Abstract
The interaction of partially hydrolyzed polyacrylamide (HPAM) with dodecyl-oxypropyl-β-hydroxyl trimethyl-ammonium bromide (C12NBr) and nonyl-phenyl-oxypropyl-β-hydroxyl trimethyl-ammonium bromide (C9phNBr) in the solution was investigated by the Dissipative Particle Dynamics (DPD) method. The calculated interaction parameters between HPAM and C12NBr or C9phNBr showed that C12NBr is most likely to form polymer/surfactant complex with HPAM in contrast to C9phNBr. The experiment of binding isotherm was used to validate the DPD results via surfactant-selective electrode and equilibrium dialysis method. In DPD method, the mean square end-to-end distance <r2> of polymer chain firstly increased, then reduced, and finally increased again. In addition, some polymer/surfactant complexes were also shown. One conclusion is that mesoscopic simulation can be considered as an adjunct to experiments and provide otherwise inaccessible (or not easily accessible) information in the experiment.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The interaction between ionic surfactants and water-soluble polymers has gained a growing interest in recent years because of the various industrial applications of such systems, such as colloid stabilization and destabilization, flocculation, and biotechnology [1, 2, 3, 4, 5]. It is remarkable that the fluid-like structures of amphiphiles under appropriate conditions can form different polymer-surfactant aggregates with polymeric coils, such as molecular bottlebrush or swollen cage conformations and a necklace of micelles on a polymer backbone [6]. These complexes are also subject to intensive theoretical and experimental investigations due to their ability to exhibit unusual behavior with variation of external conditions [7, 8].
In the last decade, different polymer/surfactant systems were selected to investigate the interaction between polymer and surfactant, such as polyethylene oxide and sodium dodecyl sulfate system [9], n-dodecyldimethylamine [10] or alkyltrimethylammonium bromide [11], and polymer systems. Several molecular simulation methods [12] have been used to investigate the microstructure of polymer/surfactant aggregates, but few mesoscopic simulation [13] was used to investigate the properties of polymer/surfactant system.
In this paper, two cationic surfactants and polymer are selected to investigate the properties of polymer and surfactant system using mesoscopic simulation, i.e., the Dissipative Particle Dynamics (DPD) method. Although these surfactants, dodecyloxypropyl β-hydroxyl trimethyl-ammonium bromide (C12NBr) and nonylphenyloxypropyl β-hydroxyl trimethyl-ammonium bromide (C9phNBr), have similar structures, many properties in the solution are different, such as the lowest surface tension and critical micelle concentration (cmc). Some different micro-information is expected to be found using DPD simulation. The selected polymer is partially hydrolyzed polyacrylamide (HPAM), which is often used in oilfield development [14], and has been utilized as mobility control agents for enhanced oil recovery [15]. The investigation of the interaction between surfactant and polymer is helpful to understand the difference of macro-properties of polymer/surfactant systems, and maybe the simulation result can guide the application of HPAM and surfactants in enhanced oil recovery of oilfields.
The DPD method is an effective mesoscopic one based on solving Newton's motion equation with the Verlet algorithm [13]. In this technique surfactant molecules and polymer are described by particles that act as centers of mass, and each particle represents a large number of atoms. Using different parameters representing the liquid compressibility and mutual solubility, surfactant and polymer molecules represented by a series of particles can be introduced into this model, and the properties of complicated systems can be predicted.
The paper is organized as follows. We briefly introduce DPD simulation in the experimental and computational section. In the results and discussion section, the interaction parameters between different molecules are firstly calculated, and then the aggregated properties are investigated via DPD simulation. Some aggregates of polymer and surfactant are shown in the simulated cells. Finally, a binding isotherm from the experiment is selected to validate the DPD simulated result.
Experimental and computational section
Computational details
Dissipative Particle Dynamics (DPD) is a stochastic simulation technique introduced by Hoogerbrugge and Koelman [16, 17] to simulate complex fluid dynamical phenomena. In DPD, the fluid is essentially comprised of particles that represent fluid packets. A modified velocity-Verlet algorithm [18] is performed to integrate the Newton's equations of motion. In the scheme, the values of next position, velocity and force on a soft particle are obtained. The position r i and momentum p i of particles can be obtained using the next equations:
where w is r-dependent weight function vanishing for the distance r between the particles and w(r)=(1−r) for r<1 and w(r)=0 for r>1, α is a maximum repulsion parameter, and the random force is governed by the σ parameter [18]. Three forces are conservative force, random force, and dissipative force in the square brackets of Eq. (3). The latter forces act as heat sink and source respectively, and their combined effect is a thermostat [19]. This particular thermostat is special in which it conserves (angular) momentum.
In the present simulations we have chosen the radius of interaction, the particle mass, and the temperature as Rc=m=kT=1. Consequently, the corresponding quantities in the DPD simulation (r, v, t, ρ) are given by
The liquid compressibility is first matched in DPD simulation, which determines the free energy change associated with density fluctuations; then the mutual solubility is chosen using the Flory-Huggins χ-parameter [18]. The repulsion parameter between water particles is recommended to be set at 25 kT for density ρ=3 to match the compressibility of liquid water at room temperature [13]
where a ii is the repulsion parameter between particles of the same type. Another larger repulsion between unlike beads stronger than that between beads of the same type is used to indicate the behavior in which different type of beads usually tend to segregate, such as water molecule and a monomer of polymer. The interaction parameter between different types of beads is linearly related with the χ-parameter. And it is easy to obtain the correct DPD parameters using the next equation [13, 20]:
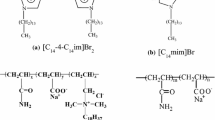
For the present application, we used a simpler possible model, i.e., C12NBr and C9phNBr molecules are showed by three beads (see Fig. 1), which are divided into one tail, one middle, and one head bead tied together by a harmonic spring; at the same time, the water molecule is shown by one bead, and HPAM polymer has 60 beads, which implies that the polymer is composed of 60 monomers. The simulations comprised of a total of 3000 beads containing surfactant, water, and polymer beads in a cubic cell of size 10×10×10 RC 3, where RC is a cut-off radius. The spring constant between different beads in the surfactant molecule is 4.0 according to [13]. The DPD steps are usually equal to 10,000 in order to obtain steady and balanceable results.
Binding isotherm
Equilibrium dialysis and surfactant-selective electrode are especially applicable for quantitative measurement of surfactant binding to polymers. Liu et al. [21, 22, 23] have used the experiment to analyze different surfactant-polymer systems via binding isotherms, and obtained some information about the interaction between polymer and surfactant. In the surfactant-polymer system, the degree of binding (β) can be calculated using the following expression:
where Cb is the concentration of bond surfactant, Cf the concentration of equilibrium surfactant, Cp the concentration of polymer residue, and Cs the total concentration of surfactant. Therefore, binding isotherms can be constructed by plotting the binding degree β vs free surfactant concentration (Cf). The concentrations of Cs and Cf can be obtained using the emf responses in surfactant solutions in the absence and presence of polymer via the equilibrium dialysis and surfactant-selective electrode method [24].
Results and discussion
The interaction parameters
The three-dimensional shape and size of surfactant molecules play a crucial role in their packing of the aggregation and indirectly determine the aggregation number and diameter of the micelle formed [25]. For the C12NBr and C9phNBr surfactant structure, it is difficult to distinguish a hydrophobic tail and a hydrophilic domain, because of complex interaction between the -CH2CHOHCH2- groups (referred to as the "elbow" region between the polar head and apolar tail) and water molecules at the origins of the tail. In our simulation, the elbow group is considered as a middle of the surfactant molecule. In addition, the -N+(CH3)3Br− group of C12NBr and C9phNBr surfactant is also selected as the hydrophilic group; thus, the C12NBr and C9phNBr surfactants have the same hydrophilic head and a middle group. In Fig. 1, the structures on the left of the first dashed line (line a) are selected as the hydrophobic group; accordingly, the structures on the right of the second dashed line (line b) is selected as the hydrophobic head and the group between two lines is considered as the middle group. In the simulation, we selected three acrylamide monomers and one acrylate monomer as a monomer of polyacrylamide. This implies that the polymer is partially hydrolyzed polyacrylamide with a 25% hydrolyzed degree.
When the surfactant molecule, the head, the middle, the tails, a monomer of polymer, and water molecule are considered as the simulated objects, the mixing free energies and interaction parameters, i.e., the Flory-Huggins χ-parameters, between two simulated objects can be obtained. The interaction free energies of mixing between different monomers E mix were calculated using the following equation:
where E ij is the interaction free energy, i.e., the free energy of the complex being composed of one molecules i and one molecule j, and Z ij is the coordinated number, i.e., the number of molecules j which can surround one molecule i in space. We must point out that the entropic value is small in contrast to the free energy; it means that the entropic term is neglected in the calculation. Thus the free energy can approximately be considered as the internal energy in the vacuum. So, in the following, we use the interaction energy represent the free energy. After the interaction energy of mixing between two polymers was calculated, the interaction parameter can be obtained via the following equation:
where Z* denotes the average coordination number and V seg the volume of one polymer segment [26]. It is essential to emphasize that different force fields can give different mixing energies. We find that only AMBER-calculated energy includes the electrostatic interaction energy and H-bonding energy, COMPASS, PCFF, or CVFF-calculated energies do not include H-bonding energy, and UFF-calculated energy does not include the electrostatic interaction energy and H-bonding energy. So AMBER force field is selected, and these mixing energies and interaction parameters can be given from Blend simulation [27].
If the interaction parameter between the water molecules is selected as the basis (i.e., equal to zero), the interaction parameters for HPAM/C12NBrand HPAM/C9phNBr systems can also be calculated using Eq. (9). Because the interaction parameters are calculated from the mixing energies, the physical meaning of the parameter is in agreement with that of the energy. Therefore, a plus value of parameters indicates that the repulsion interaction occurs between the two molecules (in contrast to that between two water molecules), while the minus means that the attraction interaction occurs. This interaction parameter implies the interaction intensity between polymer and surfactant. When the temperature is equal to 298 K, the calculated interaction parameters for HPAM/C12NBr and HPAM/C9phNBr system are −11.594 and 5.6179, respectively. By comparing the values of parameters, one conclusion is shown that the interaction between HPAM and C9phNBr is weaker than that between HPAM and C12NBr, and there should be different types of interactions for HPAM/C12NBr and HPAM/C9phNBr systems.
Other interaction parameters among the head, middle, tail, water, and a monomer of polymer at 298 K are listed in Table 1. From the parameters, the interaction information about HPAM/C12NBr or HPAM/C9phNBr system can be obtained. The molecular structures of C12NBr and C9phNBr only have different tails, so the different macro-properties should arise from the interaction between the tails and other molecules. As shown in Table 1, the interaction parameter between the tail and the monomer of polymer in the C12NBr system is smaller than the one in the C9phNBr system. It indicates that the C12NBr tail is easier than the C9phNBr tail in associating with the monomer of polymer. This result is in agreement with the conclusion above, i.e., the interaction between C12NBr and HPAM is stronger than that between C9phNBr and HPAM. Actually, we should point out that the calculated energies are different when different force fields are used. However, the values of the interaction parameters which are calculated from the energies and volume of molecule show a similar reasonable trend representing the interaction between different molecules. So, we emphasize that the parameters come from the AMBER force field.
Mean squared end-to-end distance in the surfactant/water/HPAM system
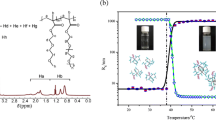
The Flory-Huggins parameters, χ ij, can be translated into the DPD parameters, a ij, using Eq. (6). When these interaction DPD parameters in Table 2 are used in DPD simulation, some properties of surfactant/water/polymer system can be obtained, such as the micelle shape, the aggregates of surfactant and polymer, the density distribution of water in the micelle, and so on. Figure 2 shows the change of the mean squared end-to-end distance <r2> with the increase of surfactant concentration. The similar curves are found in the HPAM/C12NBr and HPAM/C9phNBr systems. As shown in Fig. 2, both of the curves can be divided into four stages. As an example of C12NBr system, the first is the increasing stage of the <r2> until a maximum is obtained (from A to B); the second is the decreasing (from B to C); the third is the re-increasing from the minimum (from C to D); and the last is the steadily balance stage (from D to E).
Combining with the aggregated morphology of polymer and surfactant in DPD simulation, the following and the cartoon of the system in Fig. 3 can interpret the strange cures of <r2> in Fig. 2. In the absence of surfactant, a spherical cluster of polymer forms in aqueous solution (A in Fig. 3). When few surfactant molecules are added, they can be adsorbed around the backbone of polymer. This result makes the polymer clusters swell (B in Fig. 3), so the end-to-end distance increases (from A to B in Fig. 2). When the clusters swell to this extent, additional surfactant molecules can form some pre-micelles out of the backbone of polymer. Because the interaction between surfactant molecules is stronger than that between surfactant molecule and polymer monomer, those surfactant molecules around the polymer backbone may flee out and compress the clusters, thus the <r2> decreases rapidly (from B to C in Fig. 2). This is the second stage of curve. At the minimum of the curve(C point in Fig. 2), the surfactant concentration is considered as critical aggregation concentration (cac), at which a compressed and surrounded complex begins to form (C in Fig. 3). When surfactant molecules are continuously added to the solution, the repulsion interaction of surfactant tails on the compressed complex makes the complex re-swell (D in Fig. 3), so the end-to-end distance increases rapidly again. At the equation stage (from D to E in Fig. 2), a steady surfactant and polymer aggregate has already formed; at the same time the <r2> gets a steady value. Some experiments have proved that the polymer initially reduces in size for the poly(ethylene oxide) (PEO) and sodium dodecyl sulfate (SDS) system, and when the surfactant concentration increases beyond a certain point, the polymer swells [28, 29]. Other experiments have found similar behavior [30, 31]. Our simulation is not in agreement with these experimental results, i.e., the polymer first increases in size, then reduces, and finally increases again. The differently aggregated manner between different surfactant and polymer systems can be explained by the PEO being water soluble and the HPAM very hydrophobic. The solubility of polymer has an important role in the aggregation and makes the strong interaction between polymer and surfactant molecules occur.
As shown in Fig. 2, different minimum <r2> is shown at different concentration of surfactant for C12NBr or C9phNBr/polymer systems. A minimum value of <r2> in the C12NBr system is obtained at a lower concentration than that in the C9phNBr system. It indicates the effect of C12NBr surfactant on HPAM polymer is stronger than that of C9phNBr, which is in agreement with the meaning of the interaction parameter between HPAM and C12NBr above.
The aggregates of surfactant and polymer
From the discussion above, we know the DPD conclusion is in agreement with the experimental results. DPD can also give some aggregates using the three-dimensional cells. In the following, the C12NBr/polymer system will be selected as an example to show the aggregates of surfactant and polymer. Figure 4 shows the changed process of aggregates with the increase of the number of DPD steps. The selected system includes 2% HPAM, 8% C12NBr, and 90% water molecules, which has a smaller <r2> of HPAM (see Fig. 2). At 100 DPD steps (Fig. 4a), surfactant molecules first take on single-dispersed states, only a few molecules aggregate together due to the interaction between them, and polymer chain aggregates a cluster (the black cluster in Fig. 4a). At 200 steps (Fig. 4b), some surfactant molecules have been absorbed around the polymer cluster. It means that the interaction between HPAM and surfactant is stronger than that between surfactant and water molecules, and this interaction makes polymer and surfactant molecules aggregate together. With the increase of the number of DPD steps, surfactant molecules absorb around the polymer continuously. As shown in Fig. 4c (at 500 steps), surfactant molecules can also form pre-micelle in the solution in addition to aggregating around polymer cluster. Hoverer, a steady complex of surfactant and polymer finally forms (Fig. 4d, at 10,000 steps). These phenomena indicate that in the present of polymer, C12NBr molecules prefer to aggregating around the polymer coil, not in the solution when the concentration of C12NBr is around cac, and also prove that the interaction between surfactant and polymer is essential to form aggregates of surfactant and polymer. One conclusion is that the DPD method can give the dynamic process of aggregates in the surfactant/polymer system, and 10,000 DPD steps can give a steady and balanceable figure. So, the final simulation results are shown after 10,000 steps in the following.
The pictures of typical polymer conformations in the absence or presence of surfactant molecules are shown in Fig. 5. In the absence of surfactant, the polymer is coiled in aqueous solution, and forms a spherical cluster (Fig. 5a). When few surfactant molecules are added to the solution, the polymer is slightly swollen (Fig. 5b), although the concentration of surfactant is less than cac, and the molecules also aggregate around polymer coil. Apparently, surfactant molecules can make the polymer coil swell in solution because the polymer is less hydrophobic than without surfactant. On the other hand, the C12NBr molecules prefer to aggregate in water due to their spatial structure and hydrophobic properties before cac. However, with the increase of surfactant molecules, all C12NBr molecules can aggregate around the polymer, and the complex is like a droplet encapsulated by a monolayer of surfactant, confining the polymer, and the polymer may be considered as a part of polymer/surfactant complexes (Fig. 5c).
Figure 5c,d shows the aggregates of HPAM and C12NBr or C9phNBr system, respectively. Comparing Fig. 5c with Fig. 5d, a non-order aggregate of HPAM and C12NBr system and an order aggregate of HPAM and C9phNBr are found. Obviously, the interaction between the tails of surfactant and HPAM is the reason that the different aggregates are formed.
Comparing with binding isotherms of surfactants to the polymer.
In Fig. 6, a typical binding isotherm can be constructed by plotting the binding degree β vs free surfactant concentration (Cf) using surfactant-selective electrode method. Two important observations are noted for both C12NBr and C9phNBr cases: (1) the C12NBr-polymer system has two apparent transition points (T1 and T2) in the curve of binding degree. There, T1, also named cac where binding suddenly starts, is usually less than critical micelle concentration (cmc), corresponding the concentration that regular micelles start to form; and T2 is usually more than cmc, while the C9phNBr system only has a transition point T1 in the investigated concentration; (2) the cac in the C12NBr/polymer system is smaller than that in the C9phNBr system. It indicates that the binding of C12NBr is much stronger than that of C9phNBr with the same polymer. This conclusion is in agreement with the DPD results, i.e., C12NBr is more likely to bind with HPAM compared to C9phNBr.
This difference is due to the different molecular structures of the two surfactants. The C9phNBr molecule has a phenyl group in the surfactant tail, in which a conjugated π-bond is found. Thus, C9phNBr and C12NBr molecules have different electoral density in the head, the middle (-CH2CHOHCH2-) or the tail group. In addition, the polymer chain is soft, and the electoral densities in -COOH and -CONH2 groups are different. Although the total electric charge in the surfactant or polymer molecule is zero, the positive and negative electric charges would be located in different groups. Maybe C9phNBr has a smaller difference between positive and negative electric charges due to the phenyl group. At the same time, the C9phNBr molecule has a bigger surface area and volume than C12NBr. So, it is difficult for C9phNBr to contact polymer chain in the solution. These differences may result in the weak interaction between C9phNBr and HPAM.
Conclusions
The interactions between HPAM and different surfactants were performed via mesoscopic simulation method. The DPD simulation method consists of the partition of surfactant molecules containing a hydrophobic tail, a middle section, and a hydrophilic head, and the calculation of interaction parameters and the change of mean square end-to-end distance <r2>. From the simulation results and binding isotherms in the experiment, a better prediction about the interaction is obtained, i.e., C12NBr is more likely to interact with polymer compared to C9phNBr. From the simulation and experiment, one conclusion is that DPD simulation can be considered as an adjunct to experiments and provide otherwise inaccessible (or not easily accessible) information that experimentalists can use.
References
Hoff E, Nyström B, Lindman B (2001) Langmuir 17:28
De Gennes PG (1990) J Phys Chem 94:8407
Brackman JC, Engberts JBFN (1993) Chem Soc Rev 85:1
Kwak JCT (1998) Polymer-surfactant systems. Marcel Dekker, New York, vol 77
Farinato RS, Dubin PL (1999) Colloid-polymer interactions: from fundamentals to practice. Wiley, New York
Yuan SL, Xu GY, Cai ZT (2002) Acta Chim Sinica 60:585
Lee LT, Cabane B (1997) Macromolecules 30:6559
Baulin VA, Kramarenko EY, Khokhlow AR (2000) Comput Theor Polym Sci 10:165
Brackman JC (1991)Langmuir 7:469
Brackman JC, Engberts JBFN (1992) Langmuir 8:424
Hansson P, Almgren M (1996) J Phys Chem 100:9038
Wallin T, Linse P (1996) J Chem Phys 100:17873; Wallin T, Linse P (1997) J Chem Phys 101:5506; Wallin T, Linse P (1998) Langmuir14:2924
Groot RD (2000) Langmuir 16:7493
Birdi KS (1997) Handbook of surface and colloid chemistry. CRC Press
Li G, Zhai L, Xu G (2000) J Dispersion Sci Technol 21:367
Hoogerbrugge PJ, Koelman JMVA (1992) Europhys Lett 19:155
Koelman JMVA, Hoogerbrugge PJ (1993) Europhys Lett 21:363
Groot RD, Warren PB (1997) J Chem Phys 107:4423
Groot RD, Madden TJ, Tildesley DJ (1999) J Chem Phys 110:9739
Yuan SL, Cai ZT, Xu GY (2002) Acta Chim Sinica 60:241
Liu J, Taisawa N, Shirahama K, Abe H, Sakamoto K (1997) J Phys Chem B 101:7520
Liu J, Shirahama K, Miyajima T, Kwak JCT (1998) Colloid Polym Sci 276:40
Liu J, Takisawa N, Kodama H, Shirahama K (1998) Langmuir14:4489
Liu J (1998) PhD thesis. Saga University, Japan
Derecskei B, Dereskei-Kovacs A, Schelly ZA (1999) Langmuir15:1981
Yuan SL, Xu GY, Cai ZT, Jiang YS (2003) Colloid Polym Sci 281:66–72
Blanco M (1991) J Comput Chem 12:237
Claesson PM, Fielden ML, Dedinaite A, Brown W, Fundin J (1998) J Phys Chem B 102:1270
Mears SJ, Cosgrove T, Obey T, Thompson L, Howell I (1998) Langmuir 14:4997
Shimabayashi S, Uno T, Oouchi Y, Komatsu E (1997) Prog Colloid Polym Sci 106:136
Chari K, Antalek B, Lin MY, Sinha SK (1994) J Chem Phys 100:5294
Acknowledgements
The authors thank Prof. Shirahama K. (Saga University, Japan) for affording the surfactant-selective electrode and polymer film. This work was supported by the Natural Scientific Foundation of Shandong Province (No. Y2001B08) and the Scientific Foundation of Engaged-professor in Shandong University.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yuan, SL., Cai, ZT., Xu, GY. et al. Mesoscopic simulation study on the interaction between polymer and C12NBr or C9phNBr in aqueous solution. Colloid Polym Sci 281, 1069–1075 (2003). https://doi.org/10.1007/s00396-003-0881-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00396-003-0881-6