Abstract
Quantification of protein expression based on immunohistochemistry (IHC) is an important step in clinical diagnoses and translational tissue-based research. Manual scoring systems are used in order to evaluate protein expression based on staining intensities and distribution patterns. However, visual scoring remains an inherently subjective approach. The aim of our study was to explore whether digital image analysis proves to be an alternative or even superior tool to quantify expression of membrane-bound proteins. We analyzed five membrane-binding biomarkers (HER2, EGFR, pEGFR, β-catenin, and E-cadherin) and performed IHC on tumor tissue microarrays from 153 esophageal adenocarcinomas patients from a single center study. The tissue cores were scored visually applying an established routine scoring system as well as by using digital image analysis obtaining a continuous spectrum of average staining intensity. Subsequently, we compared both assessments by survival analysis as an end point. There were no significant correlations with patient survival using visual scoring of β-catenin, E-cadherin, pEGFR, or HER2. In contrast, the results for digital image analysis approach indicated that there were significant associations with disease-free survival for β-catenin, E-cadherin, pEGFR, and HER2 (P = 0.0125, P = 0.0014, P = 0.0299, and P = 0.0096, respectively). For EGFR, there was a greater association with patient survival when digital image analysis was used compared to when visual scoring was (visual: P = 0.0045, image analysis: P < 0.0001). The results of this study indicated that digital image analysis was superior to visual scoring. Digital image analysis is more sensitive and, therefore, better able to detect biological differences within the tissues with greater accuracy. This increased sensitivity improves the quality of quantification.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Proteins overexpressed on the surface of tumor cells can be selectively targeted. Epidermal growth factor receptor (EGFR) and human epidermal growth factor receptor 2 (HER2) are among the most common targeted proteins in cancer therapy. Accurate determination of HER2 or EGFR expression is essential for the prediction of a cancer patient’s response to therapy and prognosis (Montemurro and Scaltriti 2014; Fusco et al. 2013). To make early, rational, and correct clinical decisions, a reliable detection of biomarkers is essential. Particularly, for HER2 evaluation, the ability to reliably identify patients with breast cancer or gastric cancer who might benefit from trastuzumab treatment is not only important for clinical reasons (high proportion of cardiotoxicity), but also for economic ones well (about 50,000–60,000 euros per treatment/quality-life adjusted year). The definition of HER2-positive status affects the clinical- and cost-effectiveness of trastuzumab (Spackman et al. 2013; Norman et al. 2011). Manual scoring is routinely used to assess protein expression based on staining intensities and distribution patterns. However, this approach is subjective and is prone to significant intra- and interobserver variability that limits reproducibility and statistical confidence (Braun et al. 2013; Dobson et al. 2010; Mulrane et al. 2008). Developed in response to this disadvantage, digital image analysis techniques offer an objective approach and the possibility for increased sensitivity and improved accuracy (Mohammed et al. 2012b; Dobson et al. 2010).
In this study, we analyzed five membrane-binding biomarkers: HER2, which is clinically approved for therapy response prediction, and EGFR, pEGFR, β-catenin, and E-cadherin which are used in tissue-based research. We applied these five biomarkers on tissue microarrays (TMAs) containing tumor tissue from 153 esophageal adenocarcinoma patients. We compared the evaluation of immunohistochemistry (IHC) by using digital image analysis with visual scoring of these five biomarkers. Kaplan–Meier survival analysis of disease-free and overall survival as clinical end points was used to determine and compare the prognostic significance of the two approaches.
Materials and methods
Patient selection and tissue samples
All patients (n = 153) underwent primary surgical resection between 1995 and 2005 without chemotherapy or radiotherapy at the Department of Surgery, Klinikum Rechts der Isar, Technische Universität München. All cases had adenocarcinomas of the distal esophagus (Barrett’s cancer) associated with histopathologically identified Barrett’s esophagus (according to WHO 2000 criteria). Data were acquired after receiving approval from the ethics committee of the Technische Universität München. Survival analyses results were calculated on all 153 patients. The mean age at the time of surgery was 64 years, and the maximum follow-up time was 164 months (median disease-free survival time = 31 months; median overall survival time = 33 months). The results describing the clinicopathologic characteristics of the collective are presented in Table 1.
Immunohistochemistry
Preparation of TMAs was performed as previously described, generating triplicate cores with a diameter of 1.0 mm each (Berezowska et al. 2013). Serial tissue sections were cut (5 µm) and transferred to slides (Rauser et al. 2007). The sections were incubated with antibodies specific for β-catenin (1:500, BD Bioscience, Heidelberg, Germany), E-cadherin (1:1,500, BD Bioscience, Heidelberg, Germany), pEGFR (Y1086) (1:100, Invitrogen, Karlsruhe, Germany), EGFR (pharmDx™-kit, DAKO, Hamburg, Germany), and HER2 (HercepTest kit, DAKO, Hamburg, Germany). Immunohistochemical staining was performed using a Discovery XT automated stainer (Ventana, Tucson, AZ, USA). Positive and negative controls were included in each staining procedure.
Only cores with technically unequivocal staining results and with sufficient tumor content (>50 tumor cells) were used for visual scoring and image analysis.
Visual scoring
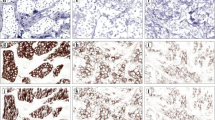
Determination of HER2 expression was assessed according to the published guidelines for routine HER2 evaluation, and each sample was classified as 0, 1+, 2+, or 3+ (Wolff et al. 2013; Ruschoff et al. 2010; Hofmann et al. 2008). The immunohistological expression of the other membrane-binding proteins (i.e., EGFR, β-catenin, E-cadherin, and pEGFR) was classified into four levels according to HER2 evaluation (Fig. 1). Cytoplasmic staining was considered a non-specific result and was not included in the scoring. The evaluation was performed by two independent pathologists (RL and AW). In the case of a disagreement, discussion was used to reach a consensus.
Digital image analysis
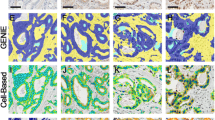
All stained slides were scanned at 20 × objective magnification using a Mirax Desk digital slide scanner (Carl Zeiss MicroImaging, Munich, Germany). Immunohistochemical membrane staining results were quantified using the commercially available image analysis software Definiens Developer XD2 (Definiens AG, Munich, Germany). This software allows to detect and quantify the immunohistochemical staining intensities in different cellular compartments (e.g., membranes) within a user-specified region of interest of the tumor cells. Algorithms were developed and modified specifically for each marker based on semantic and context-based segmentation processes, which include staining intensity, color features, shape, area, and neighborhood. The quantified parameter for EGFR, β-catenin, E-cadherin, pEGFR, and HER2 was a value representing a point on a continuous spectrum of average brown staining intensity in relative units (Fig. 2). A mean value was calculated from triplicate tissue cores from each patient.
An example of digital image analysis assessment of strong (a–c) and weak (d–f) β-catenin staining of esophageal adenocarcinoma tissue. a, d Original image, b, e detection of membrane (black), cytoplasm (green), nuclei (blue), c, f classification of membrane staining intensity (heat map for membrane staining intensity: from weak (yellow) to strong (red))
The results were correlated with patient survival times and compared with the visual scoring results.
Statistical analysis
Disease-free and overall survival rates were calculated using the Kaplan–Meier method and include median and 95 % confidence interval estimates. Survival curves were tested with the log-rank χ 2 value and Cox proportional hazards regression analysis. In each case, the cutoff point was optimized with respect to the end point. Correlation analyses were performed using the Pearson rank test. Calculations were performed using the statistical data analysis system R (‘stats’ and ‘survival’ procedures; Bell Laboratories, Muray Hill, NJ, USA) and SAS statistical software version 9.2 (SAS Institute Inc, Cary, NC, USA). All tests were two-sided, and P values <0.05 were considered to be statistically significant.
Results
Comparison of digital image analysis and visual scoring with pTNM and G status
The results for the correlation analysis of visual scoring and computer-assisted evaluation of IHC expression with pTNM and G status are summarized in Table 2. There was a significant correlation for visual scoring of E-cadherin and pEGFR1086 with pN-status (E-cadherin: P = 0.008, pEGFR: P = 0.0197) and based on image analysis with pT-status (E-cadherin: P = 0.0046, pEGFR: P = 0.0339) and pN-status (E-cadherin: P = 0.0115, pEGFR: P = 0.0125). For EGFR, the results indicated that there was a significant correlation with pT-, pN-, and pM-status based on visual scoring (pT: P = 0.0014, pN: P = 0.0125, pM: P = 0.0076) and digital image analysis (pT: P = 0.0004, pN: P = 0.0284, pM: P = 0.0119). The results for the HER2 analysis indicated that there was a significant correlation between evaluation with digital image analysis and G status.
The results for the comparison between the visually scored evaluation and the continuous values generated using digital image analysis of the membrane-binding proteins (β-catenin, E-cadherin, pEGFR, EGFR, and HER2) to determine the level of agreement between both methods are presented in Table 3. The Pearson rank correlation coefficients were 0.71, 0.68, 0.89, 0.79, and 0.74 for β-catenin, E-cadherin, pEGFR, EGFR, and HER2, respectively (a P < 0.0001 for all comparisons).
Comparison of digital image analysis with survival analysis
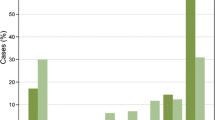
We also performed a comparative survival analysis. Based on the visual evaluation, β-catenin overexpression (score 3+) was found in 44.6 % of the patients, and no or weak expression (score 0, 1+, or 2+) in 55.4 % (Table 4). There were no significant correlations between β-catenin expression and disease-free survival (P = 0.4511) or overall survival times (P = 0.1868) (Fig. 3).
The results for HER2 and pEGFR were similar to the results for β-catenin. Using visual assessment, there were no statistically significant prognostic effects on disease-free survival (HER2: P = 0.0560; pEGFR: P = 0.2848) or overall survival times (HER2: P = 0.0640; pEGFR: P = 0.2982) (Table 5). A strong HER2 expression (score 3+) was detected in 9.4 % of the patients, but most (90.6 %) had moderate or no expression of HER2 (score 0, 1+, or 2+), and 29.4 % of the patients showed an overexpression (score 1+, 2+, or 3+) of membrane-bound pEGFR, and for 70.6 %, we found no signal (score 0) (Table 3).
Although there were no statistically significant associations with patient survival and β-catenin, pEGFR, or HER2 (P > 0.05), based on visual scoring, there was significant association between biomarker analyzed by digital image analysis and disease-free survival (β-catenin: P = 0.0125; HER2: P = 0.0096 and pEGFR: P = 0.0299) and overall survival (β-catenin: P = 0.0063; HER2: P = 0.0022 and pEGFR: P = 0.0135) (Table 5). β-Catenin and pEGFR overexpression were significantly associated with good prognosis, and HER2 overexpression was associated with a poor clinical outcome.
Based on visual evaluation, overexpression of E-cadherin was significantly positively associated with overall survival (P = 0.0145), but the association with disease-free survival was not statistically significant (P = 0.0526). An overexpression (score 3+) was present in 40.4 % of the patients, and a weak or no expression (score 0, 1+, or 2+) was present in the remaining 59.6 % of the patients. By using digital image analysis for the evaluation of E-cadherin, we were able to enhance the visually assessed level of significance for correlation with both overall survival (P = 0.0004) and disease-free survival (P = 0.0014), resulting in statistically significant good prognosis for high E-cadherin expression.
The Kaplan–Meier analysis of EGFR revealed that overexpression (score 1+, 2+, or 3+) was characterized by a poorer prognosis, in terms of overall survival (P = 0.0091) and disease-free survival (P = 0.0045). Accordingly, for 22.1 % of the patients, we observed a high expression of EGFR (score 1+, 2+, or 3+) and for 77.9 % no expression (score 0). For EGFR, the level of significance for visual evaluation was greatly increased for both disease-free survival (P > 0.0001) and overall survival (P > 0.0001) by using digital image analysis.
For each of the five membrane biomarkers, the results indicated that compared with visual scoring, biomarkers evaluated by digital image analysis had a greater association with both disease-free and overall survival. Based on digital image analysis, we obtained statistically significant results for the survival analyses of all of the five biomarkers. The results indicated that there was a positive association with clinical outcome for β-catenin, E-cadherin, and pEGFR. Overexpression of HER2 or EGFR was associated with poor prognosis.
Combinatorial survival analysis
We performed for all markers a combinatorial survival analyses of two markers each, based on computer-assisted evaluation. Therefore, patients with good prognosis for both markers or poor prognosis for both markers, respectively, were taken in one group for survival analysis. The results indicated that a combinatorial evaluation of HER2 and E-cadherin is more accurate in predicting patient overall survival (P = 0.0002) and disease-free outcomes (P = 0.0003) compared to single marker assessment (Table 5). For the remaining marker combinations, no increased significance was observed.
Discussion
There is a considerable need for standardized evaluation of biomarker expression in clinical pathology and in tissue-based research. Currently, the grading and assessment of biomarkers are usually performed using manual scoring systems. Visual evaluation of IHC remains a rather subjective process characterized by significant intra- and interobserver variability and reduced reproducibility, which may lead to inaccurate diagnosis and treatment decisions (Dolled-Filhart et al. 2010; Gudlaugsson et al. 2012; Laurinavicius et al. 2012; Adams et al. 1999). In particular, accurate determination of HER2 status by reducing the number of false positives or false negatives is critical to reliably identify patients who might benefit from trastuzumab treatment. Improvements in this area will reduce human suffering and healthcare costs (Dobson et al. 2010).
Digital image assessment has a huge potential to overcome the limitations of manual scoring systems. Numerous studies in the last two decades have compared visual and computer-assisted scoring, but most of them are limited by comparing just the level of agreement between both methods (Braun et al. 2013; Lloyd et al. 2010; Rizzardi et al. 2012; Tuominen et al. 2012; Nassar et al. 2011; Messersmith et al. 2005), although other studies compared visual scoring and IHC evaluations by digital image analysis using survival analysis, but without confirming an analytical advantage by using image analysis (Mohammed et al. 2012a, c; Ong et al. 2010; Turashvili et al. 2009). In contrary, in our approach, we investigated the association between IHC evaluation and patient outcome, and we found that biomarker evaluation using digital image analysis provided prognostic information beyond that attainable with conventional visual methods. Using image analysis, we found higher associations for biomarker expression with clinical outcome in each case (HER2, EGFR, pEGFR, β-catenin, E-cadherin).
One reason for that is that IHC measurements using digital image analysis are more precise, particularly for moderate staining intensity or heterogeneous IHC expression within the tissue. The intensity of immunoreactivity in tissue is a crucial criterion for scoring. However, visual recognition is subjective, even for the trained pathologist, and the evaluation is performed in a more approximate rather than an accurate manner (Ong et al. 2010; Dobson et al. 2010; Rimm 2006). Visual evaluation is based on distinguishing groups of similar quality, such as color intensity or size, which is prone to misclassification when these qualities slight change. Interobserver variability for HER2 classification tends to be higher when discriminating 1+ and 2+ or 2+ and 3+ cases, while negative cases are straightforward to score (Turashvili et al. 2009). Additionally, the visual approach can be biased resulting in inaccurate scoring values.
Whereas visual assessment is restricted to a pre-defined scoring system consisting of discrete values (0, 1+, 2+, 3+) for statistical analysis, image analysis produces continuous data, which considerably improves the precision of the cutoff value used for survival analysis. This difference resulted in a higher prognostic significance. Continuous variable data make statistical evaluation much more flexible and enable identification of IHC cut points of prognostic relevance that were either undetected or had a lower statistical significance when visual scores were used (Braun et al. 2013; Rizzardi et al. 2012).
Additionally, benefits of a digital image analysis-based approach include objectivity, reproducibility, and reliability of scoring results, which allows for a more robust and standardized IHC evaluation (Minot et al. 2012; Vayrynen et al. 2012; Skaland et al. 2008a).
In recent years, the capabilities of advanced, computer-assisted image analysis have improved significantly and include complex algorithms that are used to interpret images for IHC quantification (Webster and Dunstan 2014; Kayser and Kayser 2013; Foran et al. 2011; Kayser et al. 2009; Rojo et al. 2009). The results of our study confirm that digital image analysis enables investigators to become more sensitive with regard to the prognostic value and to increase the quality of IHC quantification. Using digital image analysis, we were able to identify the biological effects within the tissue much more accurately by providing important data not available when conventional visual approaches are used.
Rimm and co-workers (Welsh et al. 2011; Harigopal et al. 2010; Camp et al. 2003) also found that digital image analysis appears to be more sensitive than manual IHC analysis. In comparing HER2 IHC with fluorescence in situ hybridization (FISH), recent studies report an increased concordance with FISH, and a decrease in the number of cases interpreted as equivocal (2+) when image analysis is used (Minot et al. 2012; Dobson et al. 2010; Skaland et al. 2008b).
In practice, quality control methods are still required during the data analysis and interpretation. Digital image analysis may provide more detailed information and improve quality, but digital image analysis algorithms cannot reliably differentiate between benign and malignant lesions, or recognize artifacts, such as tissue folds, with the same precision as an experienced pathologist.
Based on the findings of this study, digital image analysis has great potential as a diagnostic support tool and may significantly improve the sensitivity and standardization of IHC evaluation.
References
Adams EJ, Green JA, Clark AH, Youngson JH (1999) Comparison of different scoring systems for immunohistochemical staining. J Clin Pathol 52(1):75–77
Berezowska S, Novotny A, Bauer K, Feuchtinger A, Slotta-Huspenina J, Becker K, Langer R, Walch A (2013) Association between HSP90 and Her2 in gastric and gastroesophageal carcinomas. PLoS ONE 8(7):e69098. doi:10.1371/journal.pone.0069098
Braun M, Kirsten R, Rupp NJ, Moch H, Fend F, Wernert N, Kristiansen G, Perner S (2013) Quantification of protein expression in cells and cellular subcompartments on immunohistochemical sections using a computer supported image analysis system. Histol Histopathol 28(5):605–610
Camp RL, Dolled-Filhart M, King BL, Rimm DL (2003) Quantitative analysis of breast cancer tissue microarrays shows that both high and normal levels of HER2 expression are associated with poor outcome. Cancer Res 63(7):1445–1448
Dobson L, Conway C, Hanley A, Johnson A, Costello S, O’Grady A, Connolly Y, Magee H, O’Shea D, Jeffers M, Kay E (2010) Image analysis as an adjunct to manual HER-2 immunohistochemical review: a diagnostic tool to standardize interpretation. Histopathology 57(1):27–38. doi:10.1111/j.1365-2559.2010.03577.x
Dolled-Filhart M, Gustavson M, Camp RL, Rimm DL, Tonkinson JL, Christiansen J (2010) Automated analysis of tissue microarrays. Methods Mol Biol 664:151–162. doi:10.1007/978-1-60761-806-5_15
Foran DJ, Chen W, Yang L (2011) Automated image interpretation computer-assisted diagnostics. Anal Cell Pathol (Amst) 34(6):279–300. doi:10.3233/ACP-2011-0046
Fusco N, Rocco EG, Del Conte C, Pellegrini C, Bulfamante G, Di Nuovo F, Romagnoli S, Bosari S (2013) HER2 in gastric cancer: a digital image analysis in pre-neoplastic, primary and metastatic lesions. Mod Pathol 26(6):816–824. doi:10.1038/modpathol.2012.228
Gudlaugsson E, Skaland I, Janssen EA, Smaaland R, Shao Z, Malpica A, Voorhorst F, Baak JP (2012) Comparison of the effect of different techniques for measurement of Ki67 proliferation on reproducibility and prognosis prediction accuracy in breast cancer. Histopathology 61(6):1134–1144. doi:10.1111/j.1365-2559.2012.04329.x
Harigopal M, Barlow WE, Tedeschi G, Porter PL, Yeh IT, Haskell C, Livingston R, Hortobagyi GN, Sledge G, Shapiro C, Ingle JN, Rimm DL, Hayes DF (2010) Multiplexed assessment of the Southwest Oncology Group-directed Intergroup Breast Cancer Trial S9313 by AQUA shows that both high and low levels of HER2 are associated with poor outcome. Am J Pathol 176(4):1639–1647. doi:10.2353/ajpath.2010.090711
Hofmann M, Stoss O, Shi D, Buttner R, van de Vijver M, Kim W, Ochiai A, Ruschoff J, Henkel T (2008) Assessment of a HER2 scoring system for gastric cancer: results from a validation study. Histopathology 52(7):797–805. doi:10.1111/j.1365-2559.2008.03028.x
Kayser G, Kayser K (2013) Quantitative pathology in virtual microscopy: history, applications, perspectives. Acta Histochem 115(6):527–532. doi:10.1016/j.acthis.2012.12.002
Kayser K, Gortler J, Bogovac M, Bogovac A, Goldmann T, Vollmer E, Kayser G (2009) AI (artificial intelligence) in histopathology–from image analysis to automated diagnosis. Folia Histochem Cytobiol 47(3):355–361. doi:10.2478/v10042-009-0087-y
Laurinavicius A, Laurinaviciene A, Dasevicius D, Elie N, Plancoulaine B, Bor C, Herlin P (2012) Digital image analysis in pathology: benefits and obligation. Anal Cell Pathol (Amst) 35(2):75–78. doi:10.3233/ACP-2011-0033
Lloyd MC, Allam-Nandyala P, Purohit CN, Burke N, Coppola D, Bui MM (2010) Using image analysis as a tool for assessment of prognostic and predictive biomarkers for breast cancer: how reliable is it? J Pathol Inform 1:29. doi:10.4103/2153-3539.74186
Messersmith W, Oppenheimer D, Peralba J, Sebastiani V, Amador M, Jimeno A, Embuscado E, Hidalgo M, Iacobuzio-Donahue C (2005) Assessment of Epidermal Growth Factor Receptor (EGFR) signaling in paired colorectal cancer and normal colon tissue samples using computer-aided immunohistochemical analysis. Cancer Biol Ther 4(12):1381–1386
Minot DM, Voss J, Rademacher S, Lwin T, Orsulak J, Caron B, Ketterling R, Nassar A, Chen B, Clayton A (2012) Image analysis of HER2 immunohistochemical staining. Reproducibility and concordance with fluorescence in situ hybridization of a laboratory-validated scoring technique. Am J Clin Pathol 137(2):270–276. doi:10.1309/AJCP9MKNLHQNK2ZX
Mohammed ZM, Edwards J, Orange C, Mallon E, Doughty JC, McMillan DC, Going JJ (2012a) Breast cancer outcomes by steroid hormone receptor status assessed visually and by computer image analysis. Histopathology 61(2):283–292. doi:10.1111/j.1365-2559.2012.04244.x
Mohammed ZM, Going JJ, McMillan DC, Orange C, Mallon E, Doughty JC, Edwards J (2012b) Comparison of visual and automated assessment of HER2 status and their impact on outcome in primary operable invasive ductal breast cancer. Histopathology 61(4):675–684. doi:10.1111/j.1365-2559.2012.04280.x
Mohammed ZM, McMillan DC, Elsberger B, Going JJ, Orange C, Mallon E, Doughty JC, Edwards J (2012c) Comparison of visual and automated assessment of Ki-67 proliferative activity and their impact on outcome in primary operable invasive ductal breast cancer. Br J Cancer 106(2):383–388. doi:10.1038/bjc.2011.569
Montemurro F, Scaltriti M (2014) Biomarkers of drugs targeting HER-family signalling in cancer. J Pathol 232(2):219–229. doi:10.1002/path.4269
Mulrane L, Rexhepaj E, Penney S, Callanan JJ, Gallagher WM (2008) Automated image analysis in histopathology: a valuable tool in medical diagnostics. Expert Rev Mol Diagn 8(6):707–725. doi:10.1586/14737159.8.6.707
Nassar A, Cohen C, Agersborg SS, Zhou W, Lynch KA, Albitar M, Barker EA, Vanderbilt BL, Thompson J, Heyman ER, Lange H, Olson A, Siddiqui MT (2011) Trainable immunohistochemical HER2/neu image analysis: a multisite performance study using 260 breast tissue specimens. Arch Pathol Lab Med 135(7):896–902. doi:10.1043/2010-0418-OAR1.1
Norman G, Rice S, Spackman E, Stirk L, Danso-Appiah A, Suh D, Palmer S, Eastwood A (2011) Trastuzumab for the treatment of HER2-positive metastatic adenocarcinoma of the stomach or gastro-oesophageal junction. Health Technol Assess 15(Suppl 1):33–42. doi:10.3310/hta15suppl1/04
Ong CW, Kim LG, Kong HH, Low LY, Wang TT, Supriya S, Kathiresan M, Soong R, Salto-Tellez M (2010) Computer-assisted pathological immunohistochemistry scoring is more time-effective than conventional scoring, but provides no analytical advantage. Histopathology 56(4):523–529. doi:10.1111/j.1365-2559.2010.03496.x
Rauser S, Weis R, Braselmann H, Feith M, Stein HJ, Langer R, Hutzler P, Hausmann M, Lassmann S, Siewert JR, Hofler H, Werner M, Walch A (2007) Significance of HER2 low-level copy gain in Barrett’s cancer: implications for fluorescence in situ hybridization testing in tissues. Clin Cancer Res : Off J Am Assoc Cancer Res 13(17):5115–5123. doi:10.1158/1078-0432.CCR-07-0465
Rimm DL (2006) What brown cannot do for you. Nat Biotechnol 24(8):914–916. doi:10.1038/nbt0806-914
Rizzardi AE, Johnson AT, Vogel RI, Pambuccian SE, Henriksen J, Skubitz AP, Metzger GJ, Schmechel SC (2012) Quantitative comparison of immunohistochemical staining measured by digital image analysis versus pathologist visual scoring. Diagn Pathol 7:42. doi:10.1186/1746-1596-7-42
Rojo MG, Bueno G, Slodkowska J (2009) Review of imaging solutions for integrated quantitative immunohistochemistry in the pathology daily practice. Folia Histochem Cytobiol 47(3):349–354. doi:10.2478/v10042-008-0114-4
Ruschoff J, Dietel M, Baretton G, Arbogast S, Walch A, Monges G, Chenard MP, Penault-Llorca F, Nagelmeier I, Schlake W, Hofler H, Kreipe HH (2010) HER2 diagnostics in gastric cancer-guideline validation and development of standardized immunohistochemical testing. Virchows Arch 457(3):299–307. doi:10.1007/s00428-010-0952-2
Skaland I, Ovestad I, Janssen EA, Klos J, Kjellevold KH, Helliesen T, Baak JP (2008a) Comparing subjective and digital image analysis HER2/neu expression scores with conventional and modified FISH scores in breast cancer. J Clin Pathol 61(1):68–71. doi:10.1136/jcp.2007.046763
Skaland I, Ovestad I, Janssen EA, Klos J, Kjellevold KH, Helliesen T, Baak JP (2008b) Digital image analysis improves the quality of subjective HER-2 expression scoring in breast cancer. Appl Immunohistochem Mol Morphol 16(2):185–190. doi:10.1097/PAI.0b013e318059c20c
Sobin L, Gospodarowicz ML, Wittekind C (2010) TNM classification of malignant tumors. Wiley, New York
Spackman E, Rice S, Norman G, Suh DC, Eastwood A, Palmer S (2013) Trastuzumab for the treatment of HER2-positive metastatic gastric cancer : a NICE single technology appraisal. Pharmacoeconomics 31(3):185–194. doi:10.1007/s40273-013-0023-z
Tuominen VJ, Tolonen TT, Isola J (2012) ImmunoMembrane: a publicly available web application for digital image analysis of HER2 immunohistochemistry. Histopathology 60(5):758–767. doi:10.1111/j.1365-2559.2011.04142.x
Turashvili G, Leung S, Turbin D, Montgomery K, Gilks B, West R, Carrier M, Huntsman D, Aparicio S (2009) Inter-observer reproducibility of HER2 immunohistochemical assessment and concordance with fluorescent in situ hybridization (FISH): pathologist assessment compared to quantitative image analysis. BMC Cancer 9:165. doi:10.1186/1471-2407-9-165
Vayrynen JP, Vornanen JO, Sajanti S, Bohm JP, Tuomisto A, Makinen MJ (2012) An improved image analysis method for cell counting lends credibility to the prognostic significance of T cells in colorectal cancer. Virchows Arch 460(5):455–465. doi:10.1007/s00428-012-1232-0
Webster JD, Dunstan RW (2014) Whole-slide imaging and automated image analysis: considerations and opportunities in the practice of pathology. Vet Pathol 51(1):211–223. doi:10.1177/0300985813503570
Welsh AW, Moeder CB, Kumar S, Gershkovich P, Alarid ET, Harigopal M, Haffty BG, Rimm DL (2011) Standardization of estrogen receptor measurement in breast cancer suggests false-negative results are a function of threshold intensity rather than percentage of positive cells. J Clin Oncol 29(22):2978–2984. doi:10.1200/JCO.2010.32.9706
Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, Allred DC, Bartlett JM, Bilous M, Fitzgibbons P, Hanna W, Jenkins RB, Mangu PB, Paik S, Perez EA, Press MF, Spears PA, Vance GH, Viale G, Hayes DF, American Society of Clinical O, College of American P (2013) Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol 31(31):3997–4013. doi:10.1200/JCO.2013.50.9984
Acknowledgments
The authors gratefully acknowledge the financial support of the Ministry of Education and Research of the Federal Republic of Germany (BMBF) (SYS-Stomach: 01ZX1310B) and the Deutsche Forschungsgemeinschaft (SFB 824 TP Z02 and WA 1656/3-1). The authors would like to thank Ulrike Buchholz, Claudia-Mareike Pflüger, and Andreas Voss for excellent technical assistance.
Conflict of interest
The authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Feuchtinger, A., Stiehler, T., Jütting, U. et al. Image analysis of immunohistochemistry is superior to visual scoring as shown for patient outcome of esophageal adenocarcinoma. Histochem Cell Biol 143, 1–9 (2015). https://doi.org/10.1007/s00418-014-1258-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00418-014-1258-2