Abstract
Soil is often collected from a suspect’s tire, vehicle, or shoes during a criminal investigation and subsequently submitted to a forensic laboratory for analysis. Plant and insect material recovered in such samples is rarely analyzed, as morphological identification is difficult. In this study, DNA barcoding was used for taxonomic identifications by targeting the gene regions known to permit discrimination in plants [maturase K (matK) and ribulose 1,5-biphosphate carboxylase (rbcL)] and insects [cytochrome oxidase subunit I (COI)]. A DNA barcode protocol suitable for processing forensic-type biological fragments was developed and its utility broadly tested with forensic-type fragments (e.g., seeds, leaves, bark, head, legs; n, 213) isolated from soils collected within Virginia, USA (n, 11). Difficulties with PCR inhibitors in plant extracts and obtaining clean Sanger sequence data from insect amplicons were encountered during protocol development; however, the final protocol produced sequences specific to the expected locus and taxa. The overall quantity and quality of DNA extracted from the 213 forensic-type biological fragments was low (< 15 ng/μL). For plant fragments, only the rbcL sequence data was deemed reliable; thus, taxonomic identifications were limited to the family level. The majority of insect sequences matched COI in both GenBank and Barcode of Life DataSystems; however, they were identified as an undescribed environmental contaminant. Although limited taxonomic information was gleaned from the forensic-type fragments processed in this study, the new protocol shows promise for obtaining reliable and specific identifications through DNA barcoding, which could ultimately enhance the information gleaned from soil examinations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Soil is an important type of forensic evidence because it is spatially variable, frequently transferred during criminal acts, and can be overlooked as evidence of the crime by the perpetrator. The examination and characterization of soil material has been completed by forensic geologists for decades, and the information gleaned is often applied to constrain the circumstances of a crime. For instance: could the soil on the suspect’s shoe have been derived from the crime scene, or does it match an alibi location? In provenance cases, where often the amount of material submitted is usually extremely limited (milligrams to a few grams), it is imperative that all components of the sample be analyzed to obtain as much probative information for investigative leads as possible. Biological material, such as plant and insect fragments, are often present in soil evidence but rarely taxonomically identified. This biological material could provide useful information, particularly in provenance cases, considering plant and insect species inhabit specific ecosystems, and may be present at specific times of the year. Traditionally, taxonomic identification of biological material is performed based on morphology. However, given that the majority of biological material observed in soil evidence is either a fragment or an incomplete specimen, morphological identification is not straightforward. In these circumstances, using DNA for identification is an attractive alternative approach, as it is present in all biological tissues and can be viable even in material not optimally preserved [1,2,3], such as would be expected in evidence samples.
DNA barcoding, which utilizes a standardized sequence of DNA, typically 400–800 base pairs in length, was coined in 2003 as a molecular approach for taxonomic identification [4]. Although DNA barcoding has received some criticism [5,6,7,8,9], it has gained broad acceptance given its application beyond taxonomy, to areas such as ecology, population genetics, and conservation [10,11,12,13,14], for monitoring and tracking invasive and economic pests [15,16,17,18,19,20,21] and in forensics [22,23,24,25]. The utility of DNA barcoding for species-level discrimination of unknown samples hinges on access and the ability to search databases of reference barcode sequences, containing relatively complete coverage of the taxa of interest. There currently are two main public databases that contain DNA barcode data, the Barcode of Life DataSystems (BOLD) [26] and GenBank [27].
A 648-bp region of the mitochondrial cytochrome oxidase subunit I (COI) gene has been adopted as the standard barcoding region for animal/insect identification [4, 28,29,30], as it has a fast mutation rate and is found in high copies within tissues. The discrimination power of this region has been evaluated in more than 10,000 peer review articles and provides species-level resolution among vertebrates [31, 32] and invertebrates [33,34,35,36,37]. As COI evolves too slowly to facilitate species-level discrimination among plants, the Consortium for the Barcode of Life (CBOL) recommended the scientific community adopt a 2-locus barcode for discrimination among land plants: ribulose 1,5-biphosphate carboxylase (rbcL) and maturase K (matK) both from the plastid genome [38]. The rationale surrounding the use of a 2-locus barcode is that although rbcL is more straightforward to amplify and sequence than matK, the level of resolution is limited (i.e., order and family as opposed to genus and species, respectively). Unlike rbcL, there is no universal primer pair to facilitate the amplification of matK across plants; thus, the taxonomic information obtained from rbcL can prove very useful for choosing the appropriate matK primers to ensure successful amplification (especially prudent when dealing with unknown material). In some plant groups however, species discrimination using only these two markers is not possible, so a range of supplementary markers are often required to increase the level of species resolution (e.g., intergenic spacers trnH-psbA, atpF-atpH, and psbK-psbl and gene regions rpoB and rpoC1) [39, 40].
Current efforts that have used molecular-based approaches such as DNA barcoding to document the biodiversity within a soil sample have primarily been focused on a bulk metagenomic approach [41,42,43,44,45,46,47,48,49,50]. Using conserved primers for the desired barcode regions, individual taxa can be amplified and sequenced simultaneously (i.e., massively parallel sequencing) from a single bulk soil extraction. Although a metagenomic approach facilitates the collection of large amounts of data from potentially highly degraded samples, the current usefulness of this technique mainly lies with cross-sample comparisons; the operational taxonomic units (OTUs) identified in the unknown sample are compared to a series of knowns, to determine the level of similarity. Additionally, a large amount of soil (at least 100 mg) is needed for a DNA extraction [46], which would be problematic in forensic applications where sample mass is often limited and non-consumption analysis is preferred.
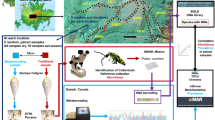
To enhance the forensic examination of soils, this study focused on developing a protocol for obtaining DNA barcode data from individual biological fragments isolated from forensic-type soil samples. Although protocols for obtaining DNA barcode data from both plants and insects have been well developed by the scientific community [29], these methods have been primarily optimized for fresh, pristine samples. Using these methods as a starting point, a DNA barcoding protocol was developed to work with both “new” and “old” biological material. The broad utility of the developed method was tested using fragments (n, 213) isolated from 11 soil samples collected from within Virginia, USA, which represent varied geology and ecohabitats. This paper outlines (1) the challenges with developing a protocol to obtain barcode data from forensic-type biological material, (2) the types of plant and insect fragments that are commonly recovered with surface soil samples (e.g., seeds, rootlets, legs, or heads), (3) whether such fragments contain viable DNA, (4) whether the appropriate DNA barcode regions could be amplified and sequenced using traditional Sanger methods, and (5) the level of taxonomic identification possible from barcode data when using public sequence databases (BOLD and GenBank).
Materials and methods
The protocol outlined below was originally developed and tested using two types of samples for both plants and insects: (1) new, fresh, and intact tissue collected immediately prior to extraction (surrogate positive control) and (2) old, fragmented tissue recovered from surface soil samples, which had been exposed to environmental conditions likely for several months (see Online Resource 1 for examples of old fragments).
DNA extraction
To remove any remaining soil particulates or fungal contaminants from the old samples, each fragment was submerged in a 5% bleach solution for 5 min, and subsequently washed three times with purified water [51]. After washing, each fragment was left to dry overnight in a drying cabinet (lid of the 1.5 mL centrifuge tube was left open). The length of each fragment, along with the dry weight of the plant fragments, was recorded. The insect fragments were not weighed given their extremely small size. Photographs were taken of each individual fragment using a Nikon D90 camera, to permit subsequent categorization. The total genomic DNA was isolated using the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) and the DNeasy Blood and Tissue DNA Purification Kit (Qiagen), for plant and insect fragments, respectively. To facilitate straightforward homogenization of the tissue, each fragment was snap frozen using liquid nitrogen and ground to a fine powder using a disposable mortar and pestle. The manufacturer’s protocols were followed for extraction with one exception: the DNA was eluted into two eluates of 50 μL of AE Buffer as opposed to one eluate of 100 μL, to increase the final DNA concentration.
Characterizing DNA quantity and purity
The quantity and purity of the extracted DNA was assessed using the Nanodrop ND-1000 (Thermo Scientific, Wilmington, DE, USA). AE Buffer was used to calibrate the blank of the instrument, and 1.5 μL of DNA eluate was used to obtain a reading. The quantity of DNA in each sample was recorded (ng/μL) along with the absorbance at descriptive wavelengths: 230 nm for phenols and humic acid; 260 nm for nucleic acids; and 280 nm for carbohydrates, proteins, and RNA.
Amplification
All amplifications were performed on a GeneAmp PCR System 9700 Thermal Cycler (Applied Biosystems, Foster City, CA, USA) using the primer pairs given in Table 1 and the cycling conditions outlined in Online Resource 2. Initially, all primer pairs were tested using a 20μL reaction mix containing: 0.4 μM of each primer, 2.5 mM MgCl2, 0.5 mM of each dNTP (Applied Biosystems), 5 U of AmpliTaq GOLD™ (Applied Biosystems), and 2 μL of genomic DNA (2 μL of nuclease free water for the negative control and 2 μL of the new extract as a surrogate positive control). In additional experiments, KAPA3G Plant DNA polymerase (KAPA Biosystems, Wilmington, MA, USA), 2× KAPA Taq DNA polymerase (KAPA Biosystems), Q5® Hot Start High-Fidelity DNA polymerase (New England BioLabs Inc. [NEB], Ipswich, MA, USA), and the Q5® High-Fidelity DNA polymerase (NEB) were tested using the manufacturer’s suggested reaction mix constituents. Inhibitor removal steps or alternate PCR constituents were examined in some experiments and included betaine (Sigma-Aldrich [B-2754], St Louis, MO, USA), final concentrations of 1–2 M; polyvinylpyrrolidone (PVP; Sigma-Aldrich [P-5288]), final concentrations of 1–3% v/v; and dimethyl sulfoxide (DMSO; Sigma-Aldrich [D8418]), final concentrations of 3–10% v/v. Purification of extracted DNA was tested with the PowerClean® Pro DNA Cleanup Kit (Mo Bio Laboratories, Inc., Carlsbad, CA, USA), Agencourt® AMPure XP Reagent (Beckman Coulter, Inc., Brea, CA, USA) and an ammonium acetate precipitation (Sigma-Aldrich [A2706]).
Amplicon screening and purification
A total of 5 μL of PCR product and 1 μL of 6× loading dye (Promega, Madison, WI, USA) were loaded into a single well of a 1.2% agarose gel. To facilitate size quantitation of amplicons, 10 μL of 1 kbp DNA Ladder (Bioline, Taunton, MA, USA) was also run. Each gel was subjected to electrophoresis prior to ethidium bromide staining and visualization under ultraviolet light. ExoSAP-IT® (USB® Products, Cleveland, OH, USA), which digests any unincorporated primer and dNTPs, was used to purify amplicons. A total of 1 μL of ExoSAP-IT® was combined with every 5 μL of PCR product and incubated as per the manufacturer’s instructions. Purified samples were quantitated using the Agilent 2100 Bioanalyzer and the Agilent DNA 1000 kit (Agilent Technologies, Santa Clara, CA, USA) following the manufacturer’s protocol.
Sequencing and data analysis
Sequencing of ExoSAP-IT®-treated PCR products was performed using the ABIPRISM® BigDye™ Terminator Cycle Sequencing Kits (v3.1 for plant amplicons and v1.1 for insect amplicons) (Applied Biosystems). Each sequencing reaction contained 10 ng of purified PCR product, 3.9 μL of BigDye™ Ready Reaction Mix, and 0.175 μM of the appropriate forward amplification primer. Samples were subjected to the following cycling conditions on a GeneAmp 9700 Thermal Cycler: plant amplicons, 1× 96 °C for 1 min and 25× 96 °C for 15 s, 50 °C for 1 s, and 60 °C for 1 min; 4 °C hold and insect amplicons, 1× 96 °C for 1 min and 25× 96 °C for 15 s, 50 °C for 1 s, and 60 °C for 4 min; 4 °C hold. Individual sequencing reactions were purified using Centri-Sep™ strip columns (Princeton Separations, Freehold, NJ, USA) following the manufacturer’s protocol.
The sequencing products were separated using an ABI 3130xl Genetic Analyzer (Applied Biosystems), and Sequence Analysis 5.2 software (Applied Biosystems) was used for basecalling. Each sequence was manually edited using 4Peaks (Nucleobytes, Amsterdam, the Netherlands) to check for base ambiguities and to remove the primer sequences. The resulting edited nucleotide sequence was subjected to a nucleotide BLAST search (blastn, searching the “other” nucleotide collection database; available at http://blast.ncbi.nlm.nih.gov) and also searched against the appropriate BOLD database (available at www.boldsystems.org) to obtain a taxonomic identification.
Broad assessment of the protocol
Once a protocol had been developed to work with both new and old samples, the broad utility of the protocol was tested on ~ 200 individual plant and insect fragments, isolated from 11 different soil samples collected within Virginia, USA (Online Resource 3). Soil was collected 0 to 3 cm below the litter layer, with the biological fragments isolated from the samples likely exposed to environmental conditions for several months (as soil collections were made in early winter and early spring, times separated from the major deposition of plant litter). For both COI and rbcL, when an amplicon for the entire barcode region could not be obtained, the “mini” primer pair was tested (primers fall within the entire barcode region) (Table 1). Given there is not a published mini primer pair for matK, a nested PCR, in which 3 μL of the initial amplification reaction mix was used as the DNA template rather than genomic DNA, was implemented using an internal primer pair (Table 1).
Results and discussion
Inhibition with plant extracts
During protocol development, amplicons of the expected size were only observed on agarose gels from the new plant extract, regardless of the primer pair used. It was possible that the DNA from the old fragment was highly degraded, such that even amplification of the smallest plant region (rbcL mini, ~ 230 bp) was not possible. However, as inhibitors such as polyphenolic/aromatic compounds, polysaccharides, and humic acid are common in plant material [57] and are known to interfere with PCR both directly and indirectly [58,59,60,61], it was also possible that such compounds were co-isolated. To confirm whether inhibitors were present in the old plant extract, an inhibition assay was performed using different sources of untreated DNA: (1) only new plant, (2) only old plant, (3) both new and old plant (with the final amount of DNA from both extracts being equal), and (4) negative control (nuclease free water). In the presence of the old DNA, the new DNA failed to amplify the fragment of interest, confirming the presence of inhibitors (Table 2). Three different strategies were used to address inhibition: incorporation of a second round of DNA purification, altering the constituents in PCR, and using an alternate specialized polymerase. Downstream efficacy was assessed using the inhibition assay outlined above for two different-sized fragments (~ 850 bp matK and ~ 230 bp rbcL mini).
DNA purification
The efficacy of three DNA purification methods was tested individually for removing inhibitors from only the old plant extract (Table 2): (1) the PowerClean® Pro DNA Cleanup Kit, which utilizes a patented Inhibitor Removal Technology® to remove challenging impurities; (2) the Agencourt® AMPure XP Reagent, which uses magnetic bead technology to isolate all genomic DNA greater than 100 bp in length; and (3) ammonium acetate, to precipitate any polyphenolics and polysaccharides in the extract [62]. For each purification method, extracts obtained from a single old plant were purified in triplicate following the manufacturer’s protocol (methods 1 and 2) and as described by Miller [62] (method 3). All three methods were successful in removing the inhibitors present in the old extracts (Table 2, DNA purification panel). In instances where an amplicon was observed in the new and old reaction but not for the old alone, this was suggestive of degraded DNA (as seen by the absence of the large matK amplicon in Table 2).
Modifying the PCR constituents
Additional experiments were performed to examine whether it were possible to suppress the activity of the inhibitors during PCR by modifying the constituents in the reaction mix. The commonly employed approach to lessen the impact of PCR inhibitors by reducing the volume of DNA extract added to the reaction mix [57] yielded no improvement in this study (volume of input DNA extract was decreased by ~ 10-fold; results not shown).
The addition of betaine, polyvinylpyrrolidone (PVP), and dimethyl sulfoxide (DMSO) to the reaction mix was investigated at a range of concentrations, which overlapped the levels previously documented to be effective in suppressing inhibitors (outlined in Table 2, PCR constituents panel) [57,58,59, 63,64,65]. Only PVP suppressed the inhibitors present in the old plant. In most instances, adding DMSO or betaine to the PCR reaction did not address the impact of inhibitors present in the old plant extract at any concentration; in some cases, DMSO had a negative impact on the new plant, by suppressing the amplification of both matK and the rbcL mini fragments.
Specialized polymerase
The KAPA3G Plant DNA polymerase is a high-efficiency polymerase formulated to improve tolerance to PCR inhibitors such as polyphenolics and polysaccharides and has previously permitted successful amplification with challenging samples [66, 67]. Therefore, we assessed the performance of this polymerase on the untreated/unpurified old plant extract. With the manufacturer’s suggested constituents for a 25-μL reaction and using the previously optimized cycling conditions (Online Resource 2), successful amplification of both the matK and rbcL mini barcode regions was achieved for the old plant (Table 2). The amount of product obtained when using the KAPA3G Plant DNA polymerase was far greater for both the new and old plant when compared to that obtained when using AmpliTaq GOLD™ (Fig. 1). Additionally, the KAPA3G Plant DNA polymerase provided strong and reproducible PCR amplifications for all of the plant primer pairs (Fig. 2). Given that the KAPA3G Plant DNA polymerase is not reported to repair DNA, amplification of the long matK fragment in the old extract (which likely contains only a few full-length, intact templates) may be due to the enzyme’s high efficiency. The resulting sequence data from amplicons generated using KAPA3G Plant DNA polymerase for all regions (matK, rbcL and rbcL mini) were clean but also matched the expected locus and taxa in GenBank and BOLD. Considering these results, the KAPA3G Plant DNA polymerase was used for amplifications in the broad assessment of the protocol, which utilized fragments isolated from forensic-type soils that likely contain similar inhibitors and DNA of suboptimal lengths.
Amplification of the mat K barcoding region (~ 850 bp; primers mat K-KIM-1R/matK-KIM-3F) for both new and old plant fragments using AmpliTaq GOLD™ (lanes 2, 4) and the KAPA3G Plant DNA polymerase (lanes 3, 5). 1 kbp ladder shown (lane 1). Results shown are typical for those obtained from numerous experiments (n, > 10)
Plant and insect DNA barcoding region amplicons obtained using KAPA3G Plant DNA polymerase (lanes 2–5) and the Q5 Hot Start High-Fidelity DNA polymerase (lanes 6–7): (1) 1 kbp ladder; (2) ~ 850 bp mat K (primers matK-KIM-1R/matK-KIM-3F); (3) nested ~ 830 bp mat K (primers matK4La/matKMALPR1); (4) ~ 590 bp rbcL (primers rbcLa-F/rbcLa-R); (5) ~ 230 bp rbcL mini (primers rbcL1/rbcLB); (6) ~ 650 bp COI (primers LCO1490-L/HCO2198-L); (7) ~ 130 bp COI mini (primers uniminibarF1/uniminibarR1); (8) 1 kbp ladder
Amplification and sequencing of the insect barcode regions
Challenges with the 648 bp COI barcode fragment
When using AmpliTaq GOLD™ to amplify the 648 bp COI barcode region using the previously published primers (Table 1) and associated cycling conditions (Online Resource 2), only a faint band from the new extract was observed on an agarose gel (band for the old extract absent). By performing an inhibition assay similar to that employed for the plant extracts, the presence of inhibitors was ruled out as the reason for the failed PCR of the old insect extract (results not shown). Thus, it was likely that the failed amplification of the old insect was due to DNA degradation or low polymerase efficiency (given that only a faint band was observed with the new extract). To address this, a nested PCR was performed using 3 μL of the previous amplification reaction as template and the same initial amplification primers and cycling conditions. This approach yielded strong, clean amplicons of the expected size on gels for both the new and old insect extracts (old amplicon shown in Fig. 3a, Agilent electropherogram).
Agilent DNA 1000 electropherograms (a, c) and Sanger sequencing electropherograms (b, d) for the old insect amplified using AmpliTaq GOLD™ in a nested PCR with a total of 80 cycles (a, b) and a non-nested PCR with Q5 Hot Start High-Fidelity DNA polymerase, using a total of 40 cycles (c, d). Agilent peaks denoted as follows: 1, lower marker; 2, the ~ 650 bp COI barcode region amplicon; 3, upper marker. The X-axis of the Agilent electropherograms is not linear, and the Y-axis reflects the relative concentration of the amplicons
Upon sequencing the nested COI amplicons, high background noise or mixed reads were observed in the sequence electropherograms, meaning the sequence was mostly unusable (Fig. 3b). To resolve this, a range of approaches known to improve the quality of the sequence data were systematically tested, including increasing the primer annealing temperature, decreasing separately the amount of dye and primer, adding DMSO in a final concentration of 5% v/v, and sequencing with alternate primers. None of these approaches produced reliable, clean sequence data. As the peak corresponding to the nested COI amplicon appeared somewhat broad at its base in the Agilent electropherogram (Fig. 3a), it is likely that obtaining clean sequence data was impeded by additional secondary products, either a few nucleotides shorter or longer than the desired fragment. Given that a nested PCR approach was utilized to obtain amplicons from both the new and old insects using AmpliTaq GOLD™, artifacts such as these can be expected.
To obtain clean sequences, reamplification of the COI barcode region from old insect DNA was tested using different polymerases (AmpliTaq GOLD™, Q5® Hot Start High-Fidelity DNA polymerase, Q5® High-Fidelity DNA polymerase, and 2× KAPA Taq polymerase), but also with varying cycle numbers (40, 45, and 50) to increase the amount of product. When using either AmpliTaq GOLD™ or 2× KAPA Taq polymerase at best faint bands of the expected size were observed, even when 50 amplification cycles were used (results not shown). Both of the NEB High-Fidelity polymerases produced strong amplicons at all cycle numbers; however, a number of strong secondary products were also visualized for the Q5® High-Fidelity DNA polymerase. The amplicon obtained when using the Q5® Hot Start High-Fidelity DNA polymerase and 40 amplification cycles appeared as a strong band on the agarose gel (Fig. 2, Lane 6) and single peak on the Agilent after cleanup with ExoSAP-IT® (Fig. 3c), albeit in a lower concentration to that obtained with a nested PCR using AmpliTaq GOLD™ (Fig. 3a). Subsequent sequencing of this COI amplicon had limited background noise (Fig. 3d) and matched to the expected locus (COI) and insect (Danaus plexippus, monarch butterfly) in public sequence databases. To ensure clean, reproducible sequencing data when processing the forensic-type insect fragments, amplification of the COI barcode region was performed using Q5® Hot Start High-Fidelity DNA polymerase at 40 amplification cycles.
Optimizing the COI mini PCR
Considering numerous papers have reported that amplifying COI mini using the uniminibar-F1/uniminibar-R1 primer pair is challenging [68, 69], a “touch-up” PCR is suggested [29] (Online Resource 2). When using the Q5® Hot Start High-Fidelity DNA polymerase with the published cycling conditions, a strong amplicon of the expected size (~ 130 bp) was obtained from the new and old extracts, along with numerous secondary products. A set of modified cycling conditions were identified that produced a single dominant amplicon; annealing temperature in the first set of cycles was increased to 50 °C, and the extension time for all cycles was reduced to only 1 s (Fig. 2, Lane 7; Online Resource 2). The resulting sequence data was clean and reproducible for both extracts; however, given the small size of the amplicon, only ~ 100 bases could be used for downstream comparison to public databases after the removal of the primer sequence.
Utility of the developed protocol on forensic-type biological material
A summary schematic of the protocol developed to obtain DNA barcode data from forensic-type plant and insect fragments is given in Fig. 4, and protocol conditions that generate locus specific amplicons have been tabulated in Online Resource 2. The results outlined in the section below address the utility of this protocol for processing fragments isolated from soils collected across Virginia, which represent a broad range of parent soil and surface material, ecoregions, and pH (Online Resource 3).
DNA barcoding protocol developed for processing biological materials isolated from forensic-type soil samples. 1Final concentration of the reaction mix constituents and thermal cycling conditions used to amplify each of the barcoding regions are given in Online Resource 2
Characterization of fragments
Biological fragments were numerous (i.e., generally > 30) in most of the 11 soil samples; thus, a wide variety of fragment types were chosen to test the broad utility of the protocol. In total, 110 plant fragments and 103 insect fragments were processed and they were categorized as follows: plants—roots (24%), leaf (21%), branch (10%), bark (9%), entire seed (9%), casing of seed (8%), grass (2%), and other (17%); insects—unidentifiable part of exoskeleton (48%), thorax/abdomen (30%), leg (15%), head (5%), wing (1%), and spider’s web (1%). The average length of the insect fragments was far smaller than the plants, 1.8 ± 3.4 and 8.4 ± 7.3 mm, respectively. The average weight of the plant fragments was 1.8 ± 3.3 mg.
DNA quality and quantity
When only considering extracts for which the concentration was above the reliable detection limits of the Nanodrop (2 ng/μL), the average total DNA yields from plants and insects were 1.15 ± 3.7 and 0.45 ± 0.75 μg, respectively (Online Resource 4). The DNA purity of each extract was assessed based on absorption ratios at various wavelengths (A260/280 and A260/230). Unexpectedly, the insect extracts had higher levels of phenolics and humic acid, whereas the plant extracts contained considerable amounts of carbohydrates, proteins, and RNA (Online Resource 4). Researchers have documented high levels of humic acid and protein contamination either when extracting bulk soil samples [70] or individual degraded plant samples [71], using a range of commercially available kits.
PCR and sequencing success
During protocol development, we confirmed that the reaction and cycling conditions for all primer pairs were reliable and specific, as the resulting sequence data matched the expected locus and taxa in public sequence databases. Given this, if a single band the same size as the surrogate positive control (the new extract) was observed on the agarose gel, the PCR was deemed successful. Both the entire barcode primers for rbcL and COI returned a ~ 70% amplification success rate, whereas the matK barcode region was only amplified in a few samples (~ 5%; Table 3). Far greater amplification success rate for matK was observed when the nested PCR was implemented (Fig. 2, lane 3), and rbcL (full length or mini) was amplified in over 90% of fragments.
Sequencing was deemed successful when clean sequence data (> 100 bp in length) was obtained from a purified PCR amplicon. At least two-thirds of all amplicons produced useable sequence data for downstream comparisons to public databases, with the majority of sequences being over 300 bp in length (Table 3). When a sequence was unusable due to high background noise, re-sequencing was attempted using the reverse amplification primer, with varying degrees of success. No distinguishable trends were observed based on the type of fragment (e.g., plants—leaf, roots, branch, bark, seeds; insects—legs, head, exoskeleton) and PCR or sequencing success.
Assessment of public sequence databases for taxonomic identification
All plant sequences (rbcL and matK) matched the expected locus when searched against GenBank (e.g., an rbcL sequence was identified as a portion of the rbcL locus) (Table 3), a reflection of high specificity in the primers and cycling conditions. When examining the taxonomic resolution obtained with DNA barcode sequences, the majority of rbcL and matK sequences achieved a minimum of order-level discrimination, with the resulting taxonomic identifications being highly concordant between the two public databases.
In instances where both rbcL and matK data are collected from a single sample, the taxonomic identification, especially at higher levels, should be congruent. In this study, 46 samples had sequence data from both rbcL and matK; however, high discordance (~ 75%) was noted in the taxonomic identifications from the two loci. In every case, the rbcL data indicated that the fragment was a pine species (Pinus, gymnosperm), whereas the matK data suggested the origin as an oak species (Quercus, angiosperm). Considering a nested PCR was implemented for matK using angiosperm primers, it was plausible that the matK data could be misleading. To verify this hypothesis, the intergenic spacer trnH-psbA (a supplemental plant barcoding locus) was amplified and sequenced (following the protocol outlined in 29) for a subsample of the fragments in which discordance was noted. The trnH-psbA data confirmed the rbcL identifications; thus, if an amplicon is not obtained in an initial PCR with the matK-KIM primers, PCR should be performed with a primer pair degenerate to another plant group (perhaps Gym_F1A/Gym-R1A [72] for gymnosperms), instead of implementing a nested PCR. When only considering the rbcL data, family-level assignments were as follows: 58% Pinaceae (pine), 13% Fagaceae (oak/stone oak), 5% Vitaceae (grapes), 3% Brassicaceae (bittercress), and 3% Brachytheciaceae (moss), with the remaining 18% of fragments assigned to one of six other families. In this study, when using the developed DNA barcoding protocol, the level of plant biodiversity captured in the 11 soil samples was low, considering only rbcL data could be used reliably. With the analysis of more fragments, but more importantly the recovery of sequence data from the more discriminatory matK locus, better taxonomic resolution would be possible. The authors envisage limited difficulty in obtaining matK data from any fragment, when the KAPA3G Plant DNA polymerase is used in tandem with well-tested cycling conditions for alternate universal matK primer pairs (i.e., angiosperms, gymnosperms, ferns, and mosses).
When examining the insect sequence data, despite ~ 75% matching to the COI locus in GenBank, only six sequences had a match in either public database to an organism from the class Insecta (Table 3); the best match for the vast majority of COI sequences was either to a fungus, marine invertebrate, algae, or uncultured bacterium. However, for any match, the similarity statistics were on average very poor and the average e-value from BLAST searches was higher than ideal. Given the extremely small size of the starting insect material (generally < 1 mm) and the known exposure of such fragments to prolonged environmental conditions, it was not surprising there was little insect DNA remaining for analysis. If more intact or larger insect fragments were processed using the developed protocol, the proportion of COI sequences matching to the class Insecta would likely increase, providing useful information for provenance cases. It is apparent that using the presence of an amplicon of the expected size on the agarose gel as a metric for PCR success provides a misleading representation of the likely downstream success of taxonomic identification.
Conclusions
Using previously published studies as a guide, a protocol was developed that permits the collection of DNA barcode sequences from biological fragments exposed to environmental conditions. The utility of this developed protocol for taxonomic identifications was subsequently tested using 213 plant and insect fragments isolated from forensic-type soil samples collected within Virginia. Amplification and sequencing was straightforward, and the resulting sequence data matched the expected loci in public sequence databases. Despite this, the level of taxonomic discrimination was low, as a result of unreliable matK data and the absence of viable insect DNA. To capitalize on the application of this protocol for the identification of biological fragments encountered in forensic-type soil samples, further research should be focused on determining the number of fragments needed for analysis to sufficiently capture the biodiversity within a sample, along with impacts of seasonal variation. With the ever-advancing field of massively parallel sequencing (MPS), the developed protocol may need to be modified or a standardized protocol may be required to permit the collection of DNA barcode data from bulk soil samples. An MPS approach might assist with obtaining more information on the insect community, especially for samples in which individual insect fragments are very small and contain little viable DNA. However, for an MPS-based approach to be feasible within a forensic context where the evidence material is generally very limited, work is needed to optimize soil extractions for small sample amounts.
References
Post RJ, Flook PK, Millest AL (1993) Method for the preservation of insects for DNA studies. Biochem Syst Ecol 21(1):85–92. https://doi.org/10.1016/0305-1978(93)90012-G
Drábková LZ (2014) DNA extraction from herbarium specimens. Methods Mol Biol 1115:69–84. https://doi.org/10.1007/978-1-62703-767-9_4
Peterson PM, Romaschenko K, Soreng RJ (2014) A laboratory guide for generating DNA barcodes in grasses: a case study of Leptochloa s.l. (Poaceae: Chloridoideae). J Plant Taxon Geogr 69(1):1–12. https://doi.org/10.1080/00837792.2014.927555
Hebert PDN, Cywinska A, Ball SL, deWaard JR (2003) Biological identifications through DNA barcodes. Proc Royal Soc London: Biol Sci 270(1512):313–321. https://doi.org/10.1098/rspb.2002.2218
Moritz C, Cicero C (2004) DNA barcoding: promise and pitfalls. PLoS Biol 2(10):e354. https://doi.org/10.1371/journal.pbio.0020354
Will KW, Rubinoff D (2004) Myth of the molecule: DNA barcodes for species cannot replace morphology for identification and classification. Cladistics 20(1):47–55. https://doi.org/10.1111/j.1096-0031.2003.00008.x
Ebach MC, Holdrege C (2005) More taxonomy, not DNA barcoding. Bioscience 55(10):822–824. https://doi.org/10.1641/0006-3568(2005)055[0823:MTNDB]2.0.CO;2
Will KW, Mishler BD, Wheeler QD, Savolainen V (2005) The perils of DNA barcoding and the need for integrative taxonomy. Syst Biol 54(5):844–851. https://doi.org/10.1080/10635150500354878
Meier R, Shiyang K, Vaidya G et al (2006) DNA barcoding and taxonomy in Diptera: a tale of high intraspecific variability and low identification success. Syst Biol 55(5):715–728. https://doi.org/10.1080/10635150600969864
Witt JDS, Threloff DL, Hebert PDN (2006) DNA barcoding reveals extraordinary cryptic diversity in an amphipod genus: implications for desert spring conservation. Mol Ecol 15(10):3073–3082. https://doi.org/10.1111/j.1365-294X.2006.02999.x
Hajibabaei M, Singer GAC, Hebert PDN, Hickey DA (2007) DNA barcoding: how it complements taxonomy, molecular phylogenetics and population genetics. Trends Genet 23(4):167–172. https://doi.org/10.1016/j.tig.2007.02.001
Ward RD, Holmes BH, White WT, Last PR (2007) DNA barcoding Australasian chondrichthyans: results and potential uses in conservation. Mar Freshw Res 59(1):57–71. https://doi.org/10.1071/MF07148
Ardura A, Linde AR, Moreira JC, Garcia-Vazquez E (2010) DNA barcoding for conservation and management of Amazonian commercial fish. Biol Conserv 143(6):1438–1443. https://doi.org/10.1016/j.biocon.2010.03.019
Kress WJ, García-Robledo C, Uriarte M, Erickson DL (2015) DNA barcodes for ecology, evolution and conservation. Trends Ecol Evol 30(1):25–65. https://doi.org/10.1016/j.tree.2014.10.008
Ball SL, Armstrong KF (2006) DNA barcodes for insect pest identification: a test case with tussock moths (Lepidoptera: Lymantriidae). Can J Forest Res 36(2):337–350. https://doi.org/10.1139/x05-276
Saunders GW (2009) Routine DNA barcoding of Canadian Gracilariales (Rhodophyta) reveals the invasive species Gracilaria vermiculophylla in British Columbia. Mol Ecol Resour 9:140–150. https://doi.org/10.1111/j.1755-0998.2009.02639.x
Stoeckle MY, Gamble CC, Kirpekar et al (2011) Commercial teas highlight plant DNA barcode identification successes and obstacles. Sci Rep 1(1):42. https://doi.org/10.1038/srep00042
Galimberti A, De Mattia F, Losa A et al (2013) DNA barcoding as a new tool for food traceability. Food Res Int 50:55–63. https://doi.org/10.1016/j.foodres.2012.09.036
Ivanova NV, Kuzmina ML, Braukmann TWA, Borisenko AV, Zakharov EV (2016) Authentication of herbal supplements using next-generation sequencing. PLoS One 11(5):e0156426. https://doi.org/10.1371/journal.pone.0156426
Mishra P, Kumar A, Nagireddy A, Mani DN, Shukla AK, Tiwari R, Sundaresan V (2016) DNA barcoding: an efficient tool to overcome authentication challenges in the herbal market. Plant Biotechnol J 14(1):8–21. https://doi.org/10.1111/pbi.12419
Zhang T, Wang Y-J, Guo W et al (2016) DNA barcoding, species-specific PCR and real-time PCR techniques for the identification of six Trobolium pests of stored products. Sci Rep 6:28494. https://doi.org/10.1038/srep28494
Bruni I, De Mattia F, Galimberti A et al (2010) Identification of poisonous plants by DNA barcoding approach. Int J Legal Med 124(6):595–603. https://doi.org/10.1007/s00414-010-0447-3
Dalton DL, Kotze A (2011) DNA barcoding as a tool for species identification in three forensic wildlife cases in South Africa. Forensic Sci Int 207:e51–e54. https://doi.org/10.1016/j.forsciint.2010.12.017
Jones YL, Peters SM, Weland C et al (2013) Potential use of DNA barcodes in regulatory science: identification of the U.S. Food and Drug Administration’s “dirty 22,” contributors to the spread of foodborne pathogens. J Food Prot 76(1):144–149. https://doi.org/10.4315/0362-028X.JFP-12-168
Ng KKS, Lee SL, Tnah LH, Nurul-Farhanah Z, Ng CH, Lee CT, Tani N, Diway B, Lai PS, Khoo E (2016) Forensic timber identification: a case study of CITES listed species, Gonystylus bancanus (Thymelaeaceae). Forensic Sci Int Genet 23:197–209. https://doi.org/10.1016/j.fsigen.2016.05.002
Ratnasingham S, Hebert PDN (2007) BOLD: the barcode of life data system. (www.barcodinglife.org). Mol Ecol Notes 7(3):355–364. https://doi.org/10.1111/j.1471-8286.2007.01678.x
Benson DA, Karsch-Mizrachi I, Lipman DJ et al (2002) GenBank. Nucleic Acids Res 30(1):17–20
PDN H, Ratnasingham S, de Waard JR (2003) Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proceedings of the Royal Society of London. Biological Sciences 270:s96–s99
Kress WJ, Erickson DL (2012) DNA barcodes: methods and protocols. Methods Mol Biol 858:3–8. https://doi.org/10.1007/978-1-61779-591-6_1
Dasmahapatra KK, Mallet J (2006) DNA barcodes: recent successes and future prospects. Heredity 97(4):254–255. https://doi.org/10.1038/sj.hdy.6800858
Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PD (2005) DNA barcoding Australia’s fish species. Philos Trans Royal Soc Biol 360(1462):1847–1857. https://doi.org/10.1098/rstb.2005.1716
Tavares ES, Baker AJ (2008) Single mitochondrial gene barcodes reliably identify sister-species in diverse clades of birds. BMC Evol Biol 8(1):81. https://doi.org/10.1186/1471-2148-8-81
Hebert PDN, Penton EH, Burns JM, Janzen DH, Hallwachs W (2004) Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc Natl Acad Sci U S A 101(41):14812–14817. https://doi.org/10.1073/pnas.0406166101
Ball SL, Hebert PDN, Burian SK, Webb JM (2005) Biological identifications of mayflies (Epheneroptera) using DNA barcodes. J N Am Benthol Soc 24(3):508–524. https://doi.org/10.1899/04-142.1
Nelson LA, Wallman JF, Dowton M (2007) Using COI barcodes to identify forensically and medically important blowflies. Med Vet Entomol 21(1):44–52. https://doi.org/10.1111/j.1365-2915.2007.00664.x
Meiklejohn KA, Wallman JF, Dowton M (2011) DNA-based identification of forensically important Australian Sarcophagidae (Diptera). Int J Legal Med 125(1):27–32. https://doi.org/10.1007/s00414-009-0395-y
Meiklejohn KA, Wallman JF, Cameron SL, Dowton M (2012) Comprehensive evaluation of DNA barcoding of the forensically important Australian Sarcophagidae (Diptera). Invertebr Syst 26(6):515–525. https://doi.org/10.1071/IS12008
Hollingsworth PM, Forrest LL, Spouge JL et al (2009) A DNA barcode for land plants. PNAS 106:12794–12797. https://doi.org/10.1073/pnas.0905845106
Hollingsworth PM, Graham SW, Little DP (2011) Choosing and using a plant DNA barcode. PLoS One 6(5):e19254. https://doi.org/10.1371/journal.pone.0019254
Ferri G, Corradini B, Ferrari F, Santunione AL, Palazzoli F, Alu M (2015) Forensic botany II, DNA barcode for land plants: which markers after international agreement? Forensic Sci Int Genet 15:131–136. https://doi.org/10.1016/j.fsigen.2014.10.005
Epp LS, Boessenkool S, Bellemain EP et al (2012) New environmental metabarcodes for analyzing soil DNA: potential for studying past and present ecosystems. Mol Ecol 21(8):1821–1833. https://doi.org/10.1111/j.1365-294X.2012.05537.x
Taberlet P, Prud’Homme SM, Campione E et al (2012) Soil sampling and isolation of extracellular DNA from large amount of starting material suitable for metabarcoding studies. Mol Ecol 21(8):1816–1820. https://doi.org/10.1111/j.1365-294X.2011.05317.x
Cristescu ME (2014) From barcoding single individuals to metabarcoding biological communities: towards an integrative approach to the study of global biodiversity. TREE 29(10):566–571. https://doi.org/10.1016/j.tree.2014.08.001
Giampaoli S, Berti A, Di Maggio RM et al (2014) The environmental biological signature: NGS profiling for forensic comparison of soils. Forensic Sci Int 240:41–47. https://doi.org/10.1016/j.forsciint.2014.02.028
Khodakova AS, Smith RJ, Bugoyne L et al (2014) Random whole metagenomic sequencing for forensic discrimination of soils. PLoS One 9(8):e104996. https://doi.org/10.1371/journal.pone.0104996
Yang C, Wang X, Miller JA et al (2014) Using metabarcoding to ask if easily collected soil and leaf-litter samples can be used as a general biodiversity indicator. Ecol Indic 46:379–389. https://doi.org/10.1016/j.ecolind.2014.06.028
Young JM, Weyrich LS, Cooper A (2014) Forensic soil DNA analysis using high-throughput sequencing: a comparison of four molecular markers. Forensic Sci Int Genet 13:176–184. https://doi.org/10.1016/j.fsigen.2014.07.014
Orgiazzi A, Dunbar MB, Panagos et al (2015) Soil biodiversity and DNA barcodes: opportunities and challenges. Soil Biol Biochem 80:244–250. https://doi.org/10.1016/j.soilbio.2014.10.014
Jesmok EM, Hopkins JM, Foran DR (2016) Next-generation sequencing of the bacterial 16S rRNA gene for forensic soil comparison: a feasibility study. J Forensic Sci 61(3):607–617. https://doi.org/10.1111/1556-4029.13049
Burgoyne L, Koh LY, Catcheside DEA (2017) Extracting information from vehicle exteriors via soil and insect DNA. Forensic Sci Int Genet Suppl Ser 6:e593–e595. https://doi.org/10.1016/j.fsigss.2017.10.004
Meyer JM, Hoy MA (2008) Removal of fungal contaminants and their DNA from the surface of Diaphorina citri (Hemiptera: Psyllidae) prior to a molecular survey of endosymbionts. Fla Entomol 91(4):702–705. https://doi.org/10.1653/0015-4040-91.4.702
Meusnier I, Singer GAC, Landry J-F, Hickey DA, Hebert PDN, Hajibabaei M (2008) A universal DNA mini-barcode for biodiversity analysis. BMC Genomics 9(1):214. https://doi.org/10.1186/1471-2164-9-214
Levin RA, Wagner WL, Hoch PC, Nepokroeff M, Pires JC, Zimmer EA, Sytsma KJ (2003) Family-level relationships of Onagraceae based on chloroplast rbcL and ndhF data. Am J Bot 90(1):107–115. https://doi.org/10.3732/ajb.90.1.107
Little DP (2014) A DNA mini-barcode for land plants. Mol Ecol Resour 14(3):437–446. https://doi.org/10.1111/1755-0998.12194
Wojciechowski MF, Lavin M, Sanderson MJ (2004) A phylogeny of legumes (Leguminosae) based on the analysis of the plastid matK gene resolves many well-supported subclades within the family. Am J Bot 91(11):1846–1862. https://doi.org/10.3732/ajb.91.11.1846
Maurin O (2012) DNA barcoding of plants: matK primers for angiosperms. Royal Botanic Garden Edinburgh. Available at http://api.ning.com/files/3inNVyMeyJHWn4ZQeytwnpIODPXqirXGN9er*vXdBQiB1Il-SDyKKYlFEtxTQU6635m4OKaVOEWfzs8158svE8JMqemREGz3/Angiosperm_matK_RBGE_PROTOCOLv1.0.pdf. (accessed on November 21st 2017)
Schrader C, Schielke A, Ellerbroek L, Johne R (2012) PCR inhibitors—occurrence, properties and removal. J Appl Microbiol 113(5):1014–1026. https://doi.org/10.1111/j.1365-2672.2012.05384.x
Hirano M, Nakamura S, Okada M, Ueda M, Mukai R (2000) Rapid discrimination of monkey B virus from human herpes simplex viruses by PCR in the presence of betaine. J Clin Microbiol 38(3):1255–1257
Katcher HL, Schwartz I (1994) A distinctive property of Tth DNA polymerase: enzymatic amplification in the presence of phenol. BioTechniques 16(1):84–92
Sutlović D, Definis GM, Andelinovic S, Gugic D, Primorac D (2005) Taq polymerase reverse inhibition of quantitative real time polymerase chain reaction by humic acid. Croat Med J 46(4):556–562
Peist R, Honsel D, Twieling G, Löffert D (2001) PCR inhibitors in plant DNA preparations. QIAGEN News 3:7–9
Miller DN, Bryant JE, Madsen EL, Ghiorse WC (1999) Evaluation and optimization of DNA extraction and purification procedures for soil and sediment samples. Appl Environ Microb 65(11):4715–4724
Koonjul PK, Brandt WF, Farrant JM, Lindsey GG (1999) Inclusion of polyvinylpyrrolidone in the polymerase chain reaction reverses the inhibitory effects of polyphenolic contamination of RNA. Nucleic Acids Res 27(3):915–916. https://doi.org/10.1093/nar/27.3.915
Demeke T, Adams RP (1992) The effects of plant polysaccharides and buffer additives on PCR. BioTechniques 12(3):332–334
Kreader CA (1996) Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Appl Environ Microbiol 62(3):1102–1106
Muzzalupo I, Pisani F, Greco F, Chiappetta A (2015) Direct DNA amplification from virgin olive oil for traceability and authenticity. Eur Food Res Technol 241(1):151–155. https://doi.org/10.1007/s00217-015-2455-5
Nilsson M, Grånemo J, Buś MM et al (2016) Comparison of DNA polymerases for improved forensic analysis of challenging samples. Forensic Sci Int Genet 24:55–59. https://doi.org/10.1016/j.fsigen.2016.05.017
Ficetola GF, Coissac E, Zundel S et al (2010) An in silico approach for the evaluation of DNA barcodes. BMC Genomics 11:434. https://doi.org/10.1186/1471-2164-11-434
Zhan A, Bailey SA, Heath DD, Macisaac HJ (2014) Performance comparison of genetic markers for high-throughput sequencing-based biodiversity assessment in complex communities. Mol Ecol Resour 14(5):1049–1059. https://doi.org/10.1111/1755-0998.12254
Finley SJ, Lorenco N, Mulle J, Robertson BK, Javan GT (2016) Assessment of microbial DNA extraction methods of cadaver soil samples for criminal investigations. Aust J Forensic Sci 48(3):265–272. https://doi.org/10.1080/00450618.2015.1063690
Ribeiro RA, Lovato MB (2007) Comparative analysis of different DNA extraction protocols in fresh and herbarium specimens of the genus Dalbergia. Genet Mol Res 6(1):173–187
Li Y, Gao L-M, Poudel RC, Li D-Z, Forrest A (2011) High universality of matK primers for barcoding of gymnosperms. J Syst Evol 49(3):169–175
Soller DR, Reheis MC, Garrity CP, Van Sistine DR (2009) Map database for surficial materials in the conterminous United States: U.S. Geological Survey Data Series 425, scale 1:5,000,000. (available at http://pubs.usgs.gov/ds/425/)
Schweitzer PN (2011) Combined geologic map of the conterminous US derived from the USGS state geological map compilation. (available at mrdata.usgs.gov/geology/state/geol_poly.zip)
Acknowledgments
This is publication number 17-03 of the Laboratory Division of the Federal Bureau of Investigation. Names of commercial manufacturers are provided for information only and inclusion does not imply endorsement by the FBI or the US Government. The views expressed are those of the authors and do not necessarily reflect the official policy or position of the FBI or the US Government. This research was supported in part by an appointment to the Visiting Scientist Program at the FBI Laboratory Division, administered by the Oak Ridge Institute of Science and Education, through an interagency agreement between the US Department of Energy and the FBI. The authors thank JenaMarie Baldaino and Dr. Jack Hietpas for assistance with isolating biological fragments from forensic-type soils.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
ESM 1
(PDF 243 kb)
Rights and permissions
About this article
Cite this article
Meiklejohn, K.A., Jackson, M.L., Stern, L.A. et al. A protocol for obtaining DNA barcodes from plant and insect fragments isolated from forensic-type soils. Int J Legal Med 132, 1515–1526 (2018). https://doi.org/10.1007/s00414-018-1772-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00414-018-1772-1