γ subunit have similar conserved domains with kleisin-α, kleisin-γ might associate with the head domains of an SMC heterodimer in the manner similar with association between kleisin-α and SMC (Schleiffer et al. 2003).
Little information on plant condensins has been obtained compared to the rapid progresses in molecular and biochemical understanding of chromosome condensation in animals and yeasts. A few examples are that developmental function of SMC2 subunits (AtCAP-E1 and AtCAP-E2) in Arabidopsis thaliana has been reported (Liu et al. 2002; Siddiqui et al. 2003). A database search shows that A. thaliana also has condensin I and II. Thus, it is reasonable to speculate that the condensins are widely spread in the plant kingdom.
In this paper, we report the characterization and dynamic analyses of Arabidopsis condensins non-SMC subunits, AtCAP-H and AtCAP-H2. This study has elucidated that the two condensins localized differently and thus they may diverge in their functions as well. It has also been shown that the conserved domain in the middle region of AtCAP-H is essential for its localization in mitotic chromosomes.
Materials and methods
Plant materials
Arabidopsis thaliana ecotype Columbia (Col-0) was used for isolation of AtCAP-H and AtCAP-H2. Plants were grown at 22°C under a 16-h light/8 h dark regime. A. thaliana cell line T87 was obtained from the RIKEN BioResource Center (Tsukuba, Japan). The T87 cells were maintained at 23°C under continuous light. Nicotiana tabacum suspension culture cell line, Bright Yellow 2 (BY-2), was used for the localization analysis using GFP fusion genes. The BY-2 cells were subcultured weekly, 50-fold diluted with Linsmaier and Skoog (LS) medium according to Nagata et al. (1992). The cell suspension was agitated on a rotary shaker at 130 rpm at 25°C in the dark.
RT-PCR analyses
Total RNA was extracted from Arabidopsis roots, stems, leaves, flower buds, flowers, and whole plants germinated after three days using TRI REAGENT (Molecular Research Center, Inc. Cincinnati, OH, USA). The mRNAs were isolated from the total RNA samples using PolyATtract mRNA Isolation Systems (Promega). The mRNA was used for the first strand cDNA synthesis using a poly dT primer.
The cDNA fragments of AtCAP-H and AtCAP-H2 were amplified using gene specific primers (5′-GCTGCTAAACAATTTAGGAGTCTATGG-3′ and 5′-GCAGGTTGAACATCTGGACCTT-3′ for AtCAP-H, 5′-TCACACCTGGATGCTCTTCTCGC-3′ and 5′-CTACACAATCTAATTTGAGTAGTCACAAT-3′ for AtCAP-H2). RT-PCR was also performed with actin2 primers as a control (Ratcliffe et al. 2003).
Cloning of the Arabidopsis CAP-H and CAP-H2 genes
Cloning of AtCAP-H and AtCAP-H2 was performed as described by Sambrook et al. (1989). The coding sequences of AtCAP-H (At2g32590) and AtCAP-H2 (At3g16730) were amplified using gene-specific primers with the recombination site (AtCAP-H, 5′-AAAAAGCAGGCTCCATGGATGAATCCTTAACTCCAAACCC-3′ and 5′-AGAAAGCTGGGTCTCAGGCAAGGTGTATTGTTAGATCATCC-3′; AtCAP-H2, 5′-AAAAAGCAGGCTCAATGACCAGTCACGGTGGCGGG-3′ and 5′-AGAAAGCTGGGTTTCACAATTTCCCCAAAGATTTTTCGATTGAAGC-3′. The recombination sites are shown in italic). The amplified fragments were cloned into pDONE221 using a in vitro recombination system (Invitrogen). The nucleotide sequences were checked with a DNA sequencer (ABI prism 3100 Genetic analyzer, Applied Biosystems). Thereafter, cloned cDNAs were recombined into the binary vector, pGWB6, with a recombination cassette for the expression of GFP-fused proteins under the control of a CaMV35S promoter. Deletion mutants were generated with PCR primers incorporating the deletion, and the deletion of each construct was confirmed by DNA sequencing.
Transformation of Arabidopsis protoplasts
Polyethylene glycol (PEG)-mediated A. thaliana protoplast transformation was according to the method described by Leon et al. (2002) with some modifications. The T87 cells were incubated in enzyme solution containing 1% (w/v) Cellulase Onozuka R-10 (Yakult Honsha, Tokyo, Japan), 0.25% (w/v) Macerozyme R-10 (Yakult Honsha), 8 mM CaCl2, 0.5 M mannitol, pH5.5 for 2 h at room temperature under gentle shaking. Protoplasts were collected by centrifugation for 5 min at 75 g, washed twice with W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, and 5 mM glucose, pH 5.8), and resuspended in MaMg solution (0.4 M mannitol, 15 mM MgCl2, and 5 mM MES pH 5.7). After incubation on ice for 30 min, protoplasts were adjusted to 1×106 cells/ml with MaMg solution. A 100-μl of PEG solution (40% (w/v) polyethylene glycol, 0.4 M mannitol, and 0.1 M Ca(NO3)2) was added for 100 μl of plasmid-protoplasts solution and mixed gently. After incubation for 20 min at room temperature, 400 μl of W5 solution was added and the PEG solution was removed by centrifugation for 5 min at 75 g. The protoplasts were resuspended in W5 solution and left at 23°C.
Transformation of BY-2 cells
Transformation was performed using BY-2 cells 2 days after subculture as described previously (Fujimoto et al. 2004). After 3-day incubation at 25°C, the inoculated BY-2 cells were transferred into liquid LS medium containing 500 μg/ml carbenicillin and 10 μg/ml hygromycin. The resulting transformed cell lines were maintained by 50-fold dilutions at weekly intervals as described above.
Fluorescence microscopy
Two days after subculture, BY-2 cells were used for the localization analysis. DNA in the cells were visualized with 0.25 μg/ml of DAPI (4′, 6-diamidino-2-phenylindole) containing 0.01% Triton X-100. The cell images were obtained with a Zeiss Axioplan II fluorescence microscope and a cooled CCD camera (MicroMax, Roper Scientific Princeton Instruments, Trenton, NJ, USA). Further image processing and assembly were performed with the Adobe Photoshop software (Adobe System, Mountain View, CA, USA).
Results
Characterization of Arabidopsis AtCAP-H and AtCAP-H2 genes
Based on the nucleotide sequences of putative AtCAP-H (At2g32590) and AtCAP-H2(At3g16730), we designed the gene-specific primers to obtain the coding region of the genes and performed RT-PCR to amplify the cDNAs. Sequence analysis revealed that AtCAP-H contained a 2016-bp coding region with 13 exons in 3790-bp genomic sequence. AtCAP-H2 contained a 2052-bp coding region with 11 exons in 3231-bp genomic sequence. The deduced AtCAP-H and AtCAP-H2 proteins have 671 amino acids and 683 amino acids, respectively. The amino acid sequences and gene structures are shown in Fig. 1a. The nucleotide sequences have been submitted to DDBJ under accession numbers AB193292 (AtCAP-H) and AB193291 (AtCAP-H2). AtCAP-H and AtCAP-H2 belong to the kleisin superfamily and also have the relatively conserved N- and C-terminal regions (Fig. 1a, underlined). In addition, AtCAP-H has the conserved domain among CAP-H in the middle region (Fig. 1a boxed, Fig. 1b). This domain was referred to as the kleisin-γ middle (GM) domain.
Sequence analyses of AtCAP-H and AtCAP-H2. a The deduced amino acid sequences of AtCAP-H and AtCAP-H2. The N- and C-terminal conserved regions in kleisin superfamily are underlined. The conserved domain in the middle region of kleisin-γ, GM (kleisin-γ middle) domain is boxed. The structure of AtCAP-H and AtCAP-H2 genes composed of 13 exons and 12 introns, and 11 exons and 10 introns, respectively. b The alignment of GM domains of CAP-H in eukaryote. The amino acid sequences of the GM domain were extracted from CAP-H of Arabidopsis thaliana, Oryza sativa (NP 918091), Homo sapiens (NP 056156), Mus musculus (NP 659067), Xenopus laevis (AAH56095), Drosophila melanogaster (AAB40125), Shizosaccharomyces pombe (NP 587811), and Saccharomyces cerevisiae (NP 009455) (Schleiffer et al. 2003; Ono et al. 2003)
Then, we designed the genes-specific primers and analyzed the expression pattern of AtCAP-H and AtCAP-H2 using semiquantitative reverse transcriptase-mediated (RT) PCR from Arabidopsis roots, stems, leaves, flower buds, flowers, and whole plants germinated after three days. AtCAP-H expressed at higher levels in flower bud and flower than the other organs. AtCAP-H2 was not different in expression level among tissues examined (Fig. 2).
Expression of AtCAP-H and AtCAP-H2 in wild-type Columbia of Arabidopsis thaliana. Total RNA was extracted from root (L), stem (S), leaf (L), flower bud (B), flower (F), and whole plants three days after germination (3). Genomic DNA (G) was used as a control. Expression was monitored by RT-PCR. RT-PCR products were electrophoresed and stained with ethidium bromide. The PCR cycles are shown in the parentheses after the gene names
Subcellular localization of AtCAP-H and AtCAP-H2 in Arabidopsis cells
To investigate localization patterns of AtCAP-H and AtCAP-H2, transient expression of GFP-AtCAP-H and GFP-AtCAP-H2 were monitored in A. thalianaT87 cells. Approximately 0.01% of the cells showed GFP fluorescence in 1×105 cells, although about 1–10% of the cells showed GFP fluorescence in the case where only the GFP genes were transformed. As shown in Fig. 3, GFP-AtCAP-H was mainly localized in the cytoplasm while GFP-AtCAP-H2 was localized at the nucleus, especially in the nucleolus.
Dynamic localization of AtCAP-H in tobacco BY-2 cells
To investigate the dynamic localizations of AtCAP-H and AtCAP-H2, we transformed either GFP-AtCAP-H or GFP-AtCAP-H2 into bright yellow-2 (BY-2) tobacco cells. BY-2 cultured cells are suitable materials for dynamic analyses during mitosis because the chromosome size is large and the cell cycle is short. The transformed cells were selected in the media containing hygromycin. In each transformation line, almost all cells show the same GFP fluorescence pattern. The growth rate and the morphology of each cell line were almost the same as the original BY-2 cells (data not shown).
Figure 4 shows the subcellular localization of GFP-AtCAP-H at different cell stages. In interphase, GFP signals were predominantly localized in the cytoplasm of the cells transformed with GFP-AtCAP-H. This localization pattern was the same as that in Arabidopsis T87 cells. GFP-AtCAP-H was not localized at mitotic chromosomes but localized at the cytoplasm until the end of prophase. In prometaphase, the signals were detected on the chromosomes. In metaphase, almost all the signals were moved to the chromosomes and the signal intensity was rapidly increased. After chromosome segregation, a few signals were diffused into the cytoplasm but the main signal remained on the chromosomes. The signals at the chromosomes were finally diffused to the cytoplasm after cytokinesis.
Dynamic localization of AtCAP-H2 in tobacco BY-2 cells
The dynamics in localization of GFP-AtCAP-H2 are shown in Fig. 5. In interphase, GFP signals were mainly detected in the nucleolus and slightly detected in the nucleoplasm. The GFP signals were also detected on the chromatin in interphase, suggesting that AtCAP-H2 interacts with interphase chromatin. This localization pattern was also the same as that in Arabidopsis T87 cells. The signals were localized in the nucleolus until the end of prophase. The signals were moved mainly to the entire chromosomes after the nucleolus disappeared. The signal intensity on the chromosomes was weaker than that of AtCAP-H. During the chromosome segregation, the signals were equally localized on both chromatids. When the nucleolus was formed in the nucleus, the signals appeared again in the nucleolus.
Subcellular localization pattern of GFP-AtCAP-H2 in tobacco cultured cells. The lower right panels of the interphase images show the higher magnification of the same nucleus. The top column shows DNA staining with DAPI and the middle shows GFP fluorescence. The bottom column shows merged images. GFP and DAPI are shown in green and red, respectively. Scale bars 20 μm
Domain responsible for chromosome localization
AtCAP-H has two relatively conserved N- and C-terminal regions among the proteins in the kleisin superfamily. Thus, AtCAP-H was structurally differentiated into the three conserved regions such as the N-terminal region (N-region, 1–199 aa), the middle region (M-region, 200–520 aa) and the C-terminal region (C-region, 521–671 aa). In addition, among the kleisin-γ family, a highly conserved ten amino acid sequence (GM domain, 406–415 aa, WAGPDHWKYR) was found in AtCAP-H. To dissect which domains of AtCAP-H are involved in such chromosomal localization, AtCAP-H deletion mutants were fused with GFP and their localizations in transformed BY-2 cells were examined.
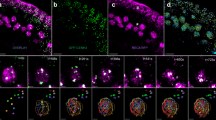
The GFP-AtCAP-H deleted with only 151 aa of the C-terminal region (GFP-NM) or deleted with 199 aa of the C-terminal region (GFP-MC) localized at most of the mitotic chromosomes but the localization is slightly loose (Fig. 6). Similar results were obtained for GFP-AtCAP-H with 321 aa of the middle region (GFP-M), although the signals were observed not only on the chromosomes but also in the cytoplasm. However, the GFP-AtCAP-H with the N-region or C-region showed no longer localization at mitotic chromosomes (GFP-N and GFP-C). Thus, the protein domains responsible for chromosomal localization would be at least in the M-region of AtCAP-H. Moreover, the deletion of the GM domain (Fig. 1) resulted in no localization of the GFP-AtCAP-H (GFP-GM del.) at mitotic chromosomes. The mutant proteins with the GM domain were localized on chromosomes in mitotic cells. Thus, the GM domain is essential for AtCAP-H to be localized on mitotic chromosomes.
Subcellular localization of GFP-AtCAP-H deletion mutant in mitosis of tobacco cultured cells. a A schematic diagram of each construct. The numbers at both ends indicate an amino acid number of AtCAP-H. b Subcellular localization pattern of AtCAP-H deletion fused to GFP. The left column shows DNA staining with DAPI and the middle shows GFP fluorescence. The right column shows merged images. GFP and DAPI are shown in green and red, respectively. In each transformation line, at least 10 mitotic cells were observed and most of them showed the same GFP fluorescent patterns. Scale bars 10 μm
Discussion
In this study, we found that the distributions of the two Arabidopsis condensin subunits, AtCAP-H and AtCAP-H2, are different in localization except for prometaphase to telophase. Subcellular localization of the condensin subunits to chromosomes during mitosis has been well known among many organisms except for plants (Swedlow and Hirano 2003). In higher eukaryotes, some studies showed that the subunits are primarily localized in the interphase nucleus (e.g., XCAP-E, hCAP-E) (Hirano and Mitchison 1994; Shimizu et al. 1998) or in the nucleolus (e.g., hCAP-H, XCAP-E, XCAP-D2) (Cabello et al. 2001; Uzbekov et al. 2003), whereas others reported cytoplasmic localization of hCAP-E, hCAP-C, hCAP-D2, and Drosophila CAP-C (Saitoh et al. 1994; Schmiesing et al. 2000; Steffensen et al. 2001; Ball et al. 2002). Recently, the spatial and temporal distributions of condensin I and II have been shown to be differently regulated during the cell cycle in HeLa cells (Ono et al. 2004). In yeast cells , the fission yeast CAP-H subunit of the condensin complex, Cnd2, functions not only in essential mitotic chromosome condensation but also in DNA repair, S phase arrest and activation of a check-point kinase, Cds1 (Aono et al. 2002).
Dynamic analyses indicated that both AtCAP-H and AtCAP-H2 were localized on chromosomes from prometaphase to the cytokinesis. However, the most remarkable difference was observed in the distribution patterns in interphase. In interphase cells, AtCAP-H is localized in the cytoplasm, showing a striking contrast to the case of AtCAP-H2 that is localized in nuclei, especially in nucleoli at interphase. In vertebrate cells, hCAP-G (subunit of condensin I) is localized in the cytoplasm of the interphase cells. After nuclear envelope breakdown, hCAP-G is moved to the chromosome, and localized on the chromosome until cytokinesis (Ono et al. 2004; Hirota et al. 2004). Therefore, the localization patterns of AtCAP-H and human condensin I are similar to each other. In addition, hCAP-H2 (subunit of condensin II) is localized in the nuclei at interphase. In prometaphase, hCAP-H2 is moved to chromosomes along with DNA, and co-localized with the DNA during the cell cycle (Ono et al. 2004; Hirota et al. 2004). The localization patterns of AtCAP-H2 and human condensin II are somewhat different, especially in interphase. In vertebrate cells, condensin II is thought to be a major form that contributes to the functional organization of the genome in interphase nucleus. Yeast condensin organizes the specialized topology of rDNA within a nucleolus and is important for silencing of the rDNA (Machin et al. 2004). Our results suggest that condensin II, rather than condensin I, mainly contributes to the organization of the genome, especially for the rDNA, in the interphase nucleus.
In the case of yeast cohesin, the connection between SMC1 and SMC3 heads linked by Scc1 is essential for the cohesion of sister chromatids. Thus, we assumed that N- and/or C-terminal regions of the AtCAP-H were important for its chromosomal localization because the triangular ring with the SMC1, SMC3, and Scc1 could trap two DNA molecules from the sister chromatid. However, we found that the M-region of AtCAP-H is sufficient for chromosomal localization, although there might be certain contribution of the other regions. Therefore, we hypothesized that the GM domain of 10 amino acid sequence in the M-region is essential for chromosomal localization. The further deletion experiment supported the hypothesis that the GM domain is essential for chromosomal localization of AtCAP-H. It is likely that kleisin-γ and SMC2-SMC4 heterodimers might associate and form a ring-like structure similar to these. The N- and C-terminal domains of Scc1 associate with the head domains of SMC1-SMC3 heterodimers and the heterodimers and kleisin-α form a ring-like structure (Arumugam et al. 2003; Gruber et al. 2003). Our results demonstrate that CAP-H could partially localize on mitotic chromosomes regardless of the deletion of N- and/or C-terminal region. This fact shows that the ring-like structure of SMC2 (CAP-E)-SMC4 (CAP-C) with CAP-H is not essential for localization of AtCAP-H on the chromosome. In addition, there is no GM domain in the other kleisin superfamily of subunit of cohesin or condensin II. Thus, the mechanism for chromosomal localization by the GM domain might be specific to CAP-H (kleisin-γ) family. The hinge conformations are remarkably different between the condensins and the cohesin SMC proteins (Anderson et al. 2002), indicating the possibility that the association manners of SMC proteins and kleisin superfamily would be different. The other possibility is that deletion of the GM domain causes unstabilization of interaction in the non-SMC subunits of the condensin I.
The condensin I and II complexes have been thought to be essential for chromosome condensation and segregation. Subcellular localization patterns of AtCAP-H and AtCAP-H2 suggest that plant condensins would have similar functions for the condensation and separation of the mitotic chromosomes in animals and yeasts. However, the localization pattern in interphase and the mechanism of chromosomal localization between them are quite different from previously reported results in other eukaryotic species. We suggest that the GM domain is a unique and conserved domain only in AtCAP-H and is essential for movement of AtCAP-H between cytoplasm and chromosomes at mitosis and thus the proper function of AtCAP-H. AtCAP-H moves from cytoplasm to chromosomes at the beginning of mitosis and moves back to cytoplasm after cytokinesis. On the other hand, AtCAP-H2 localizes at the nuclear or chromosomal regions throughout the cell cycle. The difference in localization may suggest the functional differ entiation between condensin I and II in Arabidopsis.
Abbreviations
- CAP:
-
Chromosome Associate Protein
- GFP:
-
Green fluorescent protein
- GM:
-
Kleisin-γ middle
- PEG:
-
Polyethylene glycol
- SMC:
-
Structural maintenance of chromosome
References
Anderson DE, Losada A, Erickson HP, Hirano T (2002) Condensin and cohesin display different arm conformations with characteristic hinge angles. J Cell Biol 156:419–424
Aono N, Sutani T, Tomonaga T, Mochida S, Yanagida M (2002) Cnd2 has dual roles in mitotic condensation and interphase. Nature 417:197–202
Arumugam P, Gruber S, Tanaka K, Haering CH, Mechtler K, Nasmyth K (2003) ATP hydrolysis is required for cohesin’s association with chromosomes. Curr Biol 13:1941–1953
Ball Jr AR, Schmiesing JA, Zhou C, Gregson HC, Okada Y, Doi T and Yokomori K (2002) Identification of a chromosome-targeting domain in the human condensin subunit CNAP1/hCAP-D2/Eg7. Mol Cell Biol 22:5769–5781
Bhat MA, Philp AV, Glover DM, Bellen HJ (1996) Chromatid segregation at anaphase requires the barren product, a novel chromosome-associated protein that interacts with Topoisomerase II. Cell 87:1103–1114
Cabello OA, Eliseeva E, He WG, Youssoufian H, Plon SE, Brinkley BR, Belmont JW (2001) Cell cycle-dependent expression and nucleolar localization of hCAP-H. Mol Biol Cell 12:3527–3537
Freeman L, Aragon-Alcaide L, Strunnikov A (2000) The condensin complex governs chromosome condensation and mitotic transmission of rDNA. J Cell Biol 149:811–824
Fujimoto S, Matsunaga S, Yonemura M, Uchiyama S, Azuma T, Fukui K (2004) Identification of a novel plant MAR DNA binding protein localized an chromosome surfaces. Plant Mol Biol 56:225–239
Gruber S, Haering CH, Nasmyth K (2003) Chromosomal cohesin forms a ring. Cell 112:765–777
Haering CH, Nasmyth K (2003) Building and breaking bridges between sister chromatids. Bioessays 25:1178–1191
Haering CH, Schoffnegger D, Nishino T, Helmhart W, Nasmyth K, Lowe J (2004) Structure and stability of cohesin’s Smc1-Kleisin interaction. Mol Cell 15:951–964
Hagstrom KA, Holmes VF, Cozzarelli NR, Meyer BJ (2002) C. elegans condensin promotes mitotic chromosome architecture, centromere organization, and sister chromatid segregation during mitosis and meiosis. Genes Dev 16:729–742
Hirano T, Mitchison TJ (1994) A heterodimeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell 79:449–458
Hirano T, Kobayashi R, Hirano M (1997) Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell 89:511–521
Hirota T, Gerlich D, Koch B, Ellenberg J, Peters JM (2004) Distinct functions of condensin I and II in mitotic chromosome assembly. J Cell Sci 117:6435–6445
Kimura K, Hirano T (2000) Dual roles of the 11S regulatory subcomplex in condensin functions. Proc Natl Acad Sci USA 97:11972–11977
Kimura K, Cuvier O, Hirano T (2001) Chromosome condensation by a human condensin complex in Xenopus egg extracts. J Biol Chem 276:5417–5420
Leon S, Touraine B, Briat JF, Lobreaux S (2002) The AtNFS2 gene from Arabidopsis thaliana encodes a NifS-like plastidial cysteine desulphurase. Biochem J 366:557–564
Liu C, McElver J, Tzafrir I, Joosen R, Wittich P, Patton D, Van Lammeren AA, Meinke D (2002) Condensin and cohesin knockouts in Arabidopsis exhibit a titan seed phenotype. Plant J 29:405–415
Machin F, Paschos K, Jarmuz A, Torres-Rosell J, Pade C, Aragon L (2004) Condensin regulates rDNA silencing by modulating nucleolar Sir2p. Curr Biol 14:125–130
Nagata T, Nemoto Y, Hasegawa S (1992) Tobacco BY-2 cell line as the ‘HeLa’ cell in the cell biology of higher plants. Int Rev Cytol 132:1–30
Neuwald AF, Hirano T (2000) HEAT repeats associated with condensins, cohesins, and other complexes involved in chromosome-related functions. Genome Res 10:1445–1452
Ono T, Losada A, Hirano M, Myers MP, Neuwald AF, Hirano T (2003) Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell 115:109–121
Ono T, Fang Y, Spector DL, Hirano T (2004) Spatial and temporal regulation of Condensins I and II in mitotic chromosome assembly in human cells. Mol Biol Cell 15:3296–3308
Ratcliffe OJ, Kumimoto RW, Wong BJ, Riechmann JL (2003) Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell 15:1159–1169
Saitoh N, Goldberg IG, Wood ER, Earnshaw WC (1994). ScII: an abundant chromosome scaffold protein is a member of a family of putative ATPases with an unusual predicted tertiary structure. J Cell Biol 127:303–318
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Schleiffer A, Kaitna S, Maurer-Stroh S, Glotzer M, Nasmyth K, Eisenhaber F (2003) Kleisins: a superfamily of bacterial and eukaryotic SMC protein partners. Mol Cell 11:571–575
Schmiesing JA, Gregson HC, Zhou S, Yokomori K (2000) A human condensin complex containing hCAP-C-hCAP-E and CNAP1, a homolog of Xenopus XCAP-D2, colocalizes with phosphorylated histone H3 during the early stage of mitotic chromosome condensation. Mol Cell Biol 20:6996–7006
Shimizu K, Shirataki H, Honda T, Minami S, Takai Y (1998) Complex formation of SMAP/KAP3, a KIF3A/B ATPase motor-associated protein, with a human chromosome-associated polypeptide. J Biol Chem 273:6591–6594
Siddiqui NU, Stronghill PE, Dengler RE, Hasenkampf CA, Riggs CD (2003) Mutations in Arabidopsis condensin genes disrupt embryogenesis, meristem organization and segregation of homologous chromosomes during meiosis. Development 130:3283–3295
Steffensen S, Coelho PA, Cobbe N, Vass S, Costa M, Hassan B, Prokopenko SN, Bellen H, Heck MMS, Sunkel CE (2001). A role for Drosophila SMC4 in the resolution of sister chromatids in mitosis. Curr Biol 11:295–307
Sutani T, Yuasa T, Tomonaga T, Dohmae N, Takio K, Yanagida M (1999) Fission yeast condensin complex: essential roles of non-SMC subunits for condensation and Cdc2 phosphorylation of Cut3/SMC4. Genes Dev 13:2271–2283
Swedlow JR, Hirano T (2003) The making of the mitotic chromosome: modern insights into classical questions. Mol Cell 11:557–569
Uhlmann F, Wernic D, Poupart MA, Koonin EV, Nasmyth K (2000) Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell 103:375–386
Uzbekov R, Timirbulatova E, Watrin E, Cubizolles F, Ogereau D, Gulak P, Legagneux V, Polyakov VJ, Le Guellec K, Kireev I (2003) Nucleolar association of pEg7 and XCAP-E, two members of Xenopus laevis condensin complex in interphase cells. J Cell Sci 116:1667–1678
Wignall SM, Deehan R, Maresca TJ, Heald R (2003) The condensin complex is required for proper spindle assembly and chromosome segregation in Xenopus egg extracts. J Cell Biol 161:1041–1051
Yeong FM, Hombauer H, Wendt KS, Hirota T, Mudrak I, Mechtler K, Loregger T, Marchler-Bauer A, Tanaka K, Peters JM, Ogris E (2003) Identification of a subunit of a novel Kleisin-beta/SMC complex as a potential substrate of protein phosphatase 2A. Curr Biol 13:2058–2064
Acknowledgements
We thank Akira Kawabe for valuable discussions and Joyce A. Cartagena and Reiko Isobe for technical assistances. This study was supported by a fund from the Ministry of Education, Culture, Sports, Science and Technology, Japan, to Kiichi Fukui and the Industrial Technology Research Grant Program in 2003 from the New Energy and Technology Development Organization (NEDO) of Japan.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Fujimoto, S., Yonemura, M., Matsunaga, S. et al. Characterization and dynamic analysis of Arabidopsis condensin subunits, AtCAP-H and AtCAP-H2. Planta 222, 293–300 (2005). https://doi.org/10.1007/s00425-005-1546-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-005-1546-0