Abstract
Zeins are alcohol soluble seed storage proteins synthesized within the endosperm of maize and subsequently deposited into endoplasmic reticulum (ER) derived protein bodies. The genes encoding the beta and delta zeins were previously introduced into tobacco with the expectation of improving the nutritional quality of plants (Bagga et al. in Plant Physiol 107:13, 1997). Novel protein bodies are produced in the leaves of transgenic plants accumulating the beta or delta zein proteins. The mechanism of protein body formation within leaves is unknown. It is also unknown how zeins are retained in the ER since they do not contain known ER retention motifs. Retention may be due to an interaction of zeins with an ER chaperone such as binding luminal protein (BiP). We have demonstrated protein–protein interactions with the delta zeins, beta zeins, and BiP proteins using an E. coli two-hybrid system. In this study, four putative BiP binding motifs were identified within the delta zein protein using a BiP scoring program (Blond-Elguindi et al. in Cell 75:717, 1993). These putative binding motifs were mutated and their effects on protein interactions were analyzed in both a prokaryotic two-hybrid system and in plants. These mutations resulted in reduced BiP–zein protein interaction and also altered zein–zein interactions. Our results indicate that specific motifs are necessary for BiP–delta zein protein interactions and that there are specific motifs which are necessary for zein–zein interactions. Furthermore, our data demonstrates that zein proteins must be able to interact with BiP and zeins for their stability and ability to form protein bodies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Zeins are alcohol soluble seed storage proteins located in protein bodies within the endosperm of maize. Zein proteins are differentiated into four classes based upon their mobility on SDS-PAGE gels; these classes include alpha, beta, delta and gamma (Lending and Larkins 1989). These proteins contain an N-terminal signal peptide which directs them to the endoplasmic reticulum (ER). Once the proteins are translocated into the ER, the signal peptide is cleaved and the mature proteins fold into their tertiary configuration and are subsequently deposited into ER derived protein bodies (Lending and Larkins 1989). Within the endosperm of maize, the syntheses of zein proteins are developmentally regulated and their depositions within protein bodies occur in a specific manner. The alpha and delta zeins comprise the core of the protein body with the beta and gamma zeins surrounding the periphery (Lending and Larkins 1989). The mechanism for zein protein body biogenesis within the endosperm of maize is yet to be comprehensively elucidated.
In the last decade a number of researchers have utilized the beta, delta, and gamma zeins in various transgenic plant experiments. These three classes of zeins are composed of an unusually high abundance of sulfur containing amino acids. These zeins were originally engineered into transgenic plants in the hope of increasing the methionine content of plants and thereby “balancing” the nutritional content of specific crop plants (Bagga et al. 1997; Bellucci et al. 1997). Recombinant gamma zein was engineered as a potential dietary supplement for individuals with phenylketonuria (Ems-McClung et al. 2002). More recently, the gamma zein protein was utilized in the making of a chimeric “balanced” protein named zeolin (Mainieri et al. 2004). The amino-terminal portion of the gamma zein protein was linked behind the seed storage protein phaseolin which then forms a chimeric protein rich in methionine and lysine (Mainieri et al. 2004).
Only the beta, delta, and gamma zein proteins are stable and accumulate in the leaves of transgenic tobacco plants under the control of the 35 S constitutive promoter (Bagga et al. 1995; Bellucci et al. 1997; Randall and Bagga 2000). The alpha zein protein does not accumulate to measurable levels in transgenic plants (Coleman and Herman 1996). Novel protein bodies are observed via electron microscopy analysis and immunolocalization in the leaves of transgenic plants containing the beta, delta, and gamma zein genes (Bagga et al. 1995; Bellucci et al. 1997; Randall et al. 2000, 2004). When tobacco plants containing the beta zein protein are crossed with plants containing the delta zein protein co-localization of both proteins are found in the beta zein rosette bodies (Bagga et al. 1997). This association between the beta and delta zeins results in a higher accumulation of the delta zein protein. Likewise, when plants containing the alpha zein protein are crossed with plants containing gamma zein protein, alpha zein protein is stabilized and is co-localized in gamma zein protein bodies (Coleman et al. 1996). These associations observed between the beta and delta zeins and the gamma and alpha zeins in transgenic plants have recently been verified using the yeast-two-hybrid system (Kim et al. 2002). The gamma and alpha zein proteins do not have strong interactions in the yeast two-hybrid system although domains were identified on the alpha zein which are required for interaction with the gamma zein (Kim et al. 2002). The beta and delta zein proteins interact strongly with themselves and with each other (Kim et al. 2002).
It is unknown, how the zeins are retained in the ER in maize endosperm or in transgenic leaves since there are no identifiable ER retention motifs within the zein proteins. ER resident proteins usually contain an ER retention signal such as HDEL or KDEL. Mutational experiments with the gamma zein protein suggest that a tandem repeat of the hexapeptide PPPVHL is necessary for ER retention of this gamma zein (Geli et al. 1994). However, the delta and beta zein proteins do not contain this tandem repeat. It has been suggested that the binding luminal protein (BiP) may play a role in protein body assembly and retention of zeins within the ER by their association or interaction with this chaperone protein (Bagga et al. 1997; Boston et al. 1991; Vitale and Ceriotti 2004). BiP is related to the heat shock 70 family of proteins (Hsp 70) and contains an ER retention signal at the carboxy end of the protein. BiP has a variety of functions within eukaryotic cells which include; the prevention of premature protein folding, the assembly of polypeptides, and the assistance of protein folding within the ER (Nuttall et al. 2003). BiP also binds to misfolded proteins and prevents these misfolded proteins from entering the secretory pathway via the ER-associated protein degradation (ERAD) (Molinari et al. 2002; Ng et al. 2000). BiP and prolamines (alcohol soluble, seed storage proteins) from rice do form ATP-sensitive complexes which indicate interaction between BiP and rice prolamines (Li et al. 1993). BiP was localized on the surface of prolamine protein bodies in rice (Li et al. 1993), and was also co-localized with the gamma zein protein in transgenic Arabidopsis leaves producing gamma zein protein bodies (Geli et al. 1994). BiP accumulation in transgenic plants increases in plants accumulating the beta or delta zeins (Randall et al. 2000). This immunolocalization and western data only provides evidence of an association between BiP and zeins. However, recently it was demonstrated that BiP directly interacts with a new engineered chimeric seed storage protein zeolin (Mainieri et al. 2004). Zeolin is composed of the vacuolar seed storage protein phaseolin and gamma zein (Mainieri et al. 2004).
Affinity panning of synthetic octapeptides on a bacteriophage and ATPase studies revealed that there are specific motifs to which BiP molecules bind (Blond-Elguindi et al. 1993; Knarr et al. 1999). The binding motif is described as Hy(Trp/X)HyXHyXHy, where Hy is a large hydrophobic amino acid and X is any amino acid (Knarr et al. 1999). A scoring system was developed which ranked amino acids at specific positions in a protein sequence for the probability of a BiP binding motif (Blond-Elguindi et al. 1993). This scoring system was recently employed for determining BiP binding motifs within the vacuolar seed storage protein from bean, phaseolin (Foresti et al. 2003). Experimentally, it was demonstrated that BiP does interact with phaseolin at a specific motif prior to its trimerization (Foresti et al. 2003).
In this study, four putative BiP motifs were identified in the delta zein protein. A series of mutants were generated to knockout these putative motifs then each mutagenized gene was placed into the BacterioMatch system and screened for interactions with BiP, beta zein and delta zein. This prokaryotic two-hybrid system, as mentioned above, is an effective tool that can be used to determine if proteins “interact” in E.coli; however, interaction observed in this system may not emulate what is occurring within plant systems. Therefore, each mutated delta zein gene was placed in a binary vector behind the 35 S constitutive CaMV promoter and introduced into agrobacterium. These modified genes were then studied in tobacco via agrobacterium infiltration, to determine if protein accumulation, targeting, or protein body formation is altered within plant systems. Agrobacterium infiltration is a transient method of expression which accurately reflects transgenic plant expression as confirmed by this current work and previously published by others (Voinnet et al. 2003). This report demonstrates for the first time that specific motifs within delta zein are required for the physical interaction of zeins to BiP and zein to zein proteins.
Materials and methods
Prediction of putative BiP binding motifs
The BiP Score Program designed by Blond-Elguindi et al. (1993) was utilized to predict putative BiP binding motifs within the delta zein protein. The binding motif is described as Hy(Trp/X)HyXHyXHy where Hy is a large hydrophobic aromatic amino acid and X is any amino acid.
Analysis of zein–zein and zein–BiP interactions in the BacterioMatch (Stratagene) two-hybrid system.
The mature coding region (without the signal peptide) of delta-10 kD zein was isolated by PCR amplification with primers deltaF and deltaR containing BglII and XhoI sites (Table 1). The mature coding sequence of beta-15 kD zein was isolated by PCR amplification using primers betaF and betaR (Table 1) containing BglII and XhoI sites. The mature coding regions were ligated into the pGem-T (Promega) vector and sequenced to ensure that no changes were introduced during amplification. These mature coding regions were then inserted into the BglII and XhoI sites of vector pTRG and the BamHI and XhoI sites of vector pBT of the BacterioMatch (Stratagene) two-hybrid system. The BiP (blp4 from tobacco) (Leborgne-Castel et al. 1999) coding sequence was amplified by PCR using primers bipF and bipR (Table 1) containing EcoRI and XhoI restriction sites. The 5′ and 3′ untranslated regions were removed from blp4. The addition of two extra bases prior to the start codon allows the coding region to be in-frame within the two-hybrid vectors. The BiP coding sequence was introduced into the EcoRI and XhoI sites of vectors pBT and pTRG.
The two-hybrid constructs were then co-transformed into the BacterioMatch E. coli reporter strain for protein–protein interaction analysis (Stratagene). The co-transformants were plated on LB containing 250 μg/ml carbenicillin, 15 μg/ml tetracycline, 30 μg/ml chloramphenicol, and 50 μg/ml kanamycin. Colonies which grew on these plates were then tested for β-galactosidase activity.
Beta-galactosidase assays
Single colonies were inoculated in liquid LB-CTCK (same concentrations as listed above) and grown at 30°C for 12 h with shaking. Three ml of the bacterial cells were centrifuged for 2 min at 13,000 rpm. The pellets were then resuspended in a buffer containing 60 mM sodium mono-phosphate, 40 mM sodium di-phosphate, 10 mM potassium chloride, 1 mM magnesium sulfate, and 50 mM β-mercaptoethanol. The absorbance of each culture was determined at 600 nm by a spectrophotometer and each culture was then diluted to an OD of 0.5. The cells were made permeable by the addition of 100 μl of chloroform and 50 μl of 0.1 M sodium dodecyl sulfate. The samples were incubated in a 30°C water bath for 15 min. Next 200 μl of 4 mg/ml substrate O-nitrophenyl-β-galactosidase (ONPG) in 0.1 M sodium phosphate at pH 7.0 was added to each tube and further incubated at 30°C. The amount of time required for a color change was noted and the absorbance at 420 nm and 550 nm was determined (Reynolds et al. 1997; Tottey et al. 2002) Three assays were performed for each sample and each assay was performed in triplicate.
Mutagenesis of putative BiP binding motifs
Primers were designed which would alter the four putative BiP binding motifs in delta-10 kD zein as listed in Fig. 1. The mutants were created with the use of splice-overlap-extension (SOE) PCR (Horton 1989). All primer sequences are listed in Table 1. The delta-10 kD zein primers deltaF and deltaR containing BglII and XhoI sites were utilized as the external primers in the SOE amplifications. Each SOE product was ligated into the pGem-T vector and sequenced. The resulting mutants were ligated into the BglII and XhoI sites of BacterioMatch vector pTRG. The internal SOE primers used to create mutant A were AR and AF. The internal SOE primers used to generate mutant B were BR and BF. The internal SOE primers used to generate mutant C were CR and CF. The internal SOE primers used to generate mutant D were DR and DF. Each mutated gene was ligated into pGem-T and sequenced. These genes were then introduced into pTRG for protein–protein interaction assays.
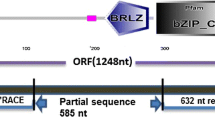
Putative BiP motifs within the δ-zein protein and their location in the predicted secondary structure for δ-zein. The top panel indicates putative BiP motifs within the δ-zein protein sequence. The BiP motifs were predicted using a scoring system developed by Blond-Elguindi et al. 1993. A score of 10 or higher indicates that there is an 80% probability of BiP binding to the sequence. The top panel also shows mutations constructed to disrupt these putative BiP binding motifs and their newly calculated scores. The bottom panel represents the predicted secondary structure of the δ-zein protein. This secondary structure was predicted using the self optimized protein modeling program (SOPM) (Geourjon and Deleage 1994). The abbreviations represent the following in the secondary structure: h is helical, c is coil, t is turn, and e is for beta sheet. The locations of the putative BiP motifs are denoted (*A−*D) on the secondary structure
Agrobacterium infiltration
Delta zein (pDSpZ) and the delta zein mutants (A, B, C, and D) were individually inserted behind the delta-10 kD zein signal peptide in the BGG-2 binary vector which contained the CaMV 35 S promoter and the Nos terminator (Rogers et al. 1987). The delta-10 kD zein signal peptide was isolated utilizing primers SpF and SpR (Table 1). The signal peptide was sequenced prior to ligation in the BglII site of BGG-2. The delta zein mutants were ligated into the BamHI site at the 3′ end of the signal peptide and the XhoI site of BGG-2. The constructs were transformed into Agrobacterium tumefaciens strain GV3101 electro competent cells and selected on LB with 50 μg/ml Kanamycin. Colonies were screened via whole cell PCR.
A. tumefaciens strain GV3101 containing the gene constructs were grown in L-broth with 50 μg/ml Kanamycin, 10 mM MES (pH 5.7), and 20 μM acetosyringone at 28°C with shaking. The bacterial cells were centrifuged at 3,000 rpm for 5 min at 4°C. The bacterial pellet was resuspended in induction medium containing 10 mM magnesium chloride, 10 mM MES (pH 5.7), and 150 μM acetosyringone to an optical density (OD600) of 0.7. The induced A. tumefaciens cultures were infiltrated into the abaxial air-spaces of Nicotiana benthamiana leaves with a 1-ml syringe (Voinnet et al. 2003). Co-infiltration was accomplished by combining an equal volume of induced A. tumefaciens delta-10 kD zein or delta-10 kD zein mutants (A, B, C, or D) to induced A. tumefaciens beta zein (pMEZ) (Bagga et al. 1995), A. tumefaciens delta zein (pMH316-18) (Hinchliffe and Kemp 2002), or to induced A. tumefaciens pWL construct. pWL is a double construct composed of the delta-18 kD zein behind the 35 S CaMV promoter and the beta zein behind the CVMV promoter. The induced A. tumefaciens both had an OD600 of 0.7.
Protein analysis
Protein was isolated from leaf tissue of infiltrated and non-infiltrated plants as described by Bagga et al. (1995). A 7-mm leaf-tissue disk was collected from the surrounding infiltration site. Zein proteins were detected after separating 25 μg of ethanol/beta-mercaptoethanol soluble protein on a 16% SDS-PAGE gel and challenging the blot with anti-delta zein polyclonal rabbit antisera (1:1000) (Kirihara et al. 1988) and anti-beta zein polyclonal rabbit antisera (1:1000) (Randall 1997).
Quantitative reverse-transcriptase polymerase chain reaction
RNA was isolated from Agrobacterium-infiltrated plants using the Qiagen RNA Easy-Prep kit. Quantification of the RNA was performed using the Ribogreen RNA quantification kit (Molecular Probes) and a Synergy HT-1 fluorescent plate reader. The first strand synthesis consisted of 100 ng of total RNA from infiltrated samples, 20 μM oligo-dT primer 5′CCAGTGAGCAGAGTGACGAGGATTTTTTTTTTTTTTTTT3′, 5 mM DTT, 0.5 mM dNTP, and 1 unit of SuperScript III Reverse Transcriptase. cDNA synthesis took place at 45°C for 45 min, followed with an extension at 70°C for 15 min.
Second strand synthesis was performed utilizing a fluorescent syber-green quantitative RT-PCR kit according to the manufacturer’s directions (MJ Research) and the Opticon II thermocycler. The forward primer for delta zein was 5′CCATGGCAGCCAAGATGCTTG3′ and reverse primer was 5′ACAGTTGCTGCAGCATCAGGG3′, these internal primers amplified a 200 bp fragment. Amplification was performed under the following reaction conditions: 95°C for 3 min, 94°C for 20 s, 67°C for 20 s, 70°C for 20 s for 40 cycles. The melting curve was analyzed from 55°C to 90°C. Amplification of actin was used as an internal control with the forward primer 5′TGGATTCTGGTGATGGTGTTAG3′ and the reverse primer of 5′ACTATACTTCCTCTCAGGTGGA3′. The same amplification reaction conditions were utilized as listed above, with the exception of a 54°C annealing temperature.
In vivo synthesis
In vivo synthesis was performed on infiltrated plants as described by Hinchliffe and Kemp (2002). Leaf-discs (7-mm diameter) were taken from infiltrated plants 96 h post-infiltration. The leaf-discs were incubated in 1 mM potassium phosphate pH 6.0, 1% sucrose, and 124 μCi L-[35S] methionine in the presence of light for 5 h. Discs were then washed in 1 mM potassium phosphate pH 6.0 and 1% sucrose. One disc was then stored for further analysis and a duplicate was chased with 6 mM L-methionine and 3 mM L-cysteine for 18 h. Alcohol (70% ethanol and 1% beta-mercaptoethanol) soluble proteins were isolated from 5 h synthesis and 18 h post-chase samples (Hinchliffe and Kemp 2002). Twenty-five micrograms of protein from each sample was separated on 16% SDS-PAGE gels. The protein was transferred to nitrocellulose and challenged with anti-delta zein polyclonal rabbit antisera (1:1000) (Kirihara et al. 1988) and anti-beta zein polyclonal rabbit antisera (1:1000) (Randall 1997). The blot was then exposed to autoradiography film.
Electron microscopy analysis
Leaves from transformed and infiltrated plants were sectioned, fixed, and immunolocalized as described by Ghoshroy and Citovsky (1998). The samples were fixed in 2.5% glutaraldehyde in cacodylate buffer and post-fixed in osmium tetra-oxide prior to dehydration in ethanol. The samples were embedded in Spurr’s resin prior to sectioning and placing on fomvar coated grids. The sections were labeled with anti-beta zein polyclonal antibodies diluted 1:100 followed by 12-nm diameter gold conjugated goat-anti-rabbit IgG diluted 1:40. The sections were also labeled with anti-delta zein polyclonal antibodies diluted 1:100 followed by 12-nm diameter gold conjugated goat-anti-rabbit IgG diluted 1:40. The grids were observed using a Hitachi H7000 transmission electron microscope.
Results
Delta zein secondary structure and putative BiP motifs
The secondary structure prediction of delta zein was accomplished using the self-optimized protein modeling program (Geourjon and Deleage 1994). The secondary structure of delta zein shown in Fig. 1, appears to consist of coils interspersed with three regions of alpha helices followed by a short beta sheet domain at the carboxy terminus. This secondary structure for delta zein is similar to the secondary structure of the clathrin adaptor core AP2 protein which is necessary for transport from the endoplasmic reticulum (determined in this study).
The scoring system developed from a BiP binding study with synthetic octapeptides was utilized to determine putative BiP binding motifs within the delta zein protein. This scoring system ranks amino acids at specific positions in a protein for the probability of BiP binding to a specific motif (Blond-Elguindi et al. 1993). The binding motif is defined as Hy(Trp/X)HyXHyXHy, where Hy is a large hydrophobic amino acid and X is any amino acid (Knarr et al. 1999). Experimental data indicates that binding motifs with scores of 10 or greater have an 80 chance of binding to BiP (Knarr et al. 1999). Four putative BiP motifs with scores greater than 10 were identified in the delta zein protein using this scoring system (Fig. 1). The first putative BiP motif designated as A on Fig. 1 is located in a coiled region on the secondary structure. The second putative BiP motif designated as B is located within an alpha helical region on the predicted secondary structure. The last two putative BiP motifs (C and D) are both located at the carboxy terminus of the delta zein protein within the beta sheet region. Mutations were designed for each putative BiP motif which would theoretically “knockout” or prevent BiP binding (Fig. 1).
Protein–protein interactions with prokaryotic two-hybrid BacterioMatch system
The coding regions for the beta zein, delta zein, and BiP (blp4 from tobacco) were introduced into the pBT and pTRG vectors of the BacterioMatch system. Protein–protein interaction analysis with the BacterioMatch system confirms recent results reported by Kim et al. that beta and delta zein have high binding affinity for each other. Interestingly, delta zein–delta zein interactions are greater than beta–delta zein interactions (Fig. 2). Protein interaction analyses with BiP and delta zein indicate a specific protein–protein interaction between these two proteins (Fig. 2).
β-galactosidase activity from protein–protein interaction assays performed in the BacterioMatch two-hybrid system. β-galactosidase activities directly correlate to the level of protein–protein interaction which occurs within this two-hybrid system. The error bars measure the standard deviation observed between the measurements. Three assays were performed for each sample and each assay was performed in triplicate. LGF and Gal11 were utilized as negative protein–protein interaction controls for each assay
Mutations were introduced in the four putative BiP motifs within the delta zein protein with the intent of disrupting BiP binding (Fig. 1). The motifs were modified to reduce their scores to less than zero. Conserved amino acids were chosen for the modifications in an attempt to minimize disruption of the tertiary structure. These delta zein genes containing the mutations were introduced into the target vector of the BacterioMatch system. Fusion protein was confirmed by western analysis for all constructs (data not shown). Protein-interaction analysis with the mutated delta zein genes resulted in decreased BiP binding in all mutants. BiP binding with mutant A was decreased by approximately 20% (Fig. 2). BiP binding with mutant B was completely eliminated. BiP binding with mutant C was decreased by approximately 40% and binding of mutant D with BiP was decreased nearly 57%. These results indicate that motifs B, C, and D are necessary for BiP and delta zein interaction.
Differences in protein interaction with beta and delta zeins were also observed with the delta zein mutants A, B, C, and D. Mutants A and B had no identifiable interaction with beta zein or delta zein. Mutant C did not interact with beta zein but had limited interaction with the delta zein protein. Mutant D interacted with beta zein and delta zein to nearly the same level as the normal delta zein. These results indicate that motif A and B are necessary for zein–zein interaction.
RNA analysis of A. tumefaciens infiltrated plants
Quantative RT-PCR was performed to determine the relative delta zein transcript levels within the infiltrated plants. Actin levels were also determined for each sample as an internal control. The average ratio of actin versus delta zein levels from three different infiltration experiments are shown for each infiltrated plant in Fig. 3a. These results show that transcript synthesis and stability does not decrease by the mutations.
RNA and in vivo synthesis data (a). Reverse-transcriptase PCR of Agrobacterium-infiltrated plants. Quantative RT-PCR was performed using primers for delta-zein. Primers for exon 2 of tobacco actin were also utilized as an internal control. The average ratios of actin to delta-zein transcript from three separate infiltration experiments are indicated in the graph. The error bars indicate the standard deviation from the three separate quantitative PCR runs. Delta zein transcript is present in the δ zein control along with mutation A,B,C, and D. No product is observed in the non-infiltrated (NI) control. (b) In vivo synthesis of plants infiltrated with δ zein (pDSpZ) and mutants A, B, C, and D performed in the presence of 35S methionine for 5 h. Alcohol/beta-mercaptoethanol soluble protein synthesis bands corresponding to δ zein and mutants A, B, C, and D are visualized following exposure to autoradiography film. NI is the non-infiltrated control at time 0 h and 5 h of in vivo synthesis
In vivo synthesis of A. tumefaciens infiltrated plants
In vivo synthesis was performed with leaf-discs of infiltrated plants 96 h post infiltration. Leaf-discs were incubated with 35S methionine in the presence of light for 5 h. Duplicate leaf-discs were then chased with cold [L]-methionine and [L]-cysteine for 18 h. After 5 h synthesis in the presence of methionine, alcohol and beta-mercaptoethanol soluble protein bands corresponding to delta zein mutants A, B, C, and D (Fig. 3b) were visualized. No bands were detected in the non-infiltrated samples at time point zero or 5 h (Fig. 3b). Following 18 h cold chase with [L]-methionine and [L]-cysteine only delta zein protein was visualized, no corresponding protein bands were visualized for mutants A, B, C, and D (data not shown). This data suggests that the mutants are rapidly degraded and turned over in vivo.
Protein analysis after Agrobacterium infiltration
In vivo analysis of each delta zein mutant was performed in leaf tissue of Nicotiana benthamiana via A. tumefaciens infiltration. A. tumefaciens containing delta zein (pDSpZ), and mutants A, B, C, and D were infiltrated into leaf tissue. The leaf tissue was analyzed for the presence of delta zein by western analysis. Delta zein (pDSpZ) is stable and accumulates in infiltrated plants as previously shown in transgenic plants (Randall et al. 2000). However, mutants A, B, C, and D do not accumulate to any detectable level in infiltrated plants (Fig. 4a). Infiltration of A. tumefaciens containing the beta zein gene (pMEZ) was performed. Western analysis indicates that beta zein is stable and accumulates in infiltrated plants as previously reported in transgenic plant work (Bagga et al. 1995). Co-infiltration of the beta zein with both the delta-10 kD zein and the delta-18 kD zein was performed. Western analysis demonstrates that co-infiltration with the beta zein stabilizes the delta-10 kD and delta-18 kD zein proteins and they accumulate to higher levels as previously reported for stable transformed plants (Fig. 4b, c) (Bagga et al. 1997; Hinchliffe and Kemp 2002). Co-infiltration of the beta zein with each mutant was performed to determine if beta zein could stabilize the mutant proteins as demonstrated for the normal delta zein proteins. Western analysis of the mutants co-infiltrated with beta zein reveals that mutant D is stabilized upon co-infiltration with the beta zein protein. This stabilization of mutant D is due to the direct interaction of mutant D with the beta zein protein. No detectable amounts of mutants A, B, or C co-infiltrated with beta zein were visualized.
Western analysis of Agrobacterium-infiltrated plants. (a) Twenty five micrograms of ethanol soluble protein isolated from plants infiltrated with δ zein (pDSpZ) and mutants A, B, C, and D was separated on a 16% SDS-PAGE gel prior to transfer to nitrocellulose and immunodetected using 1:1000 dilutions of anti-β zein and anti-δ zein antiserums. Lane NI is the non-infiltrated control. Lane M is a broad range molecular weight marker. δ zein protein accumulates in leaf tissue. No zein protein bands are visualized in the infiltrated plants containing mutants A, B, C or D. (b) Twenty five micrograms of ethanol soluble protein isolated from plants co-infiltrated with β zein was separated on a 16% SDS-PAGE gel prior to transfer to nitrocellulose and immunodetected using 1:1000 dilutions of anti-β zein and anti-δ zein antiserums. Lane M is the low molecular range marker. Lane β is protein from a plant infiltrated with β zein. Lanes δ, A, B, C, and D are samples that were co-infiltrated with β zein. δ zein bands are observed the in normal δ zein (pDSpZ) lane and in mutant D. Protein bands corresponding to δ zein are not observed in mutants A, B, and C. (c) Twenty five micrograms of ethanol soluble protein isolated from co-infiltrated plants was separated on a 16% SDS-PAGE gel prior to transfer to nitrocellulose and immunodetected using 1:1000 dilutions of anti-β zein and anti-δ zein antiserums. Lane M is the low molecular range marker. Lane δ* zein is protein from leaf tissue infiltrated with the δ-18 kD zein. Lane δ*+β is protein from leaf tissue infiltrated with the δ-18 kD zein and β zein (pMEZ). Lane δ*+δ is protein from leaf tissue infiltrated with the δ-18 kD zein and δ-10 kD zein. Lane ω is protein from leaf tissue infiltrated with the pWL construct. Lane ω+δ is protein from leaf tissue co-infiltrated with pWL and δ-10 kD zein. Lane ω+C is protein from leaf tissue co-infiltrated with pWL and mutant C. Lane ω+A is protein from leaf tissue co-infiltrated with pWL and mutant A
Co-infiltration of the delta-10 kD zein with the delta-18 kD zein was performed. Surprisingly, stabilization and higher accumulation of the delta-18 kD zein was observed by western analysis. Co-infiltration of delta-18 kD zein with mutant C (which exhibits interaction with the delta-10 kD zein in the BacterioMatch system (Fig. 2)) was performed. However, the delta-18 kD zein protein is not stable when expressed alone (Hinchliffe and Kemp 2002) and extremely low levels of delta-18 kD zein plant are observed by western analysis in these co-infiltrated plants and no detectable mutant C protein is visualized (data not shown). To overcome the stability issue of the delta-18 kD zein protein the construct pWL which contains both the delta-18 kD zein and the beta zein was utilized. This construct results in the accumulation of higher amounts of stable delta-18 kD zein protein and beta zein protein (Fig. 4; Klypin et al., in progress). Co-infiltration of mutant C and mutant A with pWL was performed. Western analysis indicates that mutant C is stabilized and now accumulates to high levels in the presence of the stabilized delta-18 kD zein (Fig. 4c). This is due to the direct interaction with mutant C and the delta-18 kD zein protein (Fig. 2). No detectable protein was visualized for mutant A co-infiltrated with pWL (Fig. 4c) or mutant B (data not shown).
Electron microscopy analysis of infiltrated plants
Electron microscopy analysis was performed on leaves of infiltrated plants to determine if protein bodies are formed within the plants. Protein bodies are produced in leaves of infiltrated plants containing the delta zein gene and the co-infiltrated genes (mutant D+pMEZ and mutant C+pWL) (Fig. 5). The protein bodies in these infiltrated plants resemble protein bodies previously reported in the leaves of transgenic plants (Bagga et al. 1995; Randall et al. 2000). These protein bodies, like other transgenic zein protein bodies, have a surrounding membrane network (Fig. 5). Not surprisingly, A. tumefaciens was observed attached to the cell wall of a plant cell producing zein protein bodies (Fig. 5a). The leaves co-infiltrated with the beta zein and mutant D zein form protein bodies as those previously observed in transgenic tobacco leaves co-expressing beta and delta zein proteins. The mutant D zein proteins and beta zein proteins were immunolocalized within the protein bodies using both the anti-delta zein antibodies and anti-beta zein antibodies (Fig. 5b,c).
Electron microscopy analysis of leaves co-infiltrated with beta zein (pMEZ) and mutant D. Leaf tissue was fixed, labeled for immunolocalization of zein proteins and stained. (a) Leaf tissue of co-infiltrated beta + mutant D. Agrobacterium (labeled A) is attached to the cell wall of a cell with protein bodies. The scale bar denotes 0.4 μm. (b) Immunolocalization of mutant D zein within the leaves of co-infiltrated beta + mutant D plants was performed using rabbit anti-delta zein antibodies 1:100 followed by labeling with 12-nm gold conjugated goat-anti-rabbit IgG antibodies (1:40). The mutant D zein protein was immunolocalized within the protein bodies. The scale bar denotes 0.25 μm. (c) Immunolocalization of the beta zein proteins within the leaves of co-infiltrated beta + mutant D was performed using rabbit anti-β zein antibodies diluted 1:100 followed by labeling with 12 nm gold conjugated goat-anti-rabbit IgG (1:40). The scale bar denotes 0.18 μm. The arrows indicate gold particle labeling within the protein bodies. Mitochondria are labeled MT. Cell walls are labeled CW. Protein bodies are labeled PB. Chloroplasts are labeled CP
The leaves co-infiltrated with the pWL gene construct and mutant C zein form spherical protein bodies as previously observed in transgenic plants accumulating the delta zein protein (data not shown) (Randall et al. 2000). The mutant C zein protein along with the delta-18 kD zein protein was immunolocalized within the bodies using the anti-delta zein antibodies and the beta zein protein was immunolocalized using the anti-beta zein antibodies (data not shown). In contrast, the mutant A construct results in no detectable protein accumulation (Fig. 4) and no observable protein bodies (data not shown).
Discussion
In the last several years, it has become increasingly apparent that protein–protein interactions are the determinants for function and cell structure within all eukaryotic and prokaryotic organisms. Although the paradigm that structure determines function is currently being replaced with binding determines function (Van Regenmortal 2002), the two actually go hand in hand. It is, in fact, the basis of protein structure which determines protein interaction and binding. Therefore, modifying protein structure can obviously alter these protein–protein interactions and hence modify cellular function and structure.
An association between BiP and zein protein bodies is well established in both maize endosperm and transgenic plants (Bagga et al. 1997; Bellucci et al. 2000; Boston et al. 1991; Geli et al. 1994). However, direct protein–protein interactions and their responsible domains between the zeins and BiP have not been described. Our recent experiments using an E.Coli two-hybrid system give the first direct evidence that BiP (blp4 from tobacco) does indeed interact with both the delta and beta zeins (Fig. 2). In this study, four putative BiP binding motifs were identified within the delta zein protein using a BiP scoring program (Blond-Elguindi et al. 1993). The first two BiP binding motifs (A and B) are located on the N-terminal region of the delta zein protein (Fig. 1). The third and fourth BiP binding motifs (C and D) are located near the C-terminal region of the protein (Fig. 1). These putative binding motifs were mutated and their effects on protein interactions were analyzed in both a prokaryotic two-hybrid system and in plants.
The data obtained with the mutants demonstrates that there are three distinct binding motifs within the delta zein protein. The first binding motif is located at the N-terminal region of the delta zein protein is composed of both motif A and motif B and is necessary for beta and delta zein and delta and delta zein interactions. Beta and delta zein protein interactions are completely eliminated when mutations are introduced in motif A and motif B (mutants A and B) (Fig. 2, summary Table 2). Motif B specifically, appears to be necessary for BiP interaction with delta zein as mutations in motif B (mutant B) completely eliminates interaction with BiP (Fig. 2, summary Table 2). There is no detectable protein accumulation in plant tissue infiltrated with mutants A and B. This lack of protein accumulation is not due to a change in RNA stability as transcript levels in infiltrated plants are comparable between the delta zein (pDspZ) and the mutants (Fig. 3a). In vivo synthesis with 35S methionine indicates that the lack of protein accumulation in the mutants is not due to reduced protein synthesis but due to increased degradation and protein turnover.
The second binding domain is motif C which is located at the C-terminus of the delta zein protein. Motif C is responsible for interaction of the delta zein protein with the beta zein protein. This was demonstrated by the fact that the mutation introduced in motif C (mutant C) eliminates interaction with beta zein and has a moderately reduced interaction with delta zein (Fig. 2). Unlike mutant C, mutant D exhibits no change in its interaction with beta and delta zeins. As with mutants A and B, leaves infiltrated with mutants C or D alone have no detectable delta zein protein accumulation. However, mutant C is stabilized within the leaves of tobacco plants when it is co-infiltrated with the delta-18 kD zein (pWL). However, it is important to note that this stabilization is due to its specific interaction with the delta-18 kD zein which accumulates within the leaf tissue since mutant C is unable to interact with the beta zein (Fig. 2). Co-infiltration of mutant C with the beta zein does not result in stabilization which is also not unexpected as mutant C no longer interacts with the beta zein. Alternatively, our data also demonstrates that the delta-18 zein protein can be stabilized by the un-mutated delta-10 zein protein. Likewise, when mutant D is co-infiltrated with the beta zein gene (pMEZ) the mutant D protein is stabilized and accumulates to detectable levels within the leaves (Fig. 4, summary Table 2). This is not surprising since mutating motif D did not interfere with its interactions with the beta or delta zein proteins (Fig. 2). Electron microscopy analysis indicates that this mutant protein is localized within protein bodies. These co-infiltration stabilization findings confirm the interactions observed within the two-hybrid system.
The third binding motif is located at the C-terminal region of the delta zein and is necessary for BiP–delta zein interaction. This binding domain consists of both motifs C and D which interact with BiP but has limited interaction with beta and delta zeins (Fig. 2). Although mutants C and D may be able to interact with BiP to some degree in vivo, this interaction is not enough to stabilize the delta zein protein. This is presumably due to the fact that BiP is unable to retain these mutants within the ER. However, as described above, these mutants can be rescued by their interaction with a wild type zein whose ability to bind to BiP has not been disrupted.
It appears that in order for the delta zeins to be stable and produce protein bodies they must first be able to interact with BiP for retention within the ER. In earlier work, a KDEL motif was engineered behind the gamma zein which resulted in an increase in gamma zein accumulation in transgenic plants (Bellucci et al. 2000). Interestingly, BiP accumulation did not increase in these gamma-KDEL zein transgenic plants presumably because it is no longer required for ER retention (Bellucci et al. 2000). Previously, it was speculated that the gamma zein tandem repeat PPPVHL which is necessary for retention in the ER may bind to BiP (Geli et al. 1994). We have analyzed the gamma zein sequence for putative BiP binding motifs and it appears that this repeat may indeed be part of a BiP binding motif (data not shown). Recent experiments with the N-terminal portion of the gamma zein indicate that this tandem repeat is amphipathic and may be a trans-membrane domain (Kogan et al. 2004). If this tandem repeat is a trans-membrane domain this could account for the retention of the gamma zein within the ER. Recently the chimeric protein zeolin, composed of phaseolin and gamma zein, was shown to have direct interaction with BiP (Mainieri et al. 2004). However, which component of the chimeric protein is directly interacting with BiP has yet to be determined (Mainieri et al. 2004).
The second requirement necessary for the stability of delta zeins and their ability to produce protein bodies is their capability to interact with themselves or with another zein protein (i.e. beta). This result mirrors the interaction of alpha zein with the gamma zein protein. For example, the alpha zein protein does not accumulate to detectable levels in transgenic plants (Coleman et al. 1996) although when we analyzed this protein for putative BiP motifs it does seem to contain approximately 15 putative BiP motifs (data not shown). This lack of stability with the alpha zein is probably due to its inability to interact with itself (Kim et al. 2002). However, when the alpha zein protein is synthesized in the presence of gamma zein protein it is stabilized and accumulates in transgenic plants.
In this study, we have shown that there are specific motifs required for zein–zein interaction and BiP–zein interaction. However, we can not rule out the possibility that there are other proteins that are involved in protein body biogenesis and hence other binding partners to the delta zein protein. Identifying these other proteins and their interactions will greatly contribute to the understanding of protein body biogenesis.
Abbreviations
- BiP:
-
Binding luminal protein
- ER:
-
Endoplasmic reticulum
References
Bagga S, Adams H, Kemp JD, Sengupta-Gopalan C (1995) Accumulation of 15-kilodalton zein in novel protein bodies in transgenic tobacco. Plant Physiol 107:13–23
Bagga S, Adams HP, Rodriguez FD, Kemp JD, Sengupta-Gopalan C (1997) Coexpression of the maize delta-zein and beta-zein genes results in stable accumulation of delta-zein in endoplasmic reticulum-derived protein bodies formed by beta-zein. Plant Cell 9:1683–96
Bellucci M, Lazzari B, Viotti A, Arcioni S (1997) Differential expression of g-zein gene in Medicago sativa, Lotus corniculatus and Nicotiana tabacum. Plant Sci 127:161–169
Bellucci M, Alpini A, Paolocci F, Cong L, Arcioni S (2000) Accumulation of maize g-zein and g-zein:KDEL to high levels in tobacco leaves and differential increase of BiP synthesis in transformants. Theor Appl Genet 101:796–804
Blond-Elguindi S, Cwirla SE, Dower WJ, Lipshutz RJ, Sprang SR, Sambrook JF, Gething MJ (1993) Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of BiP. Cell 75:717–728
Boston RS, Fontes EB, Shank BB, Wrobel RL (1991) Increased expression of the maize immunoglobulin binding protein homolog b-70 in three zein regulatory mutants. Plant Cell 3:497–505
Coleman CE, Herman EM, Takasaki K, Larkins BA (1996) The maize gamma-zein sequesters alpha-zein and stabilizes its accumulation in protein bodies of transgenic tobacco endosperm. Plant Cell 8:2335–2345
Ems-McClung SC, Benmoussa M, Hainline BE (2002) Mutational analysis of the maize gamma zein C-terminal cysteine residues. Plant Sci 2002:131–141
Foresti O, Frigerio L, Holkeri H, deVirgilio M, Vavassori S, Vitale A (2003) A phaseolin domain involved directly in trimer assembly is a determinant for binding by the chaperone BiP. The Plant Cell 15:2464–2475
Geli MI, Torrent M, Ludevid D (1994) Two structural domains mediate two sequential events in [gamma]-zein targeting: protein endoplasmic reticulum retention and protein body formation. Plant Cell 6:1911–1922
Geourjon C, Deleage G (1994) SOPM: a self optimized prediction method for protein secondary structure prediction. Protein Eng 7:157–164
Ghoshroy S, Citovsky V (1998) Preservation of plant cell ultrastructure during immunolocalization of virus particles. J Virol Methods 74:223–229
Hinchliffe DJ, Kemp JD (2002) b-Zein protein bodies sequester and protect the 18-kDa d-zein protein from degradation. Plant Sci 163:741–752
Horton R (1989) Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61–68
Kim CS, Woo Ym YM, Clore AM, Burnett RJ, Carneiro NP, Larkins BA (2002) Zein protein interactions, rather than the asymmetric distribution of zein mRNAs on endoplasmic reticulum membranes, influence protein body formation in maize endosperm. Plant Cell 14:655–72
Kirihara JA, Hunsperger JP, Mahoney WC, Messing JW (1988) Differential expression of a gene for a methionine-rich storage protein in maize. Mol Gen Genet 211:477–484
Knarr G, Modrow S, Todd A, Gething MJ, Buchnert J (1999) BiP-binding sequences in HIV gp160. J Biol Chem 274:29850–29857
Kogan, MJ, Lopez O, Cocera M, Lopez-Iglesias C, De La Maza A, Giralt E (2004) Exploring the interaction of the surfactant N-terminal domain of gamma-zein with soybean phosphatidylcholine liposomes. Biopolymers 73(2):258–268
Leborgne-Castel N, Jelitto-Van Dooren EPWM, Crofts AR, Denecke J (1999) Overexpression of BiP in tobacco alleviates endoplasmic reticulum stress. Plant Cell 11:459–469
Lending CR, Larkins BA (1989) Changes in the zein composition of protein bodies during maize endosperm development. Plant Cell 1:1011–1023
Li X, Wu Y, Zhang DZ, Gillikin JW, Boston RS, Franceschi VR, Okita TW (1993) Rice prolamine protein body biogenesis: a BiP mediated Process
Mainieri D, Rossi M, Archiniti M, Bellucci M, De Marchis F, Vavassori S, Pompa A, Arcioni S, Vitale A (2004) Zeolin. A new recombinant storage protein constructed using maize gamma-zein and bean phaseolin. Plant Physiol 136:3447–3456
Molinari M, Galli C, Piccaluga V, Pieren M, Paganetti P (2002) Sequential assistance of molecular chaperones and transient formation of covalent complexes during protein degradation from the ER. J Cell Biol 158:247–257
Ng DTW, Spear ED, Walter P (2000) The Unfolded Protein Response regulates multiple apsects of secretory and membrane protein biogenesis and endoplasmic reticulum quality control. J Cell Biol 150:77–88
Nuttall J, Vitale A, Frigerio L (2003) C-terminal extension of phaseolin with a short methionine-rich sequence can inhibit trimerisation and result in high instability. Plant Mol Biol 51:885–894
Randall J (1997) Targeting and stabilization of 10 kD and 15 kD zeins in transgenic tobacco. NMSU
Randall J, Bagga S, Adams H, Kemp JD (2000) A modified 10-kDa zein protein produces two morphologically distinct protein bodies in transgenic tobacco. Plant Sci 150:21–28
Randall J, Sutton D, Ghoshroy S, Bagga S, Kemp JD (2004) Co-ordinate expression of b- and d-zeins in transgenic tobacco. Plant Sci 167:267–372
Reynolds A, Lundbland V, Dorris D, Keaveney M (1997) Assay for b-galactosidase in liquid culture. Current protocols in molecular biology. Wiley, New York, pp 13.6.1–13.6.6
Rogers S, Klee H, Horsch R, Fraley R (1987) Improved vectors for plant transformation: expression cassette vectors and new identifiable markers. Meth Enzymol 153:253–263
Tottey S, Rondet SAM, Borrelly G, Robinson PJ, Rich PR, Robinson NJ (2002) A copper metallochaperone for photosynthesis and respiration reveals metal-specific targets, interaction with an importer, and alternative sites for copper acquisition. J Biol Chem 277:5490–5497
Van Regenmortal MHV (2002) A paradigm shift is needed in proteomics: structure determines function should be replaced by binding determines function. J Mol Recognit 15:349–351
Vitale A, Ceriotti A (2004) Protein Quality control mechanisms and protein storage in the endoplasmic reticulum. A conflict of interests?. Plant Physiol 136:3420–3426
Voinnet O, Rivas S, Mestre P, Baulcombe D (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33:949–956
Acknowledgments
The authors would like to thank Jennifer Jackson for her superb technical support of this work. The authors also thank Lorenzo Aleman for the actin primers and his critical reading of the manuscript. The authors express their gratitude to Dr. Soumitra Ghoshroy for his assistance with the EM work. The authors thank Dr. Suman Bagga for the providing the pMEZ construct, Dr. Doug Hinchliffe for providing the pMH316-18 construct, and Dr. Jose Louis Ortoga for the oliga dT primer. The authors would also like to thank Dr. Sylvie Blond-Elguindi for the BiP program, Dr. Denecke for the blp4 gene, Dr. Boston for the BiP polyclonal antibodies, and Dr. Messing for the δ zein polyclonal antibodies. The research was supported by special grant 2003-34250-13279.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Randall, J.J., Sutton, D.W., Hanson, S.F. et al. BiP and zein binding domains within the delta zein protein. Planta 221, 656–666 (2005). https://doi.org/10.1007/s00425-005-1482-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-005-1482-z