Abstract
This study aims to investigate the effects of soil as a microbial source on the assemblage of the endophytic bacterial communities in rice roots. Rice seedlings were grown hydroponically with the addition of a permeable bag filled with one of five soil types collected from different geographical locations in Japan. After 3 and 6 weeks, the endophytic bacterial communities in rice roots were analyzed using the Illumina Miseq-based 16SrRNA gene amplicon sequencing method. The results showed that the bacterial community in the soils added as a microbial source differed among the soil types, which affected the bacterial community in the hydroponic solution and consequently reflected in the endophytic bacterial community assemblage. Bacterial diversity and richness differed significantly with respect to the microbial sources. As a result, a conserved group of 16 endophytic bacterial taxa at the genus level, dominated by Burkholderia-Caballeronia-Paraburkholderia, and independent of the soil type were shared across all microbial sources, thereby underlining the ability of rice plants to selectively recruit their endophytic inhabitants. Altogether, this study demonstrates the importance of the microbial source as a crucial driving force for the formation of the endophytic bacterial communities in rice roots.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Soil is regarded as a major environmental source of plant-associated bacteria, because it provides a tremendously diverse ecosystem for a multitude of microorganisms (Mahaffee and Kloepper 1997; Rasche et al. 2006; van Overbeek and van Elsas 2008; Long et al. 2010; Lundberg et al. 2012). According to Dwevedi et al. (2017) and Vieira et al. (2020), soil ecosystems are spatially structured by a combination of physicochemical characteristics such as texture, salinity, acidity, waterlogging, compaction, pore network, and air, water, and carbon contents. Consequently, heterogeneous microbial habitats formed by the collective effects of these characteristics result in diverse soil bacterial communities (Bach et al. 2018). In other words, soils with different properties and/or conditions are likely to differ in their microbial community assemblies.
For instance, Dwevedi et al. (2017) revealed that soil microorganisms are physiologically active in moist soil conditions, but remain dormant in dry soil conditions, thus suggesting that soils in the wet state can harbor greater microbial activities than in the dry state. In addition, soils with higher microbial diversities exhibit the potential for greater microbial activities such as humus formation, nutrient cycling, aggregate formation, and stabilization (Dwevedi et al. 2017; He et al. 2019). Furthermore, Dwevedi et al. (2017) reported that the abundance of minerals, organic matter, and microbial decomposers are indicative of soil health, while Megías and Müller (2010) revealed that decomposers like bacteria and fungi are some of the key drivers of soil suitability for plant growth through nutrient cycling and humus formation. Holistically speaking, microorganisms are a very important component of soils and are thus referred to as the “biological engine of the earth” (Haygarth and Ritz 2009).
Many research investigations have revealed multifaceted interactions between terrestrial plants, the soil, and the soil microbiome, and these complex interactions still form an important aspect of research today. For instance, as plants grow, they exert bio-physicochemical effects around their roots, thereby influencing the spatial structure of the soil. As a result of the interaction, microbial communities are formed in their rhizosphere (Breidenbach et al. 2016). Additionally, several other environmental factors also influence the soil microbial communities and alter the plant-associated microbial communities. In particular, the community formation of plant-associated microorganisms such as bacterial endophytes can be driven by several biotic and abiotic factors, which are instrumental in shaping their diversities and community compositions (Walitang et al. 2018).
Previous studies have confirmed that the characteristics exhibited by a host plant such as plant growth stage (Vendan et al. 2010; Hardoim et al. 2012), tissue (Dai et al. 2014), health (Bogas et al. 2015), nutrient status (Hameed et al. 2015), species (Ding and Melcher 2016), and genotype (Elbeltagy et al. 2000; Walitang et al. 2018; Xu et al. 2020) are all contributing factors that are influential in driving the endophytic communities.
According to Edwards et al. (2019) and Mano and Morisaki (2008) endophytic bacterial communities of rice are largely soil-derived. In other studies, soil type has proven to be key in driving the endophytic bacterial communities in rice. For instance, using PCR-DGGE, Hardoim et al. (2011) investigated the effects of plant genotype, soil type, and nutrient use efficiency on root-associated bacterial communities of 10 rice cultivars, and thus observed alterations of the alphaproteobacteria, betaproteobacteria, and actinobacteria communities. Elsewhere, Xu et al. (2020) examined the root-associated bacterial communities of three rice cultivars cultivated in three typical paddy soils with different properties and observed a greater effect of soil type than rice cultivar on the root-associated bacterial community assembly. Soil bacteria have also been proven to show a preference for specific particle fractions in the soil, according to a study by Hemkemeyer et al. (2018), who concluded that soil particles of different sizes create distinct microenvironments that are inhabited by specific bacterial taxa, thereby influencing the spatial heterogeneity and bacterial diversity that characterize soils. Therefore, soil particle size contributes to the effects of soil physical factors on the overall soil bacterial community, which also plays a role in the complex host-microbe interactions by influencing which part of the community becomes endophytic. In addition, the interaction between plants and the soil they are grown on is an important factor, as shown by Samuel et al. (2022), who reported that the soil-root interface is influential in endophytic bacterial colonization. Lin et al. (2020) further suggested that soil interference caused as a result of mechanical disruption through management practices can affect the soil properties and influence the soil microbiome, and these effects are likely to be reflected in the endophytic community. Furthermore, Lin et al. (2020) identified soil pH, while Dang et al. (2020) found ammonium N and nitrate N as important factors that significantly affected the endophytic bacterial community. Altogether, prior research findings indicate that plant-soil interactions are complex and can impact the colonization of bacteria within plants via their influence on soil properties and plant growth, which is often linked to nutrient availability. Although previous studies have made significant progress in understanding the factors that shape endophytic bacterial communities in plants, they have mainly focused on the role of soil, without explicitly accounting for the diversity of microbial sources that are present in soil. This is due to the challenges of controlling for other factors in the soil that can equally affect endophytic bacterial colonization.
Endophytic bacteria are essential for plant growth and development and have the potential to transform agriculture into a more sustainable and eco-friendly practice. However, to better understand the direct interactions between rice plants and microbes, it is necessary to distinguish the impacts of the microbial and non-microbial soil factors. Restricting soil factors other than the microbial source is vital for achieving this goal, therefore, the use of a modified hydroponic system was employed in this study to investigate the effects of various microbial sources on the formation of endophytic bacterial communities in rice roots while minimizing the influence of soil physicochemical factors. The study hypothesized that different microbial sources would result in distinct endophytic bacterial communities. The experiment involved growing rice plants hydroponically using one of five microbial sources, including four paddy soils and one forest soil. The root-associated endophytic bacterial diversity and community compositions were analyzed and compared using 16S rRNA gene amplicon sequencing.
Materials and methods
Soil sampling and preparation
Five soil types representing different microbial sources were collected from different locations in Japan. Their physicochemical properties are summarized in Table 1. Briefly, of the five soil types used to prepare the microbial sources in this study, four (ACH, NAG, SHB, and SHD) were sampled from typical paddy fields, while the fifth soil type (SD) was from a forest area as representative of soils unrelated to paddy fields. After sampling, soil samples were air-dried for 1 week, passed through a 2 mm mesh sieve, and stored at 4 °C until the setting up of the experiment. The exchangeable cation (Ca2+, Mg2+, K+, and Na+) contents were extracted using 1 M ammonium acetate solution (pH7), and their respective concentrations were analyzed using an atomic absorption spectrometer (Hitachi Polarized Zeeman Atomic Absorption Spectrometer ZA3000, Tokyo). Soil pH(H2O) was also determined in 1:2.5 (weight/volume) ratio using a pH meter (HORIBA LAQUA F-71, Kyoto, Japan).
Pre-incubation study and experimental set-up
To prepare microbial sources, 40 g of each of the 2-mm sieved soil was weighed into a polystyrene woven bag and then submerged in a bath containing sterile water. The soil submergence was performed independently for each soil type in different baths with sterile water to avoid the risk of cross-contamination. The pre-incubation setup was kept in the dark and at room temperature for 21 days to allow for the adaptation, stabilization, and proliferation of the microbial communities in each soil under submerged anaerobic conditions, which is expected to mirror the conditions of a flooded paddy field.
Rice seeds (Oryza sativa L. Nipponbare) were de-husked, surface-sterilized as described by Asiloglu et al. (2020), germinated under sterile conditions, and allowed to grow for 14 days. Figure 1 illustrates the setup of this study. The experimental pots used in this study were prepared as previously described (Samuel et al. 2022). Briefly, each pot (13.0 cm height and 8.5 cm inner diameter) was prepared with the following features: a black polystyrene lid with a 2.5-cm-diameter opening at the center to allow the outgrowth of a rice plant; and a 50-ml column (11.0-cm height and 2.5-cm inner diameter) with four perforations (2-mm diameter, each) which was lined internally with a piece of polypropylene mesh sheet. The column was vertically attached to the inner base of the pot and lined with mesh sheets to allow water and bacteria to permeate into and out of the inner column while restricting rice roots from extending out of the column. One soil bag per experimental pot was gently transferred from each soil pre-incubated for 21 days using sterile forceps. Thereafter, 14-day-old sterile rice seedlings were transplanted into each pot, and 500 ml of sterile water (hereinafter referred to as hydroponic solution) was gently added and maintained. Since nutrient levels in the soil have been reported to have an impact on the endophytic bacterial communities in plants (Zhang et al. 2019; Ali et al. 2021), there was no nutrient supplementation in the experimental setup of this study in order to limit known soil factors that influence the endophytic bacterial community as much as possible, thereby making it possible to examine the effects of the microbial source. Rice was grown for 6 weeks in a growth chamber with the following conditions: 25/30 °C (day/night), 16 h (250 µmol m−2 s−1) of day length, and relative humidity of 70%. Furthermore, all plants were kept in the same growth chamber, where their positions were carefully randomized twice every week to ensure the evenness of lighting and relative humidity conditions until the last sampling. The experiment was conducted in triplicates (n = 3), considering 2 sampling times (3 and 6 weeks).
In addition, we carried out an ion flux assessment of each soil type used as a microbial source in this study by setting up a similar experiment simultaneously without rice plants (Fig. S1). The hydroponic solution for each microbial source was collected just after the pre-incubation and at 3 and 6 weeks. Eluted Cl−, NO3−, and SO42− were analyzed using an ion chromatograph (DKK-TOA, IA-300, Tokyo, Japan) as described by Kozaki et al. (2021), while PO43− was determined through a colorimetric method following the description in Truog (1930).
Sample collection and preparation for molecular analysis
Sampling of the rice roots, hydroponic solution, and soil in the bag was performed after 3 and 6 weeks. At each time, destructive sampling was carried out as described in Samuel et al. (2022). Briefly, rice roots were separated from the shoots using a pair of sterile scissors and washed thoroughly in running tap water. Surface sterilization of the roots was then performed by washing for 5 min in sterile water, followed by a 2-min wash in 70% EtOH, a 5-min wash in 2.5% NaOCl, another 1-min wash in 70% EtOH, and a final 5-min wash in sterile water performed twice. The whole root system of each plant was finally homogenized in liquid nitrogen. Finally, shoot dry weight was recorded for each sampled plant after oven-drying at 60 °C for 3 days.
The hydroponic solution sampling and the subsequent collection of bacterial pellets were performed according to Samuel et al. (2022). Briefly, aliquots of the hydroponic solution were centrifuged at 15,000 g for 10 min at room temperature until enough bacterial pellets were collected for DNA extraction. The soil bag content was sampled after thorough mixing and put into sterile tubes until further analysis. All samples were kept under cold storage (− 21 °C) until DNA extraction.
DNA extraction, library preparation, and sequencing
Genomic DNA from the homogenized root samples was extracted using an ISOPLANT kit (Nippon Gene, Tokyo, Japan), while an ISOIL for Beads Beating kit (Nippon Gene) was used for DNA extraction from the bacterial pellets and soil samples as previously described in Samuel et al. (2022). The DNA extracts of all samples were then used as the template for polymerase chain reaction (PCR) using the universal primer pair 515F/806R (Caporaso et al. 2011) with overhang adapter sequences for the Nextera XT index primers (Illumina, San Diego, CA). For the amplification of the V4 region of the 16S rRNA gene, PCR was performed with a final reaction volume of 25 µL. Briefly, the PCR reaction mixture consisted of 1 µL of template DNA, 0.1 µL of both forward and reverse primers (50 µM, 10P), 2.5 µL 10 × ExTaq buffer, 2.0 µL dNTPs, 0.125 µL Ex Taq (Takara Bio, Kusatsu, Japan). PCR conditions for the amplification of endophytic bacterial genes were set as follows: initial 2-min denaturation at 94 °C was followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 57 °C for 30 s and elongation at 72 °C for 20 s, and a final extension at 72 °C for 5 min. Amplification of bacterial genes from the hydroponic solution and the microbial sources was performed by an initial 2-min denaturation step at 94 °C, followed by 25 cycles of denaturation at 94 °C for 30 s, annealing at 57 °C for 30 s and elongation at 72 °C for 20 s, then a final 5-min elongation step at 72 °C. Agencourt AMPure XP (Beckman Coulter, Brea, CA) was used for the purification of amplicons according to the product’s manual. Index PCR was performed under the following thermocycling conditions: initial 3-min denaturation at 95 °C; 10 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s; and a final 5-min elongation step at 72 °C. This step was accompanied by a second purification step of the amplicons as previously described. Amplicon quantification was performed using the QuantiFluor device (Promega, Madison, WI), followed by a final pooling of equimolar concentrations of the purified amplicons. Finally, the pooled amplicons were pair-end-sequenced on the Illumina MiSeq platform at a read length of 2 × 300 bp using the MiSeq reagent kit v3.
Data processing and statistical analysis
All the procedures for processing the raw FASTQ data obtained after sequencing were performed with the exclusion of sequences identified as Archaea, Eukaryota, chloroplast, and mitochondria, and rarefaction for 10,000 random sequences per sample based on a 16S rRNA sequence length of 300 bp and quality score (Q) > 30 as detailed in Samuel et al. (2022) on the QIIME II pipeline (version 2020.8; http:// qiime2. org/) (Bolyen et al. 2019). Amplicon sequence variants (ASVs) were produced by denoising the paired-end sequences using the Divisive Amplicon Denoising Algorithm (DADA2) plugin in QIIME2 (Callahan et al. 2016). The forward and reverse sequence reads were truncated at 240 and 150 bp, while the singletons and doubletons were removed after joining the paired-end reads. Thereafter, the biological replicates were grouped using the “mean-ceiling” option of the “feature-table group” command. The SILVA SSU Ref NR 138 reference sequences were used as the benchmark for taxonomy assignment using the q2-feature-classifier plugin of QIIME2. Sequences were rarefied for 10,000 random sequences per sample. Finally, the 16S rRNA gene amplicon sequences for this study were deposited in the DNA Data Bank of Japan (DDBJ) with the accession number: DRA015220.
Plant growth performance was assessed by recording the shoot dry weight of each plant after oven-drying the fresh shoots at 60 °C until a constant weight was reached. The differences in growth performance between each treatment were analyzed by the One-way Analysis of Variance (ANOVA) method, while pairwise comparisons of their respective means were performed through a Tukey’s HSD test method (p < 0.05). Bacterial alpha diversity (Shannon, Faith phylogenetic diversity, and Pielou’s evenness) and richness (observed ASVs) were analyzed by the Kruskal–Wallis test method to better describe the bacterial communities in the soils used as microbial sources, hydroponic solutions, and rice roots, while the multiple comparisons (Dunn test with Benjamini–Hochberg correction, p < 0.05) was applied to detect the differences after the pairwise comparisons of their means. The principal coordinate analysis (PCoA) based on weighted UniFrac distance was performed using the R vegan package of the R program (version 4.0.3; https:// www.r- project. org/) (Core R Team 2019) to reveal the beta diversity of the bacterial communities in the rice roots, hydroponic solution, and the five microbial sources. Furthermore, permutational multivariate analysis of variance (PERMANOVA; permutations = 9999) was performed using the adonis function to provide a statistical description (p < 0.05) of the effects of the microbial source on the endophytic bacterial community composition of rice plants. The shared and unique bacterial endophytic bacterial taxa after 6 weeks under the influence of the different microbial sources were identified through the construction of a Venn diagram using the R VennDiagram package. Finally, one-way ANOVA test was used to examine the differences in the mean ion-flux concentrations from the five soils used as the microbial sources, after which the means were subjected to Tukey’s HSD test (p < 0.05).
Results
Growth performance of rice plants
The growth of rice plants as revealed by dry weight biomass after 6 weeks is shown in Fig. 2. Rice plants grown on soils SHB and SHD as the microbial sources recorded the highest growth among the treatment. Although SHB performed better in terms of growth compared to SHD, no statistical difference was found between them (p > 0.05). The rice plants grown on SD recorded the poorest growth among all the microbial source treatments, followed by NAG. A significant difference (p < 0.05) was observed in comparing NAG and SD with the others. These differences in dry weight biomass due to the different soils used as microbial sources were visible in appearance (Fig. S2).
Rice shoot biomass (dry weight) after 6 weeks. Standard error bars with different letters indicate statistically significant differences from each other (Tukey’s test, p < 0.05). ACH (Aichi), NAG (Nagano), SD (Forest), SHB (Shibata), and SHD (Shindori) all represent the soils used as the microbial sources in this study
Bacterial community compositions
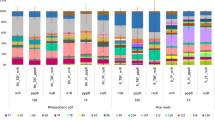
Figure 3A, which displays the relative abundances of bacterial community compositions in the soils supplied as microbial sources at the phylum level, shows that each microbial source was characterized by distinctive bacterial communities after 6 weeks, even though most of the dominant members of the bacterial phyla were present in all five microbial sources. At the initial (0 weeks, just after pre-incubation), Firmicutes was the most dominant soil bacterial phylum in all the soil types with the exception of SHD, which Chloroflexi dominated with 21.2% of the bacterial relative abundance. The dominance of Firmicutes at 0 weeks (63.4%) was reflected in the microbial sources after 3 (69.2%) and 6 weeks (60.9%), respectively in SHB. Despite the dominance of Firmicutes in ACH at 0 weeks (46.8%), Chloroflexi was enriched after 3 (25.1%) and 6 weeks (22.8%), respectively, thereby making it the most dominant bacterial group. Out of all the microbial sources, the lowest relative abundance of Chloroflexi (after 0, 3, and 6 weeks, respectively) was observed in SHB. Similarly, Acidobacteria, Actinobacterriota, Verrucomicrobiota, and Myxococcota were dominant members in ACH, NAG, and SHD, but with the least dominance in SHB. Furthermore, the lowest relative abundance of Proteobacteria in the soil types at 0 weeks was observed in ACH (3%), while the highest was observed in SD (22%). In ACH, NAG, and SHD, the proportion of Proteobacteria was decreased after 3 weeks, while in SD and SHB, the reverse was the case, with the enrichment of Proteobacteria after 6 weeks (Fig. 3A).
Changes in the relative abundance of bacterial communities at the phylum level in the soils used as microbial sources (A), hydroponic solution (B), and root endosphere (C). ACH (Aichi), NAG (Nagano), SD (Forest), SHB (Shibata), and SHD (Shindori) all represent the soils used as the microbial sources in this study
The microbial source effects observed in the soil types also manifested in the hydroponic solution as shown in Fig. 3B. The dominant members of the bacterial communities in the hydroponic solution differed in proportion among the soil types. As observed in the soil, Firmicutes was the most dominant bacterial phyla in the hydroponic solution, except for NAG (after 3 weeks), where Proteobacteria (78.3%) dominated. In ACH, NAG, and SHD, Firmicutes was enriched; however, it depleted in SD and SHB after 6 weeks. Interestingly, after 3 and 6 weeks, respectively, Firmicutes was the most abundant bacterial group in SD and SHB. Furthermore, Proteobacteria increased in relative abundance in ACH, SD, SHB, and SHD, but showed a marked decrease in NAG after 6 weeks, while Acidobacteriota and Actinobacteriota decreased in ACH and SHD, but increased in NAG, SD, and SHB after 6 weeks. Generally, bacterial groups showed differential relative abundances according to their microbial sources.
Figure 3C shows that the root endosphere compartment was predominantly occupied by Proteobacteria, which accounted for about 78.9% of total bacterial relative abundance. Although phylum Proteobacteria was the most dominant endophytic bacterial group, a notable trend of dominance at both sampling times was observed to be consistent for ACH, NAG, SD, and SHD, where they were enriched after 3 weeks and depleted after 6 weeks. The only exception to that trend was SHB in which Proteobacteria was enriched after 6 weeks. Other endophytic bacterial phyla that exhibited dominance to a lesser extent include Bacteroidota and Planctomycetota as shown in Fig. 3C. Phylum Bacteroidota was enriched in ACH, SD, and SHD, but depleted in NAG and SHB after 6 weeks, while Planctomycetota was enriched in all the rice roots after 6 weeks. At the genus level (Fig. S3), the endophytic bacterial communities showed different trends according to the microbial sources and the sampling time. For instance, Burkholderia-Caballeronia-Paraburkholderia (22.6%) was the most dominant endophytic bacterial taxa across all the microbial sources. Other less dominant endophytic bacterial genera that were present across all microbial sources, albeit in different relative abundances include Xanthomonas, Acidibacter, Variovorax, Herbaspirillum, Pantoea, Puia, and Ralstonia
Microbial diversity and richness
The alpha diversity indices for the endophytic bacterial communities are summarized in Table 2, while the results for microbial sources and hydroponic solution are provided in Table S1. NAG displayed the highest values for all alpha diversity indices in the endophytic bacterial endosphere compartment. Nevertheless, no notable differences were detected for the Shannon index and richness. Only SD demonstrated significant differences from NAG in terms of evenness (p < 0.05). Regarding Faith's phylogenetic diversity, NAG displayed significant differences only when compared to ACH and SHD (p < 0.05). Furthermore, Table 3 revealed that each of the sampled compartments exhibited varied levels of microbial diversity and richness with respect to the microbial source and sampling time. Kruskal–Wallis test results for the endophytic bacterial communities also revealed that the alpha diversity indices exhibited statistical differences with respect to their microbial sources (p < 0.05) (Table 3).
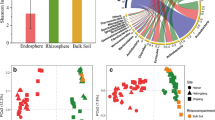
The results of the PERMANOVA analysis of the bacterial communities in all the sampled compartments based on the weighted UniFrac distance showed that the beta diversity was significantly influenced by the microbial sources. Furthermore, principal coordinate analysis (PCoA) performed based on the weighted UniFrac distances provided a visual representation of the bacterial beta diversity in the microbial source and the hydroponic solution, and the resultant effect on the assemblage of endophytic bacterial communities after 3 and 6 weeks (Fig. 4 and S4). A clear differentiation of bacterial communities was formed by the microbial sources affecting the bacterial communities in the hydroponic solution, with the communities clustering according to their respective microbial source treatments (Fig. S4) after 3 and 6 weeks respectively. A similar trend was observed with the assemblage of the endophyte communities as shown in Fig. 4 A and B, where PC1 and PC2 accounted for a combined 62.7% and 78.7% of the endophytic bacterial community dissimilarities after 3 and 6 weeks, respectively (Fig. 4 A and B).
Principal coordinate analysis (PCoA) based on the Weighted UniFrac distances showing the effects of the microbial sources on the endophytic bacterial communities formed after 3 weeks (A) and 6 weeks (B). Different colors signify the endophytic bacteria communities assembled from the different microbial sources. ACH (Aichi), NAG (Nagano), SD (Forest), SHB (Shibata), and SHD (Shindori) all represent the soils used as the microbial sources in this study
Shared and unique endophytic bacterial taxa
Venn diagram analysis at the genus level revealed the total number of bacterial genera (> 0.1%) detected in the rice roots differed after 6 weeks with 57, 128, 73, 64, and 73 bacterial genera detected in ACH, NAG, SD, SHB, and SHD, respectively (Fig. 5A). After further grouping into shared and unique groups, a total of 16 endophytic bacterial taxa were found in the roots of all the rice plants. Specifically, these shared endophytes differentially accounted for 79.2, 47.0, 78.2, 73.1, and 76.9% of the total relative abundance of endophytic bacteria in ACH, NAG, SD, SHB, and SHD, respectively. These common endophytic bacterial taxa varied in their relative abundances between the microbial source treatments, with Burkholderia-Caballeronia-Paraburkholderia being the most dominant endophytic bacterial genus in all the treatments (Fig. 5B). The number of endophytic bacterial taxa unique to each soil was highest in NAG with 45 (27.7%) taxa, while SHB had the fewest with 2 (0.6%) taxa (Fig. 5A and Table S2).
Venn diagram showing shared and unique endophytic bacterial taxa (A), and the relative abundance of the shared endophytic bacterial genera (B). Each colored oval of the Venn diagram represents the root compartment influenced by each microbial source. Values within the intersections depict shared endophytic bacterial genera, while the values outside the intersections represent unique genera. ACH (Aichi), NAG (Nagano), SD (Forest), SHB (Shibata), and SHD (Shindori) all represent the soils used as the microbial sources in this study
Ion flux assessment
The amounts of ions eluted from the soils used as microbial sources are shown in Table S3. The results showed that Cl−, NO3−, and PO43− released from the soils during the pre-incubation period were significantly higher than those released up to 6 weeks thereafter (Tukey’s test, p < 0.05). Conversely, gradual elution of SO42− was shown over time except for SD (Tukey’s test, p < 0.05).
Discussion
To confirm the hypothesis that different microbial sources generate different plant bacterial endophyte communities, rice plants were grown hydroponically using five soil types as microbial sources, and their effects on the bacterial endophyte communities were comparatively analyzed.
Microbial sources influenced bacterial endophyte assemblage
Previous investigations have revealed that the soil is the main source of plant-associated bacteria (Mano and Morisaki 2008). For instance, Zarraonaindia et al. (2015) for grapevine and Edwards et al. (2019) for rice highlighted that a complex community of microorganisms that inhabit plant roots were soil-derived. Therefore, in this study, five soils that were collected from different areas were used as bacterial sources for rice plants, and how the difference in bacterial sources manifested in the assembly of bacterial endophytes associated with the roots was investigated.
Each soil differed with regard to its soil type and chemical properties. These differences in the biogeographic locations, soil taxonomy, chemical properties, as well as land uses were possible factors that contributed to their distinct microbial community characteristics. Previous studies have given evidence of spatial variation of soil microbial distribution, which depends on environmental factors like soil properties (Griffiths et al. 2011; Prévost-Bouré et al. 2014; Kivlin and Hawkes 2016; Terrat et al. 2017). Soil properties such as N, P, and K, as well as organic matter contents, have been reported to have profound effects on the microbial communities in several paddy soils under different fertilization regimes (Luo et al. 2016). Furthermore, other factors including soil pH, moisture, temperature, C and N contents, precipitation, and vegetation can shape the soil microbial communities (Kuramae et al. 2012; Shen et al. 2013; Yuan et al. 2014; Rui et al. 2015; Lin et al. 2020).
The microbial sources were mainly dominated by Firmicutes, Chloroflexi, Proteobacteria, and Acidobacteria after pre-incubation, an outcome that was still found after each sampling period of the study. Interestingly, Firmicutes was the most dominant bacterial phylum in the microbial sources, underlining their ability to inhabit a wide range of soil environments. For instance, Bai et al. (2017) examined the microbial communities in three paddy soils and found Firmicutes as one of the dominant bacterial phyla, while Li et al. (2014) pointed out the ability of Firmicutes to survive even under extreme conditions through their sporulation activities. Jones et al. (2022) reported that Actinobacteria, Chloroflexi, Proteobacteria, and Firmicutes were persistent in dry conditions. Therefore, another possible reason for the Firmicutes-domination could be the air-drying of the used soils after sampling and prior to preincubation, which may have aided the predominance of the bacterial phylum in the microbial sources.
The distinctiveness of bacterial communities characterizing the soils used as microbial sources was well reflected in the hydroponic solution and the root endosphere of rice plants. For instance, phyla Firmicutes and Proteobacteria evidently dominated the hydroponic solution, albeit with different dominance levels. Different soil types harbored different microbial communities, probably due to different nutrient levels (Yu et al. 2021), resulting in differences in the bacterial community in the hydroponic solution as the source of the endophytes. Interestingly, even though the bacterial community composition at the phylum level observed in the hydroponic solution and their source showed significant proportional differences among the treatments, the dominant members of the endophytic bacterial community detected were similar among the rice plants from the different microbial source treatments. This may be connected to the ability of rice plants to selectively recruit their endophytic bacterial inhabitants, in this case, the dominant members detected from their rhizospheric surroundings, as reported by Hardoim et al. (2008) and Rodríguez et al. (2020). Another factor that may have contributed to the distinctiveness of the endophytic bacterial communities could be the root exudate effect. Although we speculated previously that the root exudation effect might be weakened because of the dilution in hydroponic solutions (Samuel et al. 2022), it may still play a role in the endophytic colonization of the rice roots. Therefore, considering the difference in the physicochemical properties of the soil types used as microbial sources in this study, it is likely that root exudation from the rice plants, an important colonization mechanism, influenced the endophytic community formation consequently as suggested by Ma et al. (2022).
At the genus level, the most dominant endophytic bacterial taxon detected in all the rice plants was Burkholderia-Caballeronia-Paraburkholderia, a group in the phylum Proteobacteria. They are considered to consist mostly of the plant-beneficial Burkholderia species with symbiotic and plant growth-promoting potential (Mannaa et al. 2019). Other dominant genera, Sphingomonas, Pantoea, Xanthomonas, Bradyrhizobium, and Pseudomonas, were detected as the rice-root endophytes, which include potential plant-growth-promoting species for rice (Ferrando and Scavino 2015), as well as Luteibacter known to promote root development in barley (Guglielmetti et al. 2013).
Effects of microbial source on microbial diversity and richness
The soils used as microbial sources and the hydroponic solutions exhibited different levels of bacterial diversity and richness. Particularly, the results of the chemical characterization of the soils prior to experimentation revealed higher organic matter, TC, and TN content, which may have contributed to the highest bacterial diversity recorded in the NAG microbial source. The high organic matter content of the soil may have been exploited for the C content which could be used as an energy source for the bacterial community in the NAG microbial source, hence the difference in their diversity when compared to the other microbial sources. Microbial diversity in paddy fields increases under organic matter fertilization (Kumar et al. 2017; Cui et al. 2018; Wang et al. 2022).
Table 3 revealed that the microbial sources significantly affected the bacterial community assemblages in all the sampled compartments, and this effect was observed at both sampling times. Specifically, the effect of the microbial sources on the diversity and richness of the endophytic bacterial communities in rice roots was observed after 3 and 6 weeks, respectively. These findings correspond to Nannipieri et al. (2019) who suggested that a soil microbial community shifts in response to time.
To further check the microbial source effect, principal coordinate analysis (PCoA) was performed for the endophytic bacterial communities associated with the root compartment after 3 and 6 weeks. The outcome showed dissimilar clusters of microbial communities according to their respective microbial sources, with PC1 and PC2 collectively explaining 62.7% (after 3 weeks) and 78.7% (after 6 weeks) of the bacterial community dissimilarities. Put together, the result of this study suggests that microbial sources have a significant impact on the diversity and community composition of endophytic bacteria in rice plants.
Identification of distinct endophytes independent of microbial source
A Venn diagram of shared and unique bacterial taxa at the genus level revealed that 16 bacterial genera were shared by the rice plants in all treatments with different relative abundances. Burkholderia-Caballeronia-Paraburkholderia was the most dominant endophytic bacterial genus after 6 weeks regardless of the microbial sources. Except for Variovorax, 15 of the bacterial genera shared in this study were also detected in our previous study (Samuel et al. 2022). Furthermore, previous studies have identified a distinct assemblage of plant-associated bacteria like Pantoea, Pseudomonas, and Xanthomonas (Sahu et al. 2022; Zhang et al. 2022), Bradyrhizobium, Halingium, and Sphingomonas (Li et al. 2021), Burkholderia and Herbaspirillum (Mano and Morisaki 2008), and Massilia and Ralstonia (Kataoka et al. 2018), which were all detected in this study independent of the microbial source, as members of the core endophytic bacterial communities in rice. Studies on endophytes from rice seeds have detected Pantoea, Pseudomonas, Bacillus, Sphingomonas, Xanthomonas, Metylobacterium, Stenotrophomonas, and Ochrobacterium (Kaga et al. 2009; Hardoim et al. 2012; Zhang et al. 2022), among which Zhang et al. (2022) pointed out that Pantoea and Xanthomonas are potentially vertically transmitted taxa. Pantoea and Xanthomonas were also detected as the dominant members of the endophytes in this study and were likely seed-borne species. Whether they were really seed-borne needs to be clarified in the future, but more importantly, this study showed that their relative abundance was greatly affected by the application of different soil microbial sources. This phenomenon is also indicated by Hardoim et al. (2012), while further research is needed to elucidate the mechanism.
A comparison of the number of rice endophytic bacterial taxa among the five microbial sources resulted in the highest number of 45 in NAG treatment, followed by 9 in SD treatment. Interestingly, the poorest rice growth was recorded in NAG and SD treatment. It remains unclear whether the poor rice growth was due to the high number of unique endophytic bacteria, especially in NAG treatment, or simply to low nutrient ion elution (e.g. NO3− and/or PO43−) from the soil (Table S3), which need to be elucidated in future.
Conclusion
Understanding the factors driving microbial community formation has been a topic of interest for a long time. These factors may directly or indirectly influence the formation of plant endophytic communities since their origin is widely believed to be the soil. In summary, this study aimed to clarify the effects of soils as microbial sources on endophytic bacterial colonization in rice roots, and how they differed among five soil types. Our findings sufficiently confirm the hypothesis that the microbial source is an important factor in the endophytic bacterial colonization of rice plants. Importantly, the bacterial community in the soils added as a microbial source differed among the soil types, which affected the bacterial community in the hydroponic solution and consequently reflected in the endophytic bacterial community assemblage. Although the importance of the microbial source in the build-up of the endophytic bacterial community has been shown in this study, the mechanism through which this occurs is a subject of further studies. Finally, the findings in this study could provide valuable insights that could facilitate microbial engineering for the benefit of sustainable crop production.
Data availability
The raw 16S rRNA gene amplicon sequence data for this study were deposited in the DDBJ under the following accession number: DRA015220.
References
Ali M, Ali Q, Sohail MA, Ashraf MF, Saleem MH, Hussain S, Zhou L (2021) Diversity and taxonomic distribution of endophytic bacterial community in the rice plant and its prospective. Int J Mol Sci 22:10165. https://doi.org/10.3390/ijms221810165
Asiloglu R, Shiroishi K, Suzuki K, Turgay OC, Murase J, Harada N (2020) Protist-enhanced survival of a plant growth promoting rhizobacteria, Azospirillum sp. B510, and the growth of rice (Oryza sativa L) plants. Appl Soil Ecol 154:103599. https://doi.org/10.1016/j.apsoil.2020.103599
Bach EM, Williams RJ, Hargreaves SK, Yang F, Hofmockel KS (2018) Greatest soil microbial diversity found in micro-habitats. Soil Biol Biochem 118:217–226. https://doi.org/10.1016/j.soilbio.2017.12.018
Bai R, Wang JT, Deng Y, He JZ, Feng K, Zhang LM (2017) Microbial community and functional structure significantly varied among distinct types of paddy soils but responded differently along gradients of soil depth layers. Front Microbiol 8:945. https://doi.org/10.3389/fmicb.2017.00945
Bogas AC, Ferreira AJ, Araújo WL, Astolfi-Filho S, Kitajima EW, Lacava PT, Azevedo JL (2015) Endophytic bacterial diversity in the phyllosphere of Amazon Paullinia cupana associated with asymptomatic and symptomatic anthracnose. Springerplus 4:258. https://doi.org/10.1186/s40064-015-1037-0
Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee J, Ley R, Liu YX, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson MS, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, van der Hooft JJJ, Vargas F, Vázquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan Y, Wang M, Warren J, Weber KC, Williamson CHD, Willis AD, Xu ZZ, Zaneveld JR, Zhang Y, Zhu Q, Knight R, Caporaso JG (2019) Reproducible, interactive, scalable extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. https://doi.org/10.1038/s41587-019-0209-9
Breidenbach B, Pump J, Dumont MG (2016) Microbial community structure in the rhizosphere of rice plants. Front Microbiol 6:1537. https://doi.org/10.3389/fmicb.2015.01537
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP (2016) DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. https://doi.org/10.1038/nmeth.3869
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108:4516–4522. https://doi.org/10.1073/pnas.1000080107
Core R Team (2019) A language and environment for statistical computing. R foundation for statistical computing, Vienna, Austria. https:// www.R-project.org. Accessed 20 June 2021
Cui X, Zhang Y, Gao J (2018) Long-term combined application of manure and chemical fertilizer sustained higher nutrient status and rhizospheric bacterial diversity in reddish paddy soil of Central South China. Sci Rep 8:16554. https://doi.org/10.1038/s41598-018-34685-0
Dai JX, Liu XM, Wang YJ (2014) Diversity of endophytic bacteria in Caragana microphylla grown in the desert grassland of the Ningxia Hui Autonomous Region of China. Genet Mol Res 13:2349–2358. https://doi.org/10.4238/2014.April.3.7
Dang H, Zhang T, Li G, Mu Y, Lv X, Wang Z, Zhuang L (2020) Root-associated endophytic bacterial community composition and structure of three medicinal licorices and their changes with the growing year. BMC Microbiol 20:291. https://doi.org/10.1186/s12866-020-01977-3
Ding T, Melcher U (2016) Influences of plant species, season and location on leaf endophytic bacterial communities of non-cultivated plants. PLoS ONE 11:e0150895. https://doi.org/10.1371/journal.pone.0150895
Dwevedi A, Kumar P, Kumar P, Kumar Y, Sharma YK, Kayastha AM (2017) Soil sensors: detailed insight into research updates, significance, and future prospects. In: AM Grumezescu (Ed.) New Pesticides and Soil Sensors. Academic Press 10:561–594. https://doi.org/10.1016/b978-0-12-804299-1.00016-3
Edwards J, Santos-Medellín C, Nguyen B, Kilmer J, Liechty Z, Veliz E, Ni J, Phillips G, Sundaresan V (2019) Soil domestication by rice cultivation results in plant-soil feedback through shifts in soil microbiota. Genome Biol 20:221. https://doi.org/10.1186/s13059-019-1825-x
Elbeltagy A, Nishioka K, Suzuki H, Sato T, Sato YI, Morisaki H, Mitsui H, Minamisawa K (2000) Isolation and characterization of endophytic bacteria from wild and traditionally cultivated rice varieties. Soil Sci Plant Nut 46:3. https://doi.org/10.1080/00380768.2000.10409127
Ferrando L, Scavino AF (2015) Strong shift in the diazotrophic endophytic bacterial community inhabiting rice (Oryza sativa) plants after flooding. FEMS Microbiol Ecol 91:9. https://doi.org/10.1093/femsec/fiv104
Griffiths RI, Thomson BC, James P, Bell T, Bailey M, Whiteley AS (2011) The bacterial biogeography of British soils. Env Microbiol 13:6. https://doi.org/10.1111/j.1462-2920.2011.02480.x
Guglielmetti S, Basilico R, Taverniti V, Arioli S, Piagnani C, Bernacchi A (2013) Luteibacter rhizovicinus MIMR1 promotes root development in barley (Hordeum vulgare L.) under laboratory conditions. World J Microbiol Biotechnol 29:11. https://doi.org/10.1007/s11274-013-1365-6
Hameed A, Yeh MW, Hsieh YT, Chung WC, Lo CT, Young LS (2015) Diversity and functional characterization of bacterial endophytes dwelling in various rice (Oryza sativa L.) tissues, and their seed-borne dissemination into rhizosphere under gnotobiotic P-stress. Plant Soil 394:177–197. https://doi.org/10.1007/s11104-015-2506-5
Hardoim PR, van Overbeek LS, van Elsas JD (2008) Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol 16:463–471. https://doi.org/10.1016/j.tim.2008.07.008
Hardoim PR, Andreote FD, Reinhold-Hurek B, Sessitsch A, van Overbeek LS, van Elsas JD (2011) Rice root-associated bacteria: insights into community structures across 10 cultivars. FEMS Microbiol Ecol 77:154–164. https://doi.org/10.1111/j.1574-6941.2011.01092.x
Hardoim PR, Hardoim CCP, van Overbeek LS, van Elsas JD (2012) Dynamics of seed-borne rice endophytes on early plant growth stages. PLoS ONE 7:e30438. https://doi.org/10.1371/journal.pone.0030438
Haygarth PM, Ritz K (2009) The future of soils and land use in the UK: soil systems for the provision of land-based ecosystem services. Land Use Policy 26:S187–S197. https://doi.org/10.1016/j.landusepol.2009.09.016
He H, Pang J, Wu GL, Lambers H (2019) The application potential of coal fly ash for selenium biofortification. Adv Agron 157:1–54. https://doi.org/10.1016/bs.agron.2019.05.002
Hemkemeyer M, Dohrmann AB, Christensen BT, Tebbe CC (2018) Bacterial preferences for specific soil particle size fractions revealed by community analyses. Front Microbiol 9:149. https://doi.org/10.3389/fmicb.2018.00149
IUSS Working Group WRB (2015) World Reference Base for soil resources 2014, update 2015. International soil classification system for naming soils and creating legends for soil maps. World soil resources reports No 106. FAO, Rome
Jones JM, Boehm EL, Kahmark K, Lau J, Evans S (2022) Microbial community response to drought depends on crop. Elementa 10:00110. https://doi.org/10.1525/elementa.2021.00110
Kaga H, Mano H, Tanaka F, Watanabe A, Kaneko S, Morisaki H (2009) Rice seeds as sources of endophytic bacteria. Microbes Environ 24:154–162. https://doi.org/10.1264/jsme2.ME09113
Kataoka T, Mitsunobu S, Hamamura N (2018) Influence of the chemical form of antimony on soil microbial community structure and arsenite oxidation activity. Microbes Environ 33:214–221. https://doi.org/10.1264/jsme2.ME17182
Kivlin SN, Hawkes CV (2016) Temporal and spatial variation of soil bacteria richness, composition, and function in a neotropical rainforest. PLoS ONE 11:e0159131. https://doi.org/10.1371/journal.pone.0159131
Kononov A, Hishida M, Suzuki K, Harada N (2022) Microplastic extraction from agricultural soils using canola oil and unsaturated sodium chloride solution and evaluation by incineration method. Soil Syst 6:54. https://doi.org/10.3390/soilsystems6020054
Kozaki D, Sago Y, Fujiwara T, Mori M, Kubono C, Koga T, Mitsui Y, Tachibana T (2021) Ion-exclusion/cation-exchange chromatography using dual-ion-exchange groups for simultaneous determination of inorganic ionic nutrients in fertilizer solution samples for the management of hydroponic culture. Agron J 11:1847. https://doi.org/10.3390/agronomy11091847
Kumar U, Shahid M, Tripathi R, Mohanty S, Kumar A, Bhattacharyya P, Lal B, Gautam P, Raja R, Panda BB, Jambhulkar NN, Shukla AK, Nayak AK (2017) Variation of functional diversity of soil microbial community in sub-humid tropical rice-rice cropping system under long-term organic and inorganic fertilization. Ecol Indic 73:536–543. https://doi.org/10.1016/j.ecolind.2016.10.014
Kuramae EE, Yergeau E, Wong LC, Pijl AS, Van Veen JA, Kowalchuk GA (2012) Soil characteristics more strongly influence soil bacterial communities than land-use type. FEMS Microbiol Ecol 79:12–24. https://doi.org/10.1111/j.1574-6941.2011.01192.x
Li B, Xu R, Sun X, Han F, Xiao E, Chen L, Qiu L, Sun W (2021) Microbiome-environment interactions in antimony-contaminated rice paddies and the correlation of core microbiome with arsenic and antimony contamination. Chemosphere 263:128227. https://doi.org/10.1016/j.chemosphere.2020.128227
Li C, Yan K, Tang L, Jia Z, Li Y (2014) Change in deep soil microbial communities due to long-term fertilization. Soil Biol Biochem 75:264–272. https://doi.org/10.1016/j.soilbio.2014.04.023
Lin GY, Lin CY, Chang SJ, Lin WY (2020) The dynamics of endophytic bacterial community structure in rice roots under different field management systems. Agron J 10:1623. https://doi.org/10.3390/agronomy10111623
Long HH, Sonntag DG, Schmidt DD, Baldwin IT (2010) The structure of the culturable root bacterial endophyte community of Nicotiana attenuata is organized by soil composition and host plant ethylene production and perception. New Phytol 185:554–567. https://doi.org/10.1111/j.1469-8137.2009.03079.x
Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, Tremblay J, Engelbrektson A, Kunin V, del Rio TG, Edgar RC, Eickhorst T, Ley RE, Hugenholtz P, Tringe SG, Dangl JL (2012) Defining the core Arabidopsis thaliana root microbiome. Nature 488:86–90. https://doi.org/10.1038/nature11237
Luo X, Fu X, Yang Y, Cai P, Peng S, Chen W, Huang Q (2016) Microbial communities play important roles in modulating paddy soil fertility. Sci Rep 6:20326. https://doi.org/10.1038/srep20326
Ma W, Tang S, Dengzeng Z, Zhang D, Zhang T, Ma X (2022) Root exudates contribute to belowground ecosystem hotspots: a review. Front Microbiol 13:937940. https://doi.org/10.3389/fmicb
Mahaffee WF, Kloepper JW (1997) Temporal changes in the bacterial communities of soil, rhizosphere, and endorhiza associated with field-grown cucumber (Cucumis sativus L.). Microb Ecol 34:210–213. https://doi.org/10.1007/s002489900050
Mannaa M, Park I, Seo YS (2019) Genomic features and insights into the taxonomy, virulence, and benevolence of plant-associated Burkholderia species. Int J Mol Sci 20:121. https://doi.org/10.3390/ijms20010121
Mano H, Morisaki H (2008) Endophytic bacteria in the rice plant. Microbes Environ 23:109–117. https://doi.org/10.1264/jsme2.23.109
Megías AG, Müller C (2010) Root herbivores and detritivores shape above-ground multitrophic assemblage through plant-mediated effects. J Anim Ecol 79:923–931. https://doi.org/10.1111/j.1365-2656.2010.01681.x
Nannipieri P, Penton CR, Purahong W, Schloter M, van Elsas JD (2019) Recommendations for soil microbiome analyses. Biol Fertil Soils 55:765–766. https://doi.org/10.1007/s00374-019-01409-z
Obara H, Maejima Y, Kohyama K, Ohkura T, Takata Y (2015) Outline of the comprehensive soil classification system of Japan – first approximation. Jpn Agric Res Q 49:217–226. https://doi.org/10.6090/jarq.49.217
Prévost-Bouré NC, Dequiedt S, Thioulouse J, Lelièvre M, Saby NPA, Jolivet C, Arrouays D, Plassart P, Lemanceau P, Ranjard L (2014) Similar processes but different environmental filters for soil bacterial and fungal community composition turnover on a broad spatial scale. PLoS ONE 9:e111667. https://doi.org/10.1371/journal.pone.0111667
Rasche F, Velvis H, Zachow C, Berg G, van Elsas JD, Sessitsch A (2006) Impact of transgenic potatoes expressing anti-bacterial agents on bacterial endophytes is comparable with the effects of plant genotype, soil type and pathogen infection. J Appl Ecol 43:3. https://doi.org/10.1111/j.1365-2664.2006.01169.x
Rodríguez C, Antonielli L, Mitter B, Trognitz F, Sessitsch A (2020) Heritability and functional importance of the Setaria viridis bacterial seed microbiome. Phytobiomes J 4:40–52. https://doi.org/10.1094/PBIOMES-04-19-0023-R
Rui J, Li J, Wang S, An J, Liu W, Lin Q, Yang Y, He Z, Li X (2015) Responses of bacterial communities to simulated climate changes in alpine meadow soil of the Qinghai-Tibet plateau. Appl Environ Microbiol 81:6070–6077. https://doi.org/10.1128/AEM.00557-15
Sahu KP, Kumar A, Sakthivel K, Reddy B, Kumar M, Patel A, Sheoran N, Gopalakrishnan S, Prakash G, Rathour R, Gautam RK (2022) Deciphering core phyllomicrobiome assemblage on rice genotypes grown in contrasting agroclimatic zones: implications for phyllomicrobiome engineering against blast disease. Environ Microbiol 17:28. https://doi.org/10.1186/s40793-022-00421-5
Samuel SO, Suzuki K, Asiloglu R, Harada N (2022) Soil-root interface influences the assembly of the endophytic bacterial community in rice plants. Biol Fertil Soils 58:35–48. https://doi.org/10.1007/s00374-021-01611-y
Shen C, Xiong J, Zhang H, Feng Y, Lin X, Li X, Liang W, Chu H (2013) Soil pH drives the spatial distribution of bacterial communities along elevation on Changbai Mountain. Soil Biol Biochem 57:204–211. https://doi.org/10.1016/j.soilbio.2012.07.013
Terrat S, Horrigue W, Dequietd S, Saby NPA, Lelièvre M, Nowak V, Tripied J, Régnier T, Jolivet C, Arrouays D, Wincker P, Cruaud C, Karimi B, Bispo A, Maron PA, Prévost-Bouré NC, Ranjard L (2017) Mapping and predictive variations of soil bacterial richness across France. PLoS ONE 17:e0268101. https://doi.org/10.1371/journal.pone.0186766
Truog E (1930) The determination of the readily available phosphorus of soils. Agron J 22:874–882. https://doi.org/10.2134/agronj1930.00021962002200100008x
van Overbeek L, van Elsas JD (2008) Effects of plant genotype and growth stage on the structure of bacterial communities associated with potato (Solanum tuberosum L.). FEMS Microbiol Ecol 64:283–296. https://doi.org/10.1111/j.1574-6941.2008.00469.x
Vendan RT, Yu YJ, Lee SH, Rhee YH (2010) Diversity of endophytic bacteria in ginseng and their potential for plant growth promotion. J Microbiol 48:559–565. https://doi.org/10.1007/s12275-010-0082-1
Vieira S, Sikorski J, Gebala A, Boeddinghaus RS, Marhan S, Rennert T, Kandeler E, Overmann J (2020) Bacterial colonization of minerals in grassland soils is selective and highly dynamic. Environ Microbiol 22:917–933. https://doi.org/10.1111/1462-2920.14751
Wang M, Sha C, Wu J, Li P, Tan J, Huang S (2022) Comparison of bacterial community in paddy soil after short-term application of pig manure, and the corresponding organic fertilizer. Land 11:9. https://doi.org/10.3390/land11010009
Walitang DI, Kim CG, Kim K, Kang Y, Kim YK, Sa T (2018) The influence of host genotype and salt stress on the seed endophytic community of salt-sensitive and salt-tolerant rice cultivars. BMC Plant Biol 18:51. https://doi.org/10.1186/s12870-018-1261-1
Xu Y, Ge Y, Song J, Rensing C (2020) Assembly of root-associated microbial community of typical rice cultivars in different soil types. Biol Fertil Soils 56:249–260. https://doi.org/10.1007/s00374-019-01406-2
Yu Z, Liang K, Huang G, Wang X, Lin M, Chen Y, Zhou Z (2021) Soil bacterial community shifts are driven by soil nutrient availability along a teak plantation chronosequence in tropical forests in China. Biology (basel) 10:1329. https://doi.org/10.3390/biology10121329
Yuan Y, Si G, Wang J, Luo T, Zhang G (2014) Bacterial community in alpine grasslands along an altitudinal gradient on the Tibetan Plateau. FEMS Microbiol Ecol 87:121–132. https://doi.org/10.1111/1574-6941.12197
Zarraonaindia I, Owens SM, Weisenhorn P, West K, Hampton-Marcell J, Lax S, Bokulich NA, Mills DA, Martin G, Taghavi S, van der Lelie D, Gilbert JA (2015) The soil microbiome influences grapevine-associated microbiota. Mbio 6:e02527-e2614. https://doi.org/10.1128/mBio.02527-14
Zhang X, Ma YN, Wang X, Liao K, He S, Zhao X, Guo H, Zhao D, Wei HL (2022) Dynamics of rice microbiomes reveal core vertically transmitted seed endophytes. Microbiome 10:216. https://doi.org/10.1186/s40168-022-01422-9
Zhang X, Zhong Z, Gai X, Du X, Bian F, Yang C, Gao G, Wen X (2019) Changes of root endophytic bacterial community along a chronosequence of intensively managed lei bamboo (Phyllostachys praecox) forests in subtropical China. Microorganisms 7:616. https://doi.org/10.3390/microorganisms7120616
Funding
This work was funded by JSPS KAKENHI Grant Number JP21K14952. The sequencing analysis was the result of using research equipment shared in the MEXT Project for promoting public utilization of advanced research infrastructure (Program for supporting introduction of the new sharing system) at Niigata University, Grant Number JPMXS0421100320.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval
This study does not involve animals.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Samuel, S.O., Suzuki, K., Asiloglu, R. et al. Rice endophytic communities are strongly dependent on microbial communities specific to each soil. Biol Fertil Soils 59, 733–746 (2023). https://doi.org/10.1007/s00374-023-01743-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00374-023-01743-3