Abstract
Nucleolin is a multifunctional RNA-binding protein that resides predominantly not only in the nucleolus, but also in multiple other subcellular pools in the cytoplasm in mammalian cells, and is best known for its roles in ribosome biogenesis, RNA stability, and translation. During early mitosis, nucleolin is required for equatorial mitotic chromosome alignment prior to metaphase. Using high resolution fluorescence imaging, we reveal that nucleolin is required for multiple centrosome-associated functions at the G2-prophase boundary. Nucleolin depletion led to dissociation of the centrosomes from the G2 nuclear envelope, a delay in the onset of nuclear envelope breakdown, reduced inter-centrosome separation, and longer metaphase spindles. Our results reveal novel roles for nucleolin in early mammalian mitosis, establishing multiple important functions for nucleolin during mammalian cell division.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Eukaryotic cell division is characterized by a finely orchestrated set of events that occur with a high degree of accuracy and precision to ensure the faithful segregation of chromosomes and cytoplasmic contents to the daughter cells. In vertebrate cells, the centrosomes that duplicated during S-phase lie in close apposition to the nuclear membrane (Ueda et al. 1999; Moens 1976). Mitosis commences at the G2-M cell cycle boundary with the disengagement and separation of the duplicated centrosomes along the G2/prophase nuclear envelope to position themselves at roughly opposite sides of the nucleus, concomitant with chromatin condensation inside the nucleus (Tanenbaum et al. 2010). Following centrosome separation, the nuclear envelope disintegrates as a consequence of phosphorylation and disintegration of the sub-nuclear membrane lamin meshwork and subsequent events (Peter et al. 1990, 1991; Ward and Kirschner 1990), resulting in the mixing of the cytoplasm with the nuclear content following nuclear envelope breakdown (NEB). Microtubules nucleating from the oppositely positioned centrosomes radiate in all directions; “kinetochore microtubules” are captured by the centromeric kinetochores on chromosomes, while “astral microtubules” are captured by conserved protein complexes on the plasma membrane, to generate a bipolar mitotic spindle. The mitotic spindle is used as a scaffold by microtubule-based molecular motors to align (congress) the sister chromatid pairs at the cell’s equator in metaphase (Gardner et al. 2008; Bancroft et al. 2015; Mayr et al. 2007; Auckland and McAinsh 2015; Bancroft et al. 2015; Li et al. 2007; Maiato et al. 2017), followed by kinetochore microtubule depolymerisation to segregate the sister chromatids equally to the daughter cells in anaphase (Cheerambathur et al. 2013; Kapoor 2004; Raaijmakers and Medema 2014; Saunders et al. 1995). The fidelity of early mitotic events is indispensable for generating two euploid daughter cells and is governed by several classes of molecules (Marumoto et al. 2003; Biggins and Walczak 2003; Michaelis et al. 1997; Nigg 2001).
Nucleolin is a well-studied RNA-binding protein that resides prominently not only within interphase nucleoli, but also in smaller pools in the nucleoplasm (Sapp et al. 1986; Lapeyre et al. 1987; Jordan 1987), the cytoplasm (Schwab and Dreyer 1997), and the inner and outer leaflets of the plasma membrane (Hovanessian et al. 2000; Sinclair and O'Brien 2002; Chen et al. 2008; Inder et al. 2009; Otake et al. 2007; Soundararajan et al. 2008; Soundararajan et al. 2009). Nucleolin has been best characterized for its roles in RNA binding and stability, ribosome biogenesis, and mRNA translation (Ginisty et al. 1999; Tajrishi et al. 2011; Tuteja and Tuteja 1998). Nucleolin is also known to reside at interphase centrosomes, wherein it is required for centrosome-based microtubule nucleation (Gaume et al. 2015). Nucleolin has been reported to be directly required for the mitotic process of equatorial chromosome congression to form the metaphase plate, as well as in maintaining mitotic spindle pole integrity (Ma et al. 2007; Ugrinova et al. 2007). Despite the documentation of its multifaceted functions in interphase, other major mitotic roles for nucleolin have not yet been reported.
Here, we reveal the requirement of nucleolin for multiple early mitotic functions in mammalian cells. We show its requirement in maintaining centrosome attachment to the G2/prophase nuclear membrane, for optimal inter-centrosome separation following disengagement at prophase, for the timely onset of NEB, and for maintaining metaphase spindle length. Our observations reveal multifaceted early mitotic functions of nucleolin.
Results
Nucleolin is required to maintain prophase centrosome attachment to the nuclear envelope
In order to examine whether nucleolin is required for centrosome attachment to the nuclear envelope (NE), we depleted nucleolin from HeLa cells stably expressing end-binding protein 1 (EB1) tagged to GFP (EB1-GFP) to visualize centrosomes and histone 2B-mCherry (H2B-mCherry, to visualize chromosomes) using treatment with sequence-specific siRNAs. We confirmed specific depletion of nucleolin through immunoblotting with a nucleolin-specific antibody using either a single siRNA against human nucleolin (“NCL”, Fig. 1A) or a pool of multiple anti-nucleolin siRNAs (“NCL pool”, Fig. 1B) and ascertained robust protein depletion through densitometric quantification of the Western blot band intensities (Fig. 1A, B). We synchronized the cells at the G2/prophase transition using thymidine treatment and release (Chen and Deng 2018). Live cell, time-lapse fluorescence confocal imaging of the treated cells revealed the position of the centrosomes to be closely attached to the G2 NE in control cells, while treatment with nucleolin siRNA led to a marked dissociation of the centrosome(s) from the NE (shown for the NCL siRNA, Fig. 1C). Quantification of this phenotype revealed that the average centrosome-NE distance, measured at the maximal distance from the NE in every cell analyzed by time-lapse live-cell imaging, increased significantly upon nucleolin depletion both by “NCL” (Fig. 1D) and by “NCL pool” (Fig. 1G). We quantified the fraction of cells showing centrosomes detached from the NE and observed an increase in this population by approximately 3-fold compared to control cells (Fig. 1E, H respectively for “NCL” and “NCL pool”). Closer analysis revealed that a significant proportion of mitotic cells showed a large centrosome displacement (> 2 μm) from the NE, a distance limit that was rarely breached in control cells (Fig. 1F, I respectively for “NCL” and “NCL pool”). Overall, these results confirmed a novel and specific function for nucleolin in maintaining the attachment of G2 centrosomes to the NE in mammalian cells.
Nucleolin depletion leads to centrosome-NE attachment defects. A Western blot and densitometric quantification showing the depletion of nucleolin (NCL) from HeLa cells stably expressing H2B-mCherry (red, chromatin in C) and EB1-GFP (green, microtubule plus ends in C), under the indicated conditions of control (luciferase siRNA) or NCL siRNA. B Western blot and densitometric quantification showing the depletion of nucleolin from HeLa cells stably expressing H2B-mCherry (red, chromatin in C) and EB1-GFP (green, microtubule plus ends in C), under the indicated conditions of control (untransfected) or NCL pool siRNA. C Representative stills from time-lapse videos of the cells treated in A. Time-stamps (min) included in the images. Arrowheads, centrosomes. The boundary of the red stain indicated the NE. D, G Maximum centrosome-NE distance measured from analysis of live cell movies of the cells depicted in A (D) and B (G) after the indicated siRNA treatments. 90 G2 cells counted over three independent experiments per condition for D, and 60 G2 cells counted over two independent experiments per condition for G. E, H Quantification of the fraction (%) of G2 cells showing at least one centrosome detached from the NE upon NCL siRNA treatment (E) and NCL pool siRNA treatment (H). F, I Quantification of the fraction (%) of G2 cells showing centrosomes separated from the NE for at least 2 μm upon NCL siRNA treatment (F) and NCL pool siRNA treatment (I). IB, immunoblot. GAPDH, loading control. Error bars, mean +/− SEM. *P < 0.05, **P < 0.01, ****P < 0.0001. (D, G: Mann-Whitney test; E, F, H, I: unpaired t-test). Scale bar = 25 μm
Nucleolin is required for inter-centrosome separation at the G2 phase
The two duplicated centrosomes normally stay together until just prior to NEB, when they disengage and separate from each other along the nuclear envelope, enabling the eventual formation of a bipolar spindle to facilitate chromosome segregation (M. E. Tanenbaum and Medema 2010). Upon nucleolin siRNA treatment, we observed through time-lapse fluorescence confocal imaging that the two G2/prophase centrosomes untethered from each other normally, but failed to separate completely from each other (Fig. 2A). We measured the inter-centrosome distance immediately after nuclear envelope breakdown (NEB), by which point further centrosome movement along the NE is not possible (Toso et al. 2009). Quantitative analysis of our live cell confocal imaging data revealed that the average inter-centrosome distance at G2/prophase reduced significantly upon nucleolin siRNA (NCL) treatment (Fig. 2B). These results suggested that nucleolin is required to ensure the proper separation of G2/prophase centrosomes along the NE. Nucleolin has been shown to be required for maintaining a bipolar spindle in metaphase and to prevent centrosomal fragmentation (Ma et al. 2007; Ugrinova et al. 2007). Our observations suggest that nucleolin may also facilitate the formation of a proper mitotic spindle by ensuring the optimal separation of the duplicated G2/prophase centrosomes.
Nucleolin depletion leads to reduced inter-centrosome separation following NEB. A Representative stills from time-lapse videos of HeLa cells stably expressing H2B-mCherry (red, chromatin) and EB1-GFP (green, microtubule plus ends) upon treatment with the NCL siRNA. Arrowheads, centrosomes. B Inter-centrosome linear distance measured from analysis of live cell movies of the cells depicted in A after the indicated siRNA treatments. 90 G2 cells counted over three independent experiments per condition. Error bars = mean +/− SEM. ****P < 0.0001. (B: unpaired t-test). Scale bar = 10 μm
Nucleolin is required for ensuring timely nuclear envelope breakdown
Inter-centrosome separation at prophase to position the two centrosomes at opposite sides of the nucleus is closely followed by the onset of NEB, so as to enable the formation of a bipolar mitotic spindle (Agircan et al. 2014; van Heesbeen et al. 2013). We analyzed the live cell movies of HeLa cells stably expressing EB1-GFP::H2B-mCherry to examine whether nucleolin is required for the timely onset and completion of NEB following separation. We observed that treatment with nucleolin siRNA led to a modest but consistent delay in the time taken from initial centrosome-centrosome disengagement to the dissolution of the nuclear envelope, a phenotype that was reproducibly observed using both NCL and NCL pool siRNAs (Figs. 3A, B—“NCL”; E—“NCL pool”). Similarly, nucleolin siRNA treatment also led to a delay in the time taken to complete NEB, characterized by the disappearance of any remnants of a sharp boundary demarcating the chromatin from the cytoplasm (Fig. 3A, C—“NCL”; F—“NCL pool”). To examine whether nucleolin is required for the progression of NEB itself once initiated, we analyzed the duration between NEB onset and NEB end in luciferase and nucleolin depleted cells. We observed no significant changes in NEB duration in nucleolin depleted cells as compared to controls (Fig. 3D—“NCL”; G—“NCL pool”). These observations suggested that nucleolin plays a role in ensuring the timely onset of nuclear envelope disintegration at the beginning of mitosis, but not in its progression once initiated.
Nucleolin depletion leads to a delay in the onset of NEB. A Representative stills from time-lapse videos of HeLa cells stably expressing H2B-mCherry (red, chromatin) and EB1-GFP (green, microtubule plus ends), upon treatment with the indicated siRNAs. Time-stamps (min) included in the images. Arrowheads, centrosomes, arrows, NE. B, E Time taken (min) from inter-centrosome disengagement to NEB onset upon treatment with the indicated siRNAs. C, F Time taken (min) from inter-centrosome disengagement to NEB end. D, G Time taken (min) from NEB onset to NEB end upon treatment with the indicated siRNAs. 90 G2 cells counted over three independent experiments per condition for B–D, and 60 G2 cells counted over two independent experiments per condition for E–G. Error bars = mean +/− SEM. (B–G: Mann-Whitney test). *P < 0.05, ns, non-significant. Scale bar = 25 μm
Nucleolin is required for maintaining spindle length in metaphase
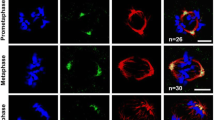
Bipolar spindle formation and mitotic spindle integrity are critical for proper chromosome segregation (van Heesbeen et al. 2014; Brouwers et al. 2017). Nucleolin is required for proper chromosome congression and maintenance of spindle pole integrity in mitosis (Ma et al. 2007; Ugrinova et al. 2007). In order to understand whether nucleolin has a role in regulating spindle length during mitosis, we depleted nucleolin in U2OS cells using sequence-specific siRNAs (NCL) and confirmed efficient knockdown by immunoblotting (Fig. 4A). We confirmed the efficiency of knockdown by quantifying protein levels in control and nucleolin depleted conditions by densitometric analysis of the respective immunoblots (Fig. 4B). We performed immunofluorescence analysis of the cells, imaged them by confocal microscopy, generated three dimensional reconstructions from the depth stacks, and measured the distance between the spindle poles from the reconstructions (Fig. 4C). Examination of the inter-centrosomal distance at metaphase revealed a significant increase in the average mitotic spindle length, as depicted by the representative linescans of the inter-centrosomal spindle axes (Fig. 4D). The fluorescence intensity profile of gamma-tubulin (centrosomes) clearly peaked at the centrosomal area, but was significantly reduced along the spindle axis. The average distance between the two gamma tubulin peaks (x-axes of the linescans in Fig. 4D) was greater upon nucleolin siRNA treatment by about 12% as compared to control treatment (Fig. 4E). These observations confirmed that nucleolin has essential roles in maintaining optimal metaphase spindle length during mammalian cell division.
Nucleolin depletion leads to elongated metaphase spindles. A Western blot showing the depletion of nucleolin from U2OS cells after the indicated siRNA treatments. IB, immunoblot. GAPDH, loading control. B Densitometric quantification of band intensities from A to show the level of protein depletion. C Representative 3-dimensional confocal immunofluorescence reconstructions of the cells in A under the indicated treatments. White spheres, centrosomes (spindle poles); red, alpha-tubulin (microtubules). D Representative linescans depicting the inter-centrosomal distance (spindle length, x-axis) under the indicated siRNA treatments. E Quantification of the average spindle length. 75 metaphase cells counted over three independent experiments per condition. Error bars = mean +/− SEM. (E: Mann-Whitney test). ***P < 0.001. Scale bar = 3 μm
Discussion
Nucleolin is a multifunctional protein that has been well studied for its functions in RNA binding, stability, translation (Ginisty et al. 1999; Tuteja and Tuteja 1998; Mongelard and Bouvet 2007) and ribosome biogenesis (Srivastava and Pollard 1999). The role of nucleolin in mitosis had been reported in ensuring equatorial chromosome alignment (congression), maintaining kinetochore-microtubule attachment stability and to a lesser extent, in maintaining centrosome integrity in mitosis (Ma et al. 2007; Ugrinova et al. 2007). Here, we report novel functions of nucleolin in the early stages of mitosis in maintaining prophase centrosome-NE tethering, inter-centrosome separation during prophase, governing the timely onset of NEB, and in maintaining metaphase spindle length. Together, these observations reveal multifaceted roles for nucleolin in mammalian mitosis.
Our results reveal that nucleolin makes a significant functional contribution in maintaining centrosome-NE attachment (Fig. 1). Cytoplasmic dynein plays an important role in this function as well, with dynein depletion leading to severe centrosome detachment from the NE (Raaijmakers et al. 2013; Kumari et al. 2021). The dynein motor is also known to exist in distinct complexes at various cellular locations (Purohit et al. 1999; Tynan et al. 2000). Taken together with the fact that nucleolin exists in multiple overlapping cellular pools as well (Berger et al. 2015), it is possible to envision a particular subcellular fraction of nucleolin, perhaps the centrosomally localized pool (Gaume et al. 2015), collaborating with cytoplasmic dynein to enable centrosome-NE tethering. Interestingly, certain dynein populations begin to decorate centrosomes only from the entry to mitosis (Horgan et al. 2011; Mahale et al. 2016a, b), suggesting the possibility of a mitotic collaboration between nucleolin and dynein to help anchor centrosomes to the NE. Indeed, nucleolin has been identified as an interactor of dynein in asynchronous mammalian cell cultures (Redwine et al. 2017).
In addition to the centrosome-NE detachment observed upon nucleolin siRNA treatment, we also observed reduced inter-centrosome separation at G2 (Fig. 2). Observations from the live imaging data showed that centrosome-centrosome disengagement occurred normally in most cells; however, the inter-centrosome distance was lower on average upon nucleolin siRNA treatment than in control cells (Fig. 2). Inter-centrosome separation is governed by the outward pushing-apart of the disengaged centrosomes due to centrosomal astral microtubules nucleating in opposing directions (van Heesbeen et al. 2013). Nucleolin has been shown to be required for centrosomal microtubule nucleation (Gaume et al. 2015), which could have a direct role in mediating centrosome separation. Nucleolin may also contribute to centrosome separation through other mechanism(s). The microtubule plus end-directed motor kinesin 5 (Eg5) plays a major role in centrosome separation by causing outward spindle microtubule sliding to separate the centrosomes (She et al. 2022; Agircan et al. 2014; Smith et al. 2011; Ferenz et al. 2010), a mechanism in which nucleolin could perhaps also be required. The demonstration of a functional or physical interaction of nucleolin with kinesin 5 would give credence to this hypothesis. Independent of Eg5, centrosome-NE-associated dynein, can also move individual centrosomes along the NE (van Heesbeen et al. 2013), as demonstrated by the depletion of either dynein, or the dynein adaptor BICD2 (Raaijmakers et al. 2012). It would be logical to surmise that nucleolin may enable NE-associated dynein function in separating the prophase centrosomes as well. The overall effect of nucleolin depletion may very well be mediated partially through both motors and will need detailed investigations to delineate.
Nucleolin is known to be present on G2 centrosomes in mammalian cells and is vital for microtubule nucleation in interphase (Gaume et al. 2015). Microtubule nucleation has been proposed to facilitate NEB by causing membrane rupture (Beaudouin et al. 2002). We observe that nucleolin depletion leads to a mild delay in the onset of NEB in HeLa cells. Cytoplasmic dynein has been reported to be a key facilitating factor of NEB (Salina et al. 2002). Certain dynein subpopulations are known to be constitutively present at mammalian cell centrosomes throughout the cell cycle, while others arrive at centrosomes at mitotic onset (Horgan et al. 2011; Mahale et al. 2016a, b). It is attractive to imagine that the function of nucleolin in enabling timely NEB onset could be in collaboration with either/both of these centrosomal dynein complexes. The cytoplasmic pool of nucleolin is likely to be the primary subpopulation responsible for both proper centrosome-NE attachment as well as timely NEB onset, since this population would have access to the centrosome and/or the NE. Elucidation of the mechanisms underlying the function of nucleolin in timely NEB onset merits an independent and detailed investigation.
We observe a requirement for nucleolin in maintaining proper mitotic spindle length, with nucleolin depletion leading to longer spindles (inter-polar distance) on average (Fig. 4). This effect is again reminiscent of dynein phenotypes, since dynein’s role in maintaining proper spindle length is well documented (Gaetz and Kapoor 2004; Gatlin and Bloom 2010; Nannas et al. 2014; Raaijmakers et al. 2013; Mahale et al. 2016a; Tanenbaum et al. 2009). It is possible to envision a scenario wherein mitotic nucleolin collaborates with mitotic dynein populations to regulate spindle length, a model consistent with nucleolin being an interactor of dynein in interphase cells. Mitotic spindle length is regulated by a balance between the opposing forces exerted on spindle microtubules by dynein and kinesin 5 (Eg5) (Ferenz et al. 2009; Tanenbaum et al. 2008). Nucleolin has also been reported to interact with multiple kinesin families in neuronal cells through its C-terminal GAR domain (Doron-Mandel et al. 2021), although not specifically yet with kinesin 5. It remains to be seen whether nucleolin also interacts with kinesin(s) to control spindle length. Further studies into the possible interplay of nucleolin with these opposing motors have the potential to reveal deeper insights into its role in regulating spindle length.
Our study reveals a broad spectrum of mitotic functions for nucleolin and opens up new questions in the field. RNA transcription and translation are known to be suppressed during mitosis (Michelotti et al. 1997; Heix et al. 1998; Sif et al. 1998; Sirri et al. 2000; Bonneau and Sonenberg 1987; Pyronnet and Sonenberg 2001; Tanenbaum et al. 2015). Nevertheless, it would be interesting to explore whether nucleolin’s RNA-binding ability is required for mitotic functions. It would be equally interesting to explore whether a potential dynein-nucleolin mitotic interaction could be required for nucleolin’s mitotic functions, especially for regulating centrosome-NE attachment. Another attractive idea is to explore whether nucleolin could serve as a linker between the two oppositely directed dynein and kinesin motors to fine-tune the balance between them during the regulation of spindle length. Indeed, it would be worthwhile to examine which of the spectrum of nucleolin’s mitotic functions may be through a collaboration with one or the other microtubule-based motors. Future investigations in these directions will clarify the mechanisms through which nucleolin performs its diverse array of mitotic functions.
Materials and methods
Cell culture, siRNA transfection, and cell synchronization
The H2B-mCherry::EB1-GFP stable cell line was a kind gift from Dr. Mahak Sharma, Indian Institute of Science Education and Research, Mohali, and U2OS cells a kind gift from Prof. Stephen J. Doxsey, University of Massachusetts Chan Medical School, Worcester, MA. Both cell lines were cultured in DMEM media (Invitrogen/Gibco) supplemented with 10% fetal bovine serum (FBS) and an antibiotic solution containing penicillin and streptomycin (HiMedia). Cells were cultured at 37 °C with 5% carbon dioxide and 95% humidity.
The siRNA sequences and respective working concentrations used in the study were Luciferase (Dharmacon; 100 nM), 5′-CAUUCUAUCCUCUAGAGGAUGUU-3′; nucleolin (NCL, Dharmacon, 100 nM), 5′-AGAGUUUGCUUCAUUCGAAUU-3′; NCL pool (35 nM, Mission esiRNA from Sigma-Aldrich, EHU080431). Individual siRNAs were transfected using Dharmafect 1 (Dharmacon) for 48 h. For late G2/prophase enrichment, HeLa and U2OS cells were arrested at the G1/S boundary by single thymidine treatment for 18 h (2.5 mM, Sigma-Aldrich), followed by a washout and release for 8 h (for HeLa cells) and 9 h (for U2OS cells).
Antibodies and chemicals
The following primary antibodies were used: nucleolin (Bethyl labs A300-709A), GAPDH (DSHB S1-1588), alpha-tubulin (Abcam ab18251), gamma tubulin (Abcam ab11316). The following antibodies were procured from Jackson Immunoresearch USA: HRP-conjugated anti-mouse (715-035-150) and anti-rabbit (711-035152) secondary antibodies for immunoblotting and fluorophore-attached anti-mouse and anti-rabbit AlexaFluor 488–conjugated secondary antibodies (715-545-150) and anti-rabbit AlexaFluor 594–conjugated secondary antibody (715-585-150) for immunofluorescence staining.
Western blotting (immunoblotting)
Cells were lysed by adding 2X Laemmli buffer containing 2-mercaptoethanol (Sigma) to the cells, followed by scraping and passing the sample through a syringe fitted with a 26-gauge needle a few times, boiled at 95 °C for 10 min, and resolved by 10% SDS-PAGE followed by transfer onto polyvinylidene difluoride membrane (PVDF, Millipore). After transfer, the membrane was blocked with 5% skimmed milk prepared in phosphate buffered saline containing 0.1% Tween 20 (PBST), followed by overnight incubation in the primary antibody at 4 °C. Following a 1 h wash after primary antibody incubation, horseradish peroxidase (HRP)-conjugated secondary antibody was incubated for 1 h at room temperature. The blot was washed before being developed for chemiluminescence signal using the Luminata Forte reagent (Millipore WBLUF0500) and captured in the ImageQuant LAS-4000 gel documentation system (GE Healthcare). The dilutions used for the various primary and secondary antibodies were as follows: NCL 1:1,000; GAPDH 1:60; anti-rabbit HRP 1:10,000; and anti-mouse HRP 1:10,000. Whole-image brightness and contrast were adjusted in Adobe Photoshop for final figure preparation.
Immunofluorescence staining
U2OS cells were cultured on glass coverslips, washed with 1x PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4, pH 7.2) before fixing them in chilled methanol. Fixed coverslips were transferred to a humidified chamber and rehydrated using 1x PBS for 10 min, followed by incubation in blocking solution (1% BSA, 1x PBS and Triton X-100) for 1 h at room temperature. Primary antibody incubation for 1 h at room temperature was followed by washing and incubation with the appropriate secondary antibodies for 1 h. Primary antibody dilutions used for immunofluorescence staining were as follows: gamma tubulin 1:500; alpha-tubulin 1:800. Secondary antibodies (listed above) were used at a dilution of 1:800. After washing twice with 1X PBSAT and once with PBS, DAPI was added at 1:10,000 dilution (from a 5 mg/ml stock solution) for 2 min, followed by washing twice with 1X PBS and once with deionized water (Millipore), and mounted on a frosted glass slide using Prolong Diamond antifade mounting medium (Invitrogen). Coverslips were dried overnight in the dark and stored at −20 °C until confocal/fluorescence imaging.
Fluorescence imaging and analysis
For live-cell imaging, HeLa cells stably expressing EB1-GFP and H2B-mCherry were maintained in DMEM supplemented with hygromycin and puromycin. Cells were grown on glass coverslips and transfected with siRNAs. Following 24 h of transfection, cells were treated with thymidine (2.5 mM) for 18 h to enrich cells at S-phase. Cells were then washed twice with PBS and incubated in DMEM for 8 h to enrich cells at the G2 phase. Set up for live-cell imaging was performed using a customized aluminum slide containing 12 mm chambers as described earlier (Mahale et al. 2016a). Time-lapse images with z-stacks containing planes 0.5 μm apart were acquired every 5 min for 12 h on a Leica TCS SP8 laser scanning optical confocal microscope using an HCX PL APO CS 63x-1.4 NA oil-immersion objective fitted in a humidified heating chamber (OkoLab) maintained at 37 °C. All image acquisition settings were kept identical for control and test samples. The Leica LASX software was used to control various imaging parameters during image acquisition as well as for post-acquisition image analysis. Fluorescence images were analyzed for measuring centrosome-NE distance, inter-centrosome distance, NEB onset, and NEB end times, starting from inter-centrosome disengagement during G2 phase using the Leica LASX offline image analysis software. Representative immunofluorescence images shown are 3D reconstructions made using the Imaris software suite (v5.7, Bitplane). Micrographs were imported into the Adobe Photoshop CS6/Photoshop v22.0 at 300-dpi resolution.
Quantification of NE-centrosome distance, inter-centrosome distance, NEB onset/end timings, and mitotic spindle length
NE-centrosome distance was measured by drawing a line connecting the nuclear envelope and centrosomes using the Leica LASX software. Inter-centrosome distances were measured by drawing a line connecting both the centrosomes at the end of prophase using the Leica LASX software. NEB onset/end timings were measured from inter-centrosome disengagement to the NEB start/end respectively in minutes, using the Leica LASX software. Spindle length was measured as the distance between the two centrosomes (gamma tubulin spots) using the Imaris software suite. Briefly, the inter-polar distance was measured by drawing spheres of 2 μm using the “spot function” to select the spindle poles and then measuring the linear distance between the two spots using the “measurement function”.
Statistical analysis
Statistical analysis and representation of the data were done using the Prism 7 software (GraphPad Software). Data was first analyzed for normality, and the appropriate downstream statistical test(s) performed as indicated in the figure legends. Data are presented as mean ± SEM. The sample sizes are specified in the figure legends for all the quantitative data. Error bars represent SEM from at least three independent experiments.
Data availability
All data is contained within this manuscript. Materials generated by the authors will be available upon request for academic researchers with applicable transfer agreements and reasonable transportation charges.
References
Agircan FG, Schiebel E, Mardin BR (2014) Separate to operate: control of centrosome positioning and separation. Philos Trans R Soc Lond B Biol Sci 369(1650). https://doi.org/10.1098/rstb.2013.0461
Auckland P, McAinsh AD (2015) Building an integrated model of chromosome congression. J Cell Sci 128(18):3363–3374. https://doi.org/10.1242/jcs.169367
Bancroft J, Auckland P, Samora CP, McAinsh AD (2015) Chromosome congression is promoted by CENP-Q- and CENP-E-dependent pathways. J Cell Sci 128(1):171–184. https://doi.org/10.1242/jcs.163659
Beaudouin J, Gerlich D, Daigle N, Eils R, Ellenberg J (2002) Nuclear envelope breakdown proceeds by microtubule-induced tearing of the lamina. Cell 108(1):83–96
Berger CM, Gaume X, Bouvet P (2015) The roles of nucleolin subcellular localization in cancer. Biochimie 113:78–85
Biggins S, Walczak CE (2003) Captivating capture: how microtubules attach to kinetochores. Curr Biol 13(11):R449–R460
Bonneau A, Sonenberg N (1987) Involvement of the 24-kDa cap-binding protein in regulation of protein synthesis in mitosis. J Biol Chem 262(23):11134–11139
Brouwers N, Mallol Martinez N, Vernos I (2017) Role of Kif15 and its novel mitotic partner KBP in K-fiber dynamics and chromosome alignment. PLoS One 12(4):e0174819. https://doi.org/10.1371/journal.pone.0174819
Cheerambathur DK, Gassmann R, Cook B, Oegema K, Desai A (2013) Crosstalk between microtubule attachment complexes ensures accurate chromosome segregation. Sci 342(6163):1239–1242. https://doi.org/10.1126/science.1246232
Chen G, Deng X (2018) Cell synchronization by double thymidine block. Bio Protoc 8(17). https://doi.org/10.21769/BioProtoc.2994
Chen X, Kube DM, Cooper MJ, Davis PB (2008) Cell surface nucleolin serves as receptor for DNA nanoparticles composed of pegylated polylysine and DNA. Molecular Ther 16(2):333–342
Doron-Mandel E, Koppel I, Abraham O, Rishal I, Smith TP, Buchanan CN, Fainzilber M (2021) The glycine arginine-rich domain of the RNA-binding protein nucleolin regulates its subcellular localization. Embo J 40(20):e107158. https://doi.org/10.15252/embj.2020107158
Ferenz NP, Gable A, Wadsworth P (2010) Mitotic functions of kinesin-5. Semin Cell Dev Biol 21(3):255–259. https://doi.org/10.1016/j.semcdb.2010.01.019
Ferenz NP, Paul R, Fagerstrom C, Mogilner A, Wadsworth P (2009) Dynein antagonizes eg5 by crosslinking and sliding antiparallel microtubules. Curr Biol 19(21):1833–1838. https://doi.org/10.1016/j.cub.2009.09.025
Gaetz J, Kapoor TM (2004) Dynein/dynactin regulate metaphase spindle length by targeting depolymerizing activities to spindle poles. J Cell Biol 166(4):465–471. https://doi.org/10.1083/jcb.200404015
Gardner MK, Bouck DC, Paliulis LV, Meehl JB, O'Toole ET, Haase J, Odde DJ (2008) Chromosome congression by kinesin-5 motor-mediated disassembly of longer kinetochore microtubules. Cell 135(5):894–906. https://doi.org/10.1016/j.cell.2008.09.046
Gatlin JC, Bloom K (2010) Microtubule motors in eukaryotic spindle assembly and maintenance. Semin Cell Dev Biol 21(3):248–254. https://doi.org/10.1016/j.semcdb.2010.01.015
Gaume X, Tassin AM, Ugrinova I, Mongelard F, Monier K, Bouvet P (2015) Centrosomal nucleolin is required for microtubule network organization. Cell Cycle 14(6):902–919. https://doi.org/10.1080/15384101.2014.1000197
Ginisty H, Sicard H, Roger B, Bouvet P (1999) Structure and functions of nucleolin. J Cell Sci 112(Pt 6):761–772
Heix J, Vente A, Voit R, Budde A, Michaelidis TM, Grummt I (1998) Mitotic silencing of human rRNA synthesis: inactivation of the promoter selectivity factor SL1 by cdc2/cyclin B-mediated phosphorylation. Embo J 17(24):7373–7381. https://doi.org/10.1093/emboj/17.24.7373
Horgan CP, Hanscom SR, McCaffrey MW (2011) Dynein LIC1 localizes to the mitotic spindle and midbody and LIC2 localizes to spindle poles during cell division. Cell Biology Int 35(2):171–178
Hovanessian AG, Puvion-Dutilleul F, Nisole S, Svab J, Perret E, Deng J-S, Krust B (2000) The cell-surface-expressed nucleolin is associated with the actin cytoskeleton. Exp Cell Res 261(2):312–328
Inder KL, Lau C, Loo D, Chaudhary N, Goodall A, Martin S, . . . Hancock JF (2009) Nucleophosmin and nucleolin regulate K-Ras plasma membrane interactions and MAPK signal transduction. J Biol Chem284(41):28410-28419 https://doi.org/10.1074/jbc.M109.001537
Jordan G (1987) At the heart of the nucleolus. Nature 329(6139):489–490
Kapoor TM (2004) Chromosome segregation: correcting improper attachment. Curr Biol 14(23):R1011–R1013. https://doi.org/10.1016/j.cub.2004.11.026
Kumari A, Kumar C, Pergu R, Kumar M, Mahale SP, Wasnik N, Mylavarapu SVS (2021) Phosphorylation and Pin1 binding to the LIC1 subunit selectively regulate mitotic dynein functions. J Cell Biol 220(12). https://doi.org/10.1083/jcb.202005184
Lapeyre B, Bourbon H, Amalric F (1987) Nucleolin, the major nucleolar protein of growing eukaryotic cells: an unusual protein structure revealed by the nucleotide sequence. Proceed Nat Academ Sci 84(6):1472–1476
Li Y, Yu W, Liang Y, Zhu X (2007) Kinetochore dynein generates a poleward pulling force to facilitate congression and full chromosome alignment. Cell Res 17(8):701–712. https://doi.org/10.1038/cr.2007.65
Ma N, Matsunaga S, Takata H, Ono-Maniwa R, Uchiyama S, Fukui K (2007) Nucleolin functions in nucleolus formation and chromosome congression. J Cell Sci 120(Pt 12):2091–2105. https://doi.org/10.1242/jcs.008771
Mahale S, Kumar M, Sharma A, Babu A, Ranjan S, Sachidanandan C, Mylavarapu SVS (2016a) The light intermediate chain 2 subpopulation of dynein regulates mitotic spindle orientation. Scientific Rep 6(1):22. https://doi.org/10.1038/s41598-016-0030-3
Mahale SP, Sharma A, Mylavarapu SV (2016b) Dynein light intermediate chain 2 facilitates the metaphase to anaphase transition by inactivating the spindle assembly checkpoint. PLoS One 11(7):e0159646
Maiato H, Gomes AM, Sousa F, Barisic M (2017) Mechanisms of chromosome congression during mitosis. Biology (Basel) 6(1). https://doi.org/10.3390/biology6010013
Marumoto T, Honda S, Hara T, Nitta M, Hirota T, Kohmura E, Saya H (2003) Aurora-A kinase maintains the fidelity of early and late mitotic events in HeLa cells. J Biol Chem 278(51):51786–51795. https://doi.org/10.1074/jbc.M306275200
Mayr MI, Hümmer S, Bormann J, Grüner T, Adio S, Woehlke G, Mayer TU (2007) The human kinesin Kif18A is a motile microtubule depolymerase essential for chromosome congression. Curr Biol 17(6):488–498. https://doi.org/10.1016/j.cub.2007.02.036
Michaelis C, Ciosk R, Nasmyth K (1997) Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91(1):35–45. https://doi.org/10.1016/s0092-8674(01)80007-6
Michelotti EF, Sanford S, Levens D (1997) Marking of active genes on mitotic chromosomes. Nature 388(6645):895–899
Moens PB (1976) Spindle and kinetochore morphology of Dictyostelium discoideum. J Cell Biol 68(1):113–122
Mongelard F, Bouvet P (2007) Nucleolin: a multiFACeTed protein. Trends Cell Biol 17(2):80–86
Nannas NJ, O'Toole ET, Winey M, Murray AW (2014) Chromosomal attachments set length and microtubule number in the Saccharomyces cerevisiae mitotic spindle. Mol Biol Cell 25(25):4034–4048. https://doi.org/10.1091/mbc.E14-01-0016
Nigg EA (2001) Mitotic kinases as regulators of cell division and its checkpoints. Nature Rev Mol Cell Biol 2(1):21–32. https://doi.org/10.1038/35048096
Otake Y, Soundararajan S, Sengupta TK, Kio EA, Smith JC, Pineda-Roman M, Fernandes DJ (2007) Overexpression of nucleolin in chronic lymphocytic leukemia cells induces stabilization of bcl2 mRNA. Blood 109(7):3069–3075
Peter M, Heitlinger E, Häner M, Aebi U, Nigg E (1991) Disassembly of in vitro formed lamin head-to-tail polymers by CDC2 kinase. EMBO J 10(6):1535–1544
Peter M, Nakagawa J, Doree M, Labbe J, Nigg E (1990) In vitro disassembly of the nuclear lamina and M phase-specific phosphorylation of lamins by cdc2 kinase. Cell 61(4):591–602
Purohit A, Tynan SH, Vallee R, Doxsey SJ (1999) Direct interaction of pericentrin with cytoplasmic dynein light intermediate chain contributes to mitotic spindle organization. J Cell Biol 147(3):481–492
Pyronnet S, Sonenberg N (2001) Cell-cycle-dependent translational control. Curr Opin Gen Dev 11(1):13–18
Raaijmakers JA, Medema RH (2014) Function and regulation of dynein in mitotic chromosome segregation. Chromosoma 123(5):407–422. https://doi.org/10.1007/s00412-014-0468-7
Raaijmakers JA, Tanenbaum ME, Medema RH (2013) Systematic dissection of dynein regulators in mitosis. J Cell Biol 201(2):201–215. https://doi.org/10.1083/jcb.201208098
Raaijmakers JA, van Heesbeen RG, Meaders JL, Geers EF, Fernandez-Garcia B, Medema RH, Tanenbaum ME (2012) Nuclear envelope-associated dynein drives prophase centrosome separation and enables Eg5-independent bipolar spindle formation. Embo J 31(21):4179–4190. https://doi.org/10.1038/emboj.2012.272
Redwine WB, DeSantis ME, Hollyer I, Htet ZM, Tran PT, Swanson SK, Florens L, Washburn MP, Reck-Peterson SL (2017) The human cytoplasmic dynein interactome reveals novel activators of motility. ELife 6:e28257. https://doi.org/10.7554/eLife.28257
Salina D, Bodoor K, Eckley DM, Schroer TA, Rattner J, Burke B (2002) Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell 108(1):97–107
Sapp M, Knippers R, Richter A (1986) DNA binding properties of a 110kDa nucleolar protein. Nucleic Acids Res 14(17):6803–6820
Saunders WS, Koshland D, Eshel D, Gibbons IR, Hoyt MA (1995) Saccharomyces cerevisiae kinesin- and dynein-related proteins required for anaphase chromosome segregation. J Cell Biol 128(4):617–624 Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/7860634
Schwab M, Dreyer C (1997) Protein phosphorylation sites regulate the function of the bipartite NLS of nucleolin. European J Cell Biol 73(4):287–297
She ZY, Zhong N, Wei YL (2022) Kinesin-5 Eg5 mediates centrosome separation to control spindle assembly in spermatocytes. Chromosoma 131(1-2):87–105. https://doi.org/10.1007/s00412-022-00772-5
Sif S, Stukenberg PT, Kirschner MW, Kingston RE (1998) Mitotic inactivation of a human SWI/SNF chromatin remodeling complex. Genes Dev 12(18):2842–2851. https://doi.org/10.1101/gad.12.18.2842
Sinclair JF, O'Brien AD (2002) Cell surface-localized nucleolin is a eukaryotic receptor for the adhesin intimin-γ of enterohemorrhagic Escherichia coliO157: H7. J Biol Chem 277(4):2876–2885
Sirri V, Roussel P, Hernandez-Verdun D (2000) In vivo release of mitotic silencing of ribosomal gene transcription does not give rise to precursor ribosomal RNA processing. J Cell Biol 148(2):259–270. https://doi.org/10.1083/jcb.148.2.259
Smith E, Hégarat N, Vesely C, Roseboom I, Larch C, Streicher H, Hochegger H (2011) Differential control of Eg5-dependent centrosome separation by Plk1 and Cdk1. Embo J 30(11):2233–2245. https://doi.org/10.1038/emboj.2011.120
Soundararajan S, Chen W, Spicer EK, Courtenay-Luck N, Fernandes DJ (2008) The nucleolin targeting aptamer AS1411 destabilizes Bcl-2 messenger RNA in human breast cancer cells. Cancer Res 68(7):2358–2365
Soundararajan S, Wang L, Sridharan V, Chen W, Courtenay-Luck N, Jones D, Fernandes DJ (2009) Plasma membrane nucleolin is a receptor for the anticancer aptamer AS1411 in MV4-11 leukemia cells. Mol Pharmacol 76(5):984–991. https://doi.org/10.1124/mol.109.055947
Srivastava M, Pollard HB (1999) Molecular dissection of nucleolin’s role in growth and cell proliferation: new insights. Faseb J 13(14):1911–1922
Tajrishi MM, Tuteja R, Tuteja N (2011) Nucleolin Commun Integ Biol 4(3):267–275. https://doi.org/10.4161/cib.4.3.14884
Tanenbaum ME, Macurek L, Galjart N, Medema RH (2008) Dynein, Lis1 and CLIP-170 counteract Eg5-dependent centrosome separation during bipolar spindle assembly. Embo J 27(24):3235–3245. https://doi.org/10.1038/emboj.2008.242
Tanenbaum ME, Macůrek L, Janssen A, Geers EF, Alvarez-Fernández M, Medema RH (2009) Kif15 cooperates with eg5 to promote bipolar spindle assembly. Curr Biol 19(20):1703–1711
Tanenbaum ME, Medema RH (2010) Mechanisms of centrosome separation and bipolar spindle assembly. Dev Cell 19(6):797–806. https://doi.org/10.1016/j.devcel.2010.11.011
Tanenbaum ME, Stern-Ginossar N, Weissman JS, Vale RD (2015) Regulation of mRNA translation during mitosis. Elife 4. https://doi.org/10.7554/eLife.07957
Toso A, Winter JR, Garrod AJ, Amaro AC, Meraldi P, McAinsh AD (2009) Kinetochore-generated pushing forces separate centrosomes during bipolar spindle assembly. J Cell Biol 184(3):365–372
Tuteja R, Tuteja N (1998) Nucleolin: a multifunctional major nucleolar phosphoprotein. Crit Rev Biochem Mol Biol 33(6):407–436
Tynan SH, Purohit A, Doxsey SJ, Vallee RB (2000) Light intermediate chain 1 defines a functional subfraction of cytoplasmic dynein which binds to pericentrin. J Biol Chem 275(42):32763–32768. https://doi.org/10.1074/jbc.M001536200
Ueda M, Schliwa M, Euteneuer U (1999) Unusual centrosome cycle in dictyostelium: correlation of dynamic behavior and structural changes. Mol Biol Cell 10(1):151–160
Ugrinova I, Monier K, Ivaldi C, Thiry M, Storck S, Mongelard F, Bouvet P (2007) Inactivation of nucleolin leads to nucleolar disruption, cell cycle arrest and defects in centrosome duplication. BMC Mol Biol 8:66. https://doi.org/10.1186/1471-2199-8-66
van Heesbeen RG, Tanenbaum ME, Medema RH (2014) Balanced activity of three mitotic motors is required for bipolar spindle assembly and chromosome segregation. Cell Rep 8(4):948–956. https://doi.org/10.1016/j.celrep.2014.07.015
van Heesbeen RGHP, Raaijmakers JA, Tanenbaum ME, Medema RH (2013) Nuclear envelope-associated dynein cooperates with Eg5 to drive prophase centrosome separation. Commun Integr Biol 6(3):e23841. https://doi.org/10.4161/cib.23841
Ward GE, Kirschner MW (1990) Identification of cell cycle-regulated phosphorylation sites on nuclear lamin C. Cell 61(4):561–577
Acknowledgements
We thank Dr. Mahak Sharma for providing the H2B-mCherry::EB1-GFP stable cell line. We thank Suraj Tewari for assistance with confocal microscopy and the Regional Centre for Biotechnology (RCB) for providing the requisite infrastructure. We are grateful to the members of the Laboratory of Cellular Dynamics, RCB for critical comments and suggestions during the study.
Funding
CK received fellowship support from the Department of Biotechnology (DBT) and the Indian Council of Medical Research (ICMR), India. This work was supported by funding to SVSM from RCB, DBT (grant number BT/PR6420/GBD/27/435/2012) and the Science and Engineering Research Board (SERB), India (grants number EMR/2016/007842 and CRG/2021/006858).
Author information
Authors and Affiliations
Contributions
Chandan Kumar (CK) and Sivaram V S Mylavarapu (SVSM) designed the experiments, analyzed the data, and prepared and edited the manuscript. CK performed experiments. SVSM conceptualized and supervised the entire study and obtained research funding.
Corresponding author
Ethics declarations
Ethical approval
This study was conducted with the approval of the Institutional Biosafety Committee (IBSC) of the Regional Centre for Biotechnology in accordance with national biosafety guidelines. This study did not require either animal or human ethics approval.
Consent to participate
Not applicable for human subjects, as no human subjects were enrolled in this study. The corresponding author obtained consent to participate in this research from all authors.
Consent for publication
Not applicable for human subjects, as no human subjects were enrolled in this study. The corresponding author obtained consent to publish this research from all authors.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
ESM 2
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kumar, C., Mylavarapu, S.V.S. Nucleolin is required for multiple centrosome-associated functions in early vertebrate mitosis. Chromosoma 132, 305–315 (2023). https://doi.org/10.1007/s00412-023-00808-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-023-00808-4