Abstract
Purpose
Genetic studies have identified the association of some single-nucleotide polymorphisms (SNPs) with polypoidal choroidal vasculopathy (PCV), but little is known about whether these SNPs are related to PCV clinical features as well. We performed this study to examine the association of 12 SNPs with PCV clinical phenotypes.
Methods
Sixty-nine PCV eyes of 69 patients were included. Genomic DNA was extracted from peripheral blood. Agilent SureSelect Human ALL Exon V6 was used to sequence the 12 SNPs previously reported to associate with PCV. Baseline best-corrected visual acuity (BCVA), sub-foveal choroidal thickness (SFCT), choroid maximum vascular diameter (MVD), choroidal vascular hyperpermeability (CVH), and greatest linear dimension (GLD) of entire lesion were measured and compared between patients of different genotypes. Fisher’s exact test and Mann-Whitney U test were mainly used to compare categorical variables and continuous variables respectively.
Results
HTRA1 rs2293870 was a protective factor of PCV or AMD in the fellow eye (P = 0.040) and was related with greater SFCT in PCV eye after multiple linear regression (P = 0.043). C3 rs17030 was associated with smaller GLD (P = 0.033). CFH rs2274700 was related to lower MVD (P = 0.043) and was a protective factor for CVH (P = 0.034).
Conclusion
Multiple PCV-associated SNPs are associated with PCV clinical phenotypes. The involvement of several synonymous SNPs calls for further research on the role of transcriptional alterations and trans-regulation of distant signaling pathways in PCV pathogenesis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Polypoidal choroidal vasculopathy (PCV), first described by Yannuzzi et al. in the 1980s [1], is characterized by orange-red lesions on fundus examination and recurrent serosanguineous pigmented epithelium detachment (PED) [2]. While it remains controversial whether PCV is a subtype of neovascular age-related macular degeneration (nAMD) or a separate clinical entity due to their differences in epidemiology, clinical features, angiographic manifestations, and clinical courses [3], some consider PCV a part of pachychoroid-related disorders, which may represent different stages of a same pathogenic process [4].
Actually, PCV itself is diverse in clinical phenotypes. Until now, no universal classification could be applied to all studies. Many investigators stratified patients into polypoidal choroidal neovascularization (CNV) and typical PCV, depending on whether feeder vessels or draining vessels of polypoidal lesions are visible in ICGA [5]. Other criteria include but are not limited to sub-foveal choroidal thickness (SFCT) over 200 μm [6], BVN morphologies with or without leakage on FA [7], etc. Although the dissection of PCV into various clinical phenotypes helps predict disease course, treatment response, and prognosis, the variety of classification methods, the unpredictable transition between subtypes, and the inevitable impact of personal experience in imaging interpretation call for a more objective and subtle classification.
The diversity of classification reflects the unclarified nature of PCV. On the basis of genome-wide association studies (GWAS) of AMD, through examination of the association between genetic markers of AMD with PCV, various studies have proved PCV associated with genes related to inflammation, complement system, extra-cellular matrix homeostasis and lipid metabolism pathways, etc. [8]. Yet it should be highlighted that results of AMD genetic studies should be carefully testified before applied to PCV, since some AMD-associated SNPs are not associated with PCV, and although some are associated with both the disorders, the strength differed significantly [9]. Besides, the association of SNPs with PCV phenotypes is worth exploring. At present, the most studied SNP is ARMS2 rs10490924, which is associated with ICGA-based classification.
Little is known about whether other SNPs are related to PCV clinical features as well. Thus, this study was performed to further examine the association of 12 PCV-associated SNPs with PCV clinical phenotypes.
Materials and methods
Sixty-nine eyes of 69 patients diagnosed as PCV at Peking Union Medical College Hospital were enrolled in this study between February 2018 to April 2018 (21 females and 48 males with a mean age of 64.8 years). For bilaterally involved patients, only one eye was randomly chosen as study eye. Inclusion criteria were as follows: (1) clear diagnosis of PCV based on hyperfluorescent dilated polypoidal lesions (“polyps”) and/or BVN lesions on ICGA, especially within the first 6 min [10]; (2) treatment-naïve or a treatment-free period for at least 6 months; (3) patients who underwent comprehensive ophthalmic examination, including BCVA, slit-lamp examination, fundus photography, fluorescein angiography (FA), ICGA, and enhanced depth imaging (EDI) spectral-domain optical coherence tomography (SD-OCT). Written informed consent was obtained from all patients. The study was approved by the institutional review board of Peking Union Medical College Hospital and was conducted in accordance with the tenets of the Declaration of Helsinki.
General information was acquired through direct or telephone interview, including name, gender, age of disease-onset, comorbidities including hypertension and diabetes mellitus, conditions of contralateral eye, and baseline BCVA. BCVA was converted to the logarithm of the minimum angle of resolution (logMAR) equivalent.
SD-OCT images were obtained using Heidelberg Spectralis HRA-OCT device (HRA2, Heidelberg Engineering, Heidelberg, Germany). Specifically, EDI mode was selected to measure SFCT and choroid MVD (maximum vascular diameter). FA and ICGA were performed in the same session to measure GLD of entire lesion [11] and determine the angiographic type of each patient. The presence or absence of choroidal vascular hyperpermeability (CVH) was evaluated using ICGA images of middle and late phases [12]. All measurements were performed separately by three independent retinal specialists (ML, XZ, and JY), and any disagreement was discussed with a senior specialist (YXC). Average value for each continuous variable was calculated and used for statistical analysis.
Sample collection, DNA genotyping, and SNP selection
Genomic DNA was extracted from peripheral blood using the Qiagen FlexiGene DNA Kit (Qiagen, Hilden, Germany) under standard procedures. Genotyping was performed using Agilent SureSelect Human ALL Exon V6 (Agilent Technologies, Santa Clara, CA).
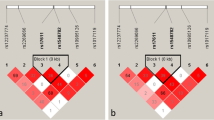
Through literature reading, we selected 12 PCV-associated SNPs of 9 genes: rs10490924 [13] and rs2736911 [8] of ARMS2, rs1049331 [14] and rs2293870 [14] of HTRA1, rs2274700 [15] and rs1065489 [16] of CFH, rs547154 [17] of C2, rs541862 [17] of CFB, rs2217332 [18] of HERPUD1, rs5882 [19] of CETP, rs17030 [20] of C3, and rs78488639 [21] of FPR1. All aforementioned SNPs were reported to relate with PCV and are detectable with Agilent probes.
Statistical analysis
Statistical analyses were performed using IBM SPSS Statistics version 22 (IBM Corp., Armonk, NY, USA). Categorical variables were compared between patients of different genotypes using Fisher’s exact test or binary logistic regression analysis. Mann-Whitney U test or one-way ANOVA was applied to compare continuous variables. Multiple linear regression was performed to assess the contributions of major risk factors to PCV phenotypes. Values of P < 0.05 were considered statistically significant.
Results
A total of 69 eyes of 69 PCV patients were included in the study. The characteristics of PCV patients, including gender, age, baseline BCVA, CVH, SFCT, comorbidities of hypertension and diabetes mellitus, and contralateral eye conditions, are summarized in Table 1. The features of selected SNPs, including located genes, base alteration, and distribution of genotypes, are summarized in Table 2.
Subjects whose genotype was TT or TG type of HTRA1 rs2293870 were more likely to develop unilateral PCV with no AMD or PCV in the contralateral eye (P = 0.040, OR = 2.917, 95% CI = 1.028–8.273; Table 3). Further analysis using genotypic model showed that TT type was more likely to develop unilateral PCV than GG type (P = 0.021, OR = 3.958, 95% CI = 1.230–12.734), but there was no significant difference between TG type and GG type (P = 0.329). Rs2293870 was also associated with greater SFCT (P = 0.022). The average SFCT for TT and GT patients was 265.73 ± 66.74 μm, compared with 230.62 ± 82.93 μm for GG patients. CFH rs1065489 and CETP rs5882 were also related to greater SFCT, but only rs2293870 remained significantly associated after multiple linear regression (P = 0.047).
MVD was associated with the minor allele of CFH rs2274700 (P = 0.043; Table 4). The average MVD of the 39 subjects who had at least one copy of minor allele (AA and AG) was 183.85 ± 52.10 μm, compared with 207.87 ± 58.04 μm for the 30 subjects homogeneous for the major allele (GG) (P = 0.043). No significant variation was detected using genotypic model. CFH rs2274700 was also identified as a protective factor from CVH (P = 0.034, OR = 0.344, 95% CI = 0.126–0.935; Table 3).
GLD was related with C3 rs17030 (P = 0.033; Table 4). Fifty-nine subjects had at least one copy of the minor allele (AA and AG). The average GLD of these subjects was 4909.63 ± 2694.85 μm. Ten subjects were homogeneous for the major allele (GG). The average GLD was 7558.00 ± 1291.37 μm. Further one-way ANOVA analysis showed that GLD of both AA and AG type was significantly smaller than GG type (4817.94 ± 2685.73, 4946.74 ± 2730.13 μm). Thus, C3 rs17030 is a protective factor from larger GLD.
No association was detected between the other SNPs and PCV clinical phenotypes in this study.
Discussion
In this study, we found that subjects with variants of HTRA1 rs2293870 were more likely to develop unilateral PCV. Rs2293870, rs5882, and rs1065489 were all associated with larger SFCT in this study, but only rs2293870 remained significantly associated after logistic regression with age. Rs2293870 is a synonymous mutation (c.108G>T) of the first exon of HTRA1, resulting in altered mRNA secondary structure, but no difference in expressional level and protein sequence [22]. Yet some studies indicated the difference of protein structure, which renders varied properties of heat-induced unfolding, trypsin accessibility, and secretion behavior [23]. Direct binding and proteolysis of transforming growth factor β1 (TGF-β1) is seen only in normal HTRA1, but not the c.108G>T isomer, which results in varied downstream TGF-β1 pathway activities. Yet it remains to be clarified how these changes in mRNA and protein secondary structure are related with PCV pathogenesis, disease laterality, and SFCT.
Our study first reported rs2274700 as a protective factor for CVH. Rs2274700 is a synonymous SNP of the tenth exon of CFH. Multiple studies have reported the association of rs2274700 with PCV, but the association seems stronger with AMD. Li et al. [24] examined 84 polymorphisms in and around CFH in AMD patients and found that the disease susceptibility was related more to certain haplotypes in a specific 32 kb region instead of SNPs. As a synonymous SNP, although the secondary structure of mRNA and CFH may slightly differ, but since rs2274700 is in linkage disequilibrium with multiple SNPs and copy number variations (CNP) in this region, it is not sound to attribute the disease susceptibility to rs2274700 itself. These variations may synergistically regulate the expression of CFH or other nearby complement genes [24]. Further detailed, in-depth genetic analysis should be carried out to dissect the association of disease susceptibility with this region.
In this study, we have proved that some of previously reported PCV-associated SNPs are also related to disease phenotypes. We first reported HTRA1 rs2293870 to be related with greater SFCT, and a protective factor for fellow eye involvement. Our results also showed that C3 rs17030 is associated with smaller GLD and CFH rs2274700 is a protective factor for CVH. These findings above indicate molecular interactions of these SNP with PCV clinical phenotypes. Although some SNPs are within intron region, transcriptional and translational alterations may occur and so does trans-regulation of distant signaling pathways. Limitations of this study include a relatively small sample size and inevitable selection bias, calling for replication studies with larger sample size and subjects from various genetic backgrounds.
In conclusions, multiple PCV-associated SNPs were associated with PCV clinical phenotypes. Further research into the molecular interactions of these key SNPs may provide valuable hints for PCV pathogenesis.
References
Yannuzzi LA, Sorenson J, Spaide RF, Lipson B (1990) Idiopathic polypoidal choroidal vasculopathy (IPCV). Retina (Philadelphia, Pa) 10(1):1–8
Cheung CMG, Lai TYY, Ruamviboonsuk P, Chen SJ, Chen Y, Freund KB, Gomi F, Koh AH, Lee WK, Wong TY (2018) Polypoidal choroidal vasculopathy: definition, pathogenesis, diagnosis, and management. Ophthalmology 125(5):708–724. https://doi.org/10.1016/j.ophtha.2017.11.019
Laude A, Cackett PD, Vithana EN, Yeo IY, Wong D, Koh AH, Wong TY, Aung T (2010) Polypoidal choroidal vasculopathy and neovascular age-related macular degeneration: same or different disease? Prog Retin Eye Res 29(1):19–29. https://doi.org/10.1016/j.preteyeres.2009.10.001
Balaratnasingam C, Lee WK, Koizumi H, Dansingani K, Inoue M, Freund KB (2016) Polypoidal choroidal vasculopathy: a distinct disease or manifestation of many? Retina (Philadelphia, Pa) 36(1):1–8. https://doi.org/10.1097/iae.0000000000000774
Yanagisawa S, Sakurada Y, Miki A, Matsumiya W, Imoto I, Honda S (2015) The association of elastin gene variants with two angiographic subtypes of polypoidal choroidal vasculopathy. PLoS One 10(3):e0120643. https://doi.org/10.1371/journal.pone.0120643
Lee WK, Baek J, Dansingani KK, Lee JH, Freund KB (2016) Choroidal morphology in eyes with polypoidal choroidal vasculopathy and normal or subnormal subfoveal choroidal thickness. Retina (Philadelphia, PA) 36:S73–S82
Tan CS, Ngo WK, Lim LW, Lim TH (2014) A novel classification of the vascular patterns of polypoidal choroidal vasculopathy and its relation to clinical outcomes. Br J Ophthalmol 98(11):1528–1533. https://doi.org/10.1136/bjophthalmol-2014-305059
Ma L, Li Z, Liu K, Rong SS, Brelen ME, Young AL, Kumaramanickavel G, Pang CP, Chen H, Chen LJ (2015) Association of genetic variants with polypoidal choroidal vasculopathy: a systematic review and updated meta-analysis. Ophthalmology 122(9):1854–1865. https://doi.org/10.1016/j.ophtha.2015.05.012
Fan Q, Cheung CMG, Chen LJ, Yamashiro K, Ahn J, Laude A, Mathur R, Mun CC, Yeo IY, Lim TH, Teo YY, Khor CC, Park KH, Yoshimura N, Pang CP, Wong TY, Cheng CY (2017) Shared genetic variants for polypoidal choroidal vasculopathy and typical neovascular age-related macular degeneration in east Asians. J Hum Genet 62(12):1049–1055. https://doi.org/10.1038/jhg.2017.83
Yannuzzi LA, Sorenson J, Spaide RF, Lipson B (2012) Idiopathic polypoidal choroidal vasculopathy (IPCV). Retina (Philadelphia, PA) 32:1–8. https://doi.org/10.1097/IAE.0b013e31823f992a
Liakopoulos S, Ongchin S, Bansal A, Msutta S, Walsh AC, Updike PG, Sadda SR (2008) Quantitative optical coherence tomography findings in various subtypes of neovascular age-related macular degeneration. Invest Ophthalmol Vis Sci 49(11):5048–5054. https://doi.org/10.1167/iovs.08-1877
Jirarattanasopa P, Ooto S, Nakata I, Tsujikawa A, Yamashiro K, Oishi A, Yoshimura N (2012) Choroidal thickness, vascular hyperpermeability, and complement factor H in age-related macular degeneration and polypoidal choroidal vasculopathy. Invest Ophthalmol Vis Sci 53(7):3663–3672. https://doi.org/10.1167/iovs.12-9619
Kondo N, Honda S, Ishibashi K, Tsukahara Y, Negi A (2007) LOC387715/HTRA1 variants in polypoidal choroidal vasculopathy and age-related macular degeneration in a Japanese population. Am J Ophthalmol 144(4):608–612. https://doi.org/10.1016/j.ajo.2007.06.003
Ng TK, Liang XY, Lai TY, Ma L, Tam PO, Wang JX, Chen LJ, Chen H, Pang CP (2016) HTRA1 promoter variant differentiates polypoidal choroidal vasculopathy from exudative age-related macular degeneration. Sci Rep 6:28639. https://doi.org/10.1038/srep28639
Huang L, Meng Q, Zhang C, Sun Y, Bai Y, Li S, Deng X, Wang B, Yu W, Zhao M, Li X (2015) Gene-gene interaction of CFH, ARMS2, and ARMS2/HTRA1 on the risk of neovascular age-related macular degeneration and polypoidal choroidal vasculopathy in Chinese population. Eye (London, England) 29(5):691–698. https://doi.org/10.1038/eye.2015.32
Huang L, Li Y, Guo S, Sun Y, Zhang C, Bai Y, Li S, Yang F, Zhao M, Wang B, Yu W, Zhao M, Khor CC, Li X (2014) Different hereditary contribution of the CFH gene between polypoidal choroidal vasculopathy and age-related macular degeneration in Chinese Han people. Invest Ophthalmol Vis Sci 55(4):2534–2538. https://doi.org/10.1167/iovs.13-13437
Nakata I, Yamashiro K, Yamada R, Gotoh N, Nakanishi H, Hayashi H, Akagi-Kurashige Y, Tsujikawa A, Otani A, Saito M, Iida T, Oishi A, Matsuo K, Tajima K, Matsuda F, Yoshimura N (2012) Significance of C2/CFB variants in age-related macular degeneration and polypoidal choroidal vasculopathy in a Japanese population. Invest Ophthalmol Vis Sci 53(2):794–798. https://doi.org/10.1167/iovs.11-8468
Jin E, Bai Y, Huang L, Zhao M, Zhang C, Zhao M, Li X (2015) Evidence of a novel gene HERPUD1 in polypoidal choroidal vasculopathy. Int J Clin Exp Pathol 8(11):13928–13944
Zhang X, Li M, Wen F, Zuo C, Chen H, Wu K, Zeng R (2013) Different impact of high-density lipoprotein-related genetic variants on polypoidal choroidal vasculopathy and neovascular age-related macular degeneration in a Chinese Han population. Exp Eye Res 108:16–22. https://doi.org/10.1016/j.exer.2012.12.005
Liu K, Lai TY, Chiang SW, Chan VC, Young AL, Tam PO, Pang CP, Chen LJ (2014) Gender specific association of a complement component 3 polymorphism with polypoidal choroidal vasculopathy. Sci Rep 4:7018. https://doi.org/10.1038/srep07018
Liang XY, Chen LJ, Ng TK, Tuo J, Gao JL, Tam PO, Lai TY, Chan CC, Pang CP (2014) FPR1 interacts with CFH, HTRA1 and smoking in exudative age-related macular degeneration and polypoidal choroidal vasculopathy. Eye (London, England) 28(12):1502–1510. https://doi.org/10.1038/eye.2014.226
Wang G, Dubovy SR, Kovach JL, Schwartz SG, Agarwal A, Scott WK, Haines JL, Pericak-Vance MA (2013) Variants at chromosome 10q26 locus and the expression of HTRA1 in the retina. Exp Eye Res 112:102–105. https://doi.org/10.1016/j.exer.2013.04.019
Friedrich U, Datta S, Schubert T, Plossl K, Schneider M, Grassmann F, Fuchshofer R, Tiefenbach KJ, Langst G, Weber BH (2015) Synonymous variants in HTRA1 implicated in AMD susceptibility impair its capacity to regulate TGF-beta signaling. Hum Mol Genet 24(22):6361–6373. https://doi.org/10.1093/hmg/ddv346
Li M, Atmaca-Sonmez P, Othman M, Branham KEH, Khanna R, Wade MS, Li Y, Liang L, Zareparsi S, Swaroop A, Abecasis GR (2006) CFH haplotypes without the Y402H coding variant show strong association with susceptibility to age-related macular degeneration. Nat Genet 38(9):1049–1054. https://doi.org/10.1038/ng1871
Funding
This study was funded by the Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (2018PT32029).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the Peking Union Medical College Hospital Review Board and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 73 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Luo, M., Zhao, X., Yang, J. et al. The association of polypoidal choroidal vasculopathy clinical phenotypes with previously reported genetic markers. Graefes Arch Clin Exp Ophthalmol 258, 1199–1203 (2020). https://doi.org/10.1007/s00417-020-04702-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00417-020-04702-y