Abstract
The present study compares the community structures of microbiota at different habitats in Japanese rice fields by comparing their phospholipid fatty acid (PLFA) compositions to understand the contribution of different habitats to microbiological diversity. The data were collected from four neighboring rice fields. Comparison was made for the PLFA compositions extracted from the floodwater, percolating water, rice soils under flooded and drained conditions, rice straw (RS) placed in flooded and drained rice soils, RS in the composting process, and RS compost placed in a flooded rice field. Average amounts of PLFAs were 33 μg L−1 in the floodwater, 17.1 μg L−1 in the percolating water from plow layers, 34.6 μg L−1 in the percolating water from subsoil layers, 108 μg g−1 dry weight basis (dw) in flooded rice soils, 382 μg g−1 dw in RS materials, 2,510 μg g−1 dw in RS composts, 2,850 μg g−1 dw in RS composts after application to a flooded rice soil, 222 μg g−1 wet weight basis (ww) in RS in drained rice soils, and 284 μg g−1 ww in RS in flooded rice soils. The total amount of PLFAs to the soil depth of 10 cm was estimated to be about 12 g m−2. The PLFA compositions were different from each other depending on the habitats. Rice soils were characterized by the predominance of actinomycetes and Gram-positive bacteria in comparison with the other habitats. In contrast, the microbial communities in the floodwater and percolating water were characterized by the predominance of Gram-negative bacteria and eukaryotes (presumably algae), and Gram-negative bacteria, respectively. The microbial community of RS materials was dominated by fungi. Gram-positive bacteria became predominant in RS after application to flooded rice soils, while RS placed in a drained rice field after harvesting rice was characterized by the predominance of Gram-negative bacteria and fungi. The community structures at respective habitats were stable and specific, irrespective of the season of sampling and the duration of decomposition of RS.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The rice field is a unique agro-ecosystem, where the field is maintained under flooded conditions during most of the period of rice cultivation, and is left under drained conditions during the off-crop season. Flooding the field results in several specific habitats for microbiota such as floodwater and the reduced soil layer (Kimura 2000). The reduced soil layer becomes aerobic after drainage for harvesting rice. Rice straw (RS) and stubble, rice rhizosphere, applied RS compost, and percolating water are also specific habitats for microbiota in the rice field ecosystem. Thus, a rice field consists of diverse habitats for microorganisms in time and space.
The phospholipid fatty acids (PLFAs) analysis characterizes the community structure of microbiota belonging not only to eubacteria, but also to eukaryotes (Zelles 1999). Phospholipid fatty acid compositions have been used as biomarkers for identifying specific groups of microorganisms (Lechevalier and Lechevalier 1988): straight, monounsaturated PLFAs for Gram-negative bacteria (Wilkinson 1988), branched-chain PLFAs for Gram-positive bacteria (O'Leary and Wilkinson 1988), methylated PLFAs for actinomycetes (Kroppenstedt 1992), 20:3ω6 and 20:4ω6 PLFAs for protozoa (Vestal and White 1989; Herrmann and Shann 1997), and straight, polyunsaturated PLFAs for eukaryotes including fungi (Lösel 1988; Zelles 1999). Our group has used this technique to elucidate microbial communities in various habitats in rice fields such as floodwater, percolating water, RS in soil, RS under the composting process, RS compost in soil, and rice soils, as shown in Table 1. These studies show the predominant members in microbial communities, relative successions, and the effects of fertilizer application on the community structure at the respective habitats. However, direct comparison was not conducted among microbiota in various habitats to understand the “whole community structures of microbiota in rice field ecosystems”. As rice field ecosystems consist of distinct habitats for organisms, which would make the ecosystem diverse as a whole, such comparison is requisite to understand the diversity of microbial worlds in the rice field ecosystem. The following question still remains unanswered: “Is the microbial diversity in rice field ecosystems mainly caused by the assemblage of various microbial habitats, by the variations of environmental conditions at the respective habitats, or by both?” The present study compares the community structures of microbiota at different habitats in rice fields by measuring their PLFA compositions to answer this question. The data sources for comparison were from the studies listed in Table 1. Flooded soil samples with several electron acceptors were also included in the present study. This experiment was conducted to examine the effects of electron acceptors on the composition of microbial communities of rice field soil (Takai and Kamura 1966; Kimura 2000).
Materials and methods
Study sites
The following four rice fields, which were located at Aichi-ken Anjo Research and Extension Center, central Japan (latitude 34°48' N, longitude 137°30' E), were subjected to PLFA composition analysis in the present study: (1) the fields under long-term fertilizer trial (LTFT), (2) A2 field, (3) D2 field, and (4) E2 field. The details of treatments including fertilizer application and management of these fields and the soil properties were described previously (Okabe et al. 2000a,b; Nakamura et al. 2003; Shimizu et al. 2002a,b; Tanahashi et al. 2004) (Table 1). The soil was an Anthraquic Yellow Soil (Oxyaquic Dystrudept).
Soil samples collected from the plow layer in the E2 field were used in a pot experiment for determining the microbial composition in the percolating water (Kimura et al. 2001a), and in laboratory experiments for examining the effects of N fertilizers and of electron acceptors for anaerobic respiration on the microbial communities of rice straw and flooded soil, respectively (Kimura et al. 2001b).
Sample collection for the determination of PLFA composition of microbiota
Methods for the collection and preparation of samples were previously described in the references listed in Table 1. In addition, flooded soil samples treated with several electron acceptors were analyzed in the present study. Soil collected from the E2 rice field on September 14, 1999 (drained conditions) was first air-dried, and then treated with glucose at the rate of 4 g kg−1 air dry soil. Then, 10 g of Ca(NO3)2, 10 g of MnO2, 30 g of ferric oxide/hydroxide precipitate, or 6 g of (NH4)2SO4 was amended as electron acceptors to 1 kg of the air-dry soil samples. Ferric oxide/hydroxide precipitate was obtained by alkalinizing Fe(NH4)2(SO4)2.6H2O solution with NaOH. The precipitate was washed several times with distilled water, and air-dried. Distilled water (15 mL) was added to treated and untreated soil (20 g) in a test tube, and the soil was submerged carefully without leaving any bubbles. Then, the test tubes were closed with a W-shaped butyl rubber stopper, and the air inside the test tube was exchanged several times with N2 gas. The soil samples thus prepared were incubated for 40 days at 24–28°C. PLFA analysis was conducted after 10 and 40 days for samples treated with ferric oxide/hydroxide precipitate, 10 and 30 days for samples treated with MnO2, 15 and 40 days for samples treated with Ca(NO3)2, 10 and 30 days for samples treated with (NH4)2SO4, and 10 and 40 days for untreated samples. The sampling periods during incubation were determined depending on the reduction conditions of the respective treatment soils.
PLFA analysis
The analysis of PLFAs was performed as reported by Okabe et al. (2000b), who modified the methods of Bligh and Dyer (1959) and White et al. (1979). The gas chromatography operation was conducted according to the method of Okabe et al. (2000b).
Fatty acids were designated based on the total number of C atoms and the number of double bonds, followed by the position of the double bond from the methyl end (aliphatic end [ω]) of the molecule. Cis and trans configurations are indicated by “c” and “t”, respectively. The prefixes “ai” and “i” refer to anteiso and iso branchings, respectively, 10Me refers to a methyl group on the tenth carbon atom from the carboxyl end of the molecule, and “cy” refers to cyclopropane fatty acids. We used the proportion (mol%) of PLFAs and Simpson's (1-D) and Shannon-Wiener's (H') diversity indexes for evaluating the diversity of PLFA composition. The indexes were calculated according to Ito and Sato (2002).
Statistical analysis
The proportions of straight, saturated PLFAs; straight, monounsaturated PLFAs; straight, polyunsaturated PLFAs; and branched-chain PLFAs in the total PLFAs extracted from respective habitats, and the degree of stress at respective habitats based on the trans/cis ratio of 16:1ω7 PLFA, were compared by nonparametric analysis with the Stat macros program for Excel (Hirota 2001). To compare the microbial communities among samples, differences in the composition of PLFAs extracted from the samples were analyzed using cluster analysis and principal component analysis (PCA). The values were normalized for the analyses. Cluster analysis was performed using JMP IN (SAS Institute, Tokyo) by the Ward method. PCA was performed using Excel Statistics 97 for Windows (SRI, Tokyo). A correlation matrix was used in the analysis. Analysis of molecular variance (AMOVA) was conducted for the comparison of the PLFA patterns of microbiota. The calculation of Φ ST and a significant test for the Φ ST value based on the permutation procedure were performed according to Excoffier et al. (1992).
Results and discussion
Amounts of PLFAs at respective habitats
Average amounts of PLFAs in aquatic habitats were 17.1–34.6 μg L−1 (Table 2). Wide variations in the amounts in the floodwater were attributed to the field management systems that included fertilizer treatments. The average amount of PLFAs in the floodwater in the A2 plots was 86.8±48.8 μg L−1 (n=10; wheat was the second crop and wheat straw was left on the field), while that in the rice fields under LTFT was 7.1±3.2 μg L−1 (n=20; the fields were left fallow during winter season). On the contrary, variations in the amount of PLFAs in flooded rice soils were smaller than those in aquatic habitats (108±33 μg g−1 dry weight basis [dw]).
Average amounts of PLFAs were 382–2,850 μg g−1 dw for RS or RS composts, and 222–284 μg g−1 wet weight basis (ww) for RS in rice soils. As wet RS samples were expected to contain about 90% of water in weight, amounts of PLFAs were considered not to be significantly different among RS composts, RS composts in soil and RS materials in soil. In addition, as the soil samples used in the present study contained about 13 g C kg−1, similar amounts of PLFAs were estimated to be present in the soil to those in RS composts and RS in soil per unit weight of organic matter (C contents of RS were supposed to be about 35%).
These average values permit a rough estimation of the total amount of PLFAs in the surface part of rice fields. Microbial biomass (g m−2) as expressed by the amounts of PLFAs was 1.64×10−3 in floodwater, 0.86×10−3 in soil water, 11.9 in plow layer soil, 1.11 in RS, and 0.57 in RS compost. Here, the depths of the floodwater and the plow layer are supposed to be 5 cm and 10 cm, respectively. In addition, the pore space and the bulk density in the plow layer are postulated to be 50% and 1.1 Mg m−3, respectively. The total amount of PLFAs in the 0–10 cm soil layer was about 12 g m−2, and it accounted for more than 99% of total microbial biomass resulting from the sum of microbial biomass values of the floodwater and the plow layer. Six tons dw ha−1 of RS roughly corresponds to a rice yield of 5 tons ha−1 under field conditions (the average yields of brown rice and RS in Japan are 5.2 and about 6 tons ha−1, respectively). These amounts correspond to about 10 tons fresh weight of RS compost. The PLFAs in RS and RS compost amounted to about 10 and 4%, respectively, of the average PLFA values of the plow layer soil. Because the weight losses of RS and RS compost after their application to the plow layer were not considered, it is estimated that more than 90% of PLFAs remain in the plow layer soil even after their application to the soil.
PLFA composition of microbiota at respective habitats
RS materials contained the largest proportion of straight, saturated PLFAs (median: 53%), which tended to decrease through the microbial decomposition of RS in the soil or in the composting process (Fig. 1). The proportions of straight, saturated PLFAs were significantly larger in the habitats with RS materials (RS in flooded rice soils, RS in drained/aerobic rice soil, RS in the composting process, and RS compost in flooded rice soil) than in habitats not treated with RS (floodwater, percolating water from the plow layer, percolating water collected from the culvert, soils under flooded conditions, and soils under drained/aerobic conditions) (P<0.001, Mann–Whitney U test). Proportions of straight, monounsaturated PLFAs fluctuated widely among the habitats from 16% of RS in flooded soils to 59% in the percolating water collected from culverts. However, aquatic environments such as floodwater and percolating water contained significantly larger proportions of unsaturated PLFAs than other environments (P<0.001, Mann–Whitney U test). As straight, monounsaturated PLFAs are generally the biomarkers of Gram-negative bacteria (Wilkinson 1988), Gram-negative bacteria were probably predominant in the aquatic environments of the rice field ecosystem. The proportions of branched-chain PLFAs showed the following order: both flooded and drained soils (43%); RS of flooded soil (39%), and RS under composting process (37%)> percolating water from plow layer (34%), and RS compost of flooded rice soil (30%)> percolating water from culvert (16%); floodwater (14%); RS in drained soil (9.4%); and RS materials (6.4%) (P<0.01, Kruskal–Wallis test and Bonferroni procedure). This indicates that Gram-positive bacteria played important roles in the soils and in the composting process of RS. The proportion of straight, polyunsaturated PLFAs, the biomarker of eukaryotes, was significantly higher in RS materials (16%) and the RS of drained rice field soil (17%) than in the other samples (2–9%) (P<0.001, Mann–Whitney U test), and this behavior was attributed to the predominant colonization of RS materials by fungi as mentioned below. However, the possibility that straight, polyunsaturated PLFAs originated from rice plants that remained undecomposed in RS materials cannot be discarded.
Proportions of straight, saturated PLFAs; straight, monounsaturated PLFAs; straight, polyunsaturated PLFAs; and branched-chain PLFAs of microbiota inhabiting at respective habitats. Boxes show 90% range (5–95%) of data. Solid and broken lines inside the boxes indicate median and lower and upper quartiles (25 and 75%) of the data, respectively
Figure 2 compares the average percentage of each PLFA in the samples among different habitats. An interesting aspect was lower percentages of 10Me-group PLFAs in floodwater and percolating water than in soil samples, which indicates that these aquatic environments do not represent good habitats for actinomycetes. On the contrary, a relatively larger proportion of 20:4ω6c PLFA was found in the floodwater and the percolating water collected from the culverts than in the other habitats, indicating the presence of protozoa. All the standard deviations of respective PLFAs were small except for 16:0, 16:1ω7c, and 18:2ω6c PLFAs, indicating that the community structure of microbiota in the rice field ecosystem is stable and site-specific, which was also revealed by cluster analysis and PCA (see below). These characteristics may have contributed to the biological diversity of the rice field ecosystem as a whole.
Statistical analyses
Cluster analysis and PCA were performed on all samples to estimate the development of specific communities of biota and the effect of field management on microbial communities of respective habitats in the rice field ecosystem. Cluster analysis firstly enabled us to divide the PLFA patterns of microbial communities in the rice field ecosystem into two groups (Fig. 3). The PLFA patterns of microbial communities of clusters A and B were significantly different (P<0.001) upon AMOVA analysis. The first group consisted of floodwater, percolating water, RS compost of flooded soil, and RS samples in flooded and drained soils. The RS samples of flooded and drained soils belonging to this group were mainly at the early stage of decomposition. The second group included soil samples, RS samples in the composting process, and degraded RS in the soil. The first group (cluster A) was further separated into two subclusters (A1 and A2): with aquatic samples in the subcluster A1 and all other samples in the subcluster A2. The second group (cluster B) was also divided into two subclusters, namely the subcluster B1 with RS samples of soils and the subcluster B2 with soil samples and RS samples in the composting process. The PLFA patterns of microbial communities of the subclusters (A1 and A2, and B1 and B2) were significantly different (P<0.001) upon AMOVA analysis.
Cluster analysis of the PLFA patterns of microbial communities in the rice field ecosystem. Arrows indicate the samples that were out of their respective subclusters. ▵ Floodwater, ▵▵ percolating water, × rice straw materials, □ rice straw in flooded soil, ▲ rice straw in drained soil, ⇐ rice straw compost, ⇐⇐ rice straw compost in flooded soil, ○ flooded soil, ○○ drained soil
Soil samples amended with several electron acceptors in the laboratory experiments were also included in the subcluster B2 because effects of the electron acceptors on the structure of microbial communities of rice field soil were small. Lueders and Friedrich (2000) also showed that the sequential reduction process had little effect on archaeal communities of rice soil in microcosm experiments. Floodwater and percolating water samples could be distinguished in the subcluster A1 (Fig. 3). Only 5 samples among 172 could not be included in the above mentioned subclusters. This analysis suggests that community structures of microbiota in the rice field ecosystem are determined predominantly by the characteristics of respective habitats and that the temporal fluctuations in the community structure at respective habitats are significantly small in comparison with the variations among the habitats.
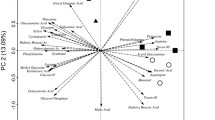
The results of PCA are shown in Figs. 4 and 5. The total percentages by the first, second, and third principal components were 19.1, 12.6 and 10.7%, respectively. PCA elucidated microbial populations that were responsible for groupings in cluster analysis. In Fig. 4, samples in cluster A are mainly located in the left hand side and those in cluster B in the right hand side, and samples belonging to subclusters B1 and B2 are distributed in the first and fourth quadrants in the same figure, respectively. On the contrary, samples distributed in subclusters A1 and A2 are mainly located in the second and third quadrants, respectively (Fig. 5). The high positive and negative loads in the first principal component included biomarkers of Gram-positive bacteria (branched-chain PLFAs) and those of Gram-negative bacteria and eukaryotes (monounsaturated and polyunsaturated PLFAs), respectively, and the high negative loads in the second principal component included biomarkers of actinomycetes and Gram-positive bacteria (10Me-group and branched-chain PLFAs). In addition, the high negative loads in the third principal component included two biomarkers of fungi (18:2ω6c and 18:1ω9c).
Distribution of the score plots of the PLFA composition in the floodwater, percolating water, RS compost, and soil samples in the coordinate system of the first and second principal components. ⋄ Floodwater, gray ♦ percolating water from plow layer, ♦ percolating water from subsoil, □ rice straw materials, gray ▪ rice straw in drained soil, ▪ rice straw in flooded soil, ▲ rice straw compost, △ rice straw compost in flooded soil, • flooded soil, ○ drained soil
Distribution of the score plots of the PLFA composition in the floodwater, percolating water, RS compost, and soil samples in the coordinate system of the first and third principal components. Symbols are the same as those in Fig. 4
RS materials used in the present study are distributed at the lower region in the third quadrant of Fig. 5, and the relative community structure was characterized by the predominance of fungi (18:2ω6c), whose proliferation was probably stimulated by dry moisture conditions. Nearly all RS samples placed in flooded rice soils are distributed in the first quadrant in Fig. 4, which differed from the other samples. The scores of the first, second, and third principal components of RS samples in flooded rice soil correlated positively (P<0.05), negatively (P<0.01), and positively (P<0.01) with the duration of placement, respectively. By considering the PLFAs with high positive loads in the first and second principal components of Fig. 4, the community structure of microbial population in RS of flooded rice soils was characterized by the predominance of Gram-positive bacteria. In contrast, RS placed in a drained rice field after rice harvest is distributed in the third quadrant of Fig. 5, and was characterized by the predominance of Gram-negative bacteria and fungi. These findings indicate that different microbial populations were responsible for RS decomposition in Japanese rice fields during the flooding and the drained-aerobic periods. Floodwater and percolating water samples are distributed mainly in the second quadrant of Fig. 5, and differed from the other samples due to the predominance of Gram-negative bacteria and eukaryotes (presumably algae), and Gram-negative bacteria, respectively.
Soil samples distributed in the fourth quadrant of Fig. 4 were characterized by the predominance of actinomycetes and Gram-positive bacteria (10Me-group and branched-chain PLFAs), irrespective of the type of samples (from the rice field or from microcosm experiments), the treatment (the addition of electron acceptors), and field conditions (flooded or drained). RS from the composting process is also located in the fourth quadrant of Fig. 4, and experienced an upper-leftward shift with the duration of composting. In contrast, RS compost samples that were placed in the flooded rice field are distributed at the center of Fig. 4, together with the decomposed RS samples of the composting process. The score plots of RS compost samples in soil did not show marked changes with the duration of placement compared to those of RS samples in flooded rice soil.
Figures 4 and 5 confirm the characteristics (stable and site-specific) of the structure of microbial communities in the rice field ecosystem, as shown by cluster analysis (see above). Another interesting aspect is the predominance of Gram-positive bacteria (branched-chain PLFAs) in the solid habitats (soils, RS and RS compost) and Gram-negative bacteria (straight, monounsaturated PLFAs) in the aquatic habitats (floodwater and percolating water).
Simpson's (1-D) and Shannon-Wiener's (H') diversity indexes of PLFA compositions were highest in samples of drained soil, flooded soil, and RS compost (0.916 to 0.931 and 2.87 to 2.96 for 1-D and H', respectively). Samples of percolating water from the plow layer, RS in flooded soil, and RS compost in flooded soil showed intermediate diversity indexes (0.887 to 0.909 and 2.75 to 2.76 for 1-D and H', respectively), whereas those of percolating water from culvert, floodwater, RS in drained soil, and RS materials were the lowest (0.780 to 0.873 and 2.08 to 2.54 for 1-D and H', respectively; P<0.01, Kruskal–Wallis test and Bonferroni procedure). This suggests that microbial diversity might have been different among the three habitats as described above. It is important to emphasize that PLFA analysis does not allow assessing the diversity at genus or species levels (Nannipieri et al. 2003).
Changes in PLFAs in RS materials
RS materials contained large proportions of 16:0 and 18:2ω6c PLFAs (Fig. 2). Thus, fungi grew preferentially in RS materials. The proportions of 16:0 and 18:2ω6c PLFAs decreased during the decomposition of RS under flooded conditions, while branched-chain PLFAs such as i15:0, i16:1 and ai15:0 PLFAs increased, which indicated that Gram-positive bacteria contributed to the decomposition of RS in the flooded rice field. Probably, a part of 16:0 PLFA originated from RS materials and may have remained in RS samples in flooded rice soils for a long time. Although lower proportions of straight, monounsaturated PLFAs also characterized RS in flooded rice soils, the PLFA patterns of microbial populations in decomposing RS under flooded conditions became similar to those of soil samples after long decomposition. In contrast, the proportions of straight, monounsaturated PLFAs increased and 18:2ω6c PLFA remained high when RS was decomposed in the drained rice field after rice harvest. Thus, different microbial communities characterized RS decomposition in the soil during the flooded rice cultivation and after rice harvest, being Gram-positive bacteria including actinomycetes predominant during the flooding and Gram-negative bacteria and fungi predominant after rice harvest.
The proportions of 16:0 and 18:2ω6c PLFAs also decreased during the composting process as RS decomposition in the soil as described above, while branched-chain PLFAs such as i15:0, i16:0, i17:0, ai15:0, ai17:0, and 10Me-group PLFAs increased (Fig. 2). Thus, Gram-positive bacteria including actinomycetes predominated in the composting process of RS. It was interesting that the PLFA patterns of microbiota in composting RS were similar to those of soil samples. This was probably because Bacillus (branched-chain PLFAs) and actinomycetes (10Me-group PLFAs) were active in the thermophilic stage of the composting process of RS (Cahyani et al. 2003).
Stressed conditions for microorganisms at respective habitats as estimated from the trans/cis ratios of 16:1ω7 PLFA
The trans/cis ratios of 16:1ω7 PLFA for the microbiota at respective habitats are reported in Table 3. The average values of the ratio ranged from 0.06 for the microbiota in the floodwater to 0.29 for microbiota of RS in flooded soils. The trans/cis ratios of 16:1ω7 PLFA were significantly different among the microbiota of the investigated habitats (RS in flooded rice soils, RS compost in flooded rice soil, flooded soils, floodwater and percolating water from plow layer soil and culvert, RS in composting process, drained soil, RS materials, and RS in drained rice soil) (P<0.01, Kruskal–Wallis test and Bonferroni procedure). The microbiota under flooded conditions showed the highest values and those in aquatic habitats the lowest values, and this indicated that microbial stress was high under flooded conditions, and low in aquatic habitats. Reduced conditions and a surplus of organic materials (substrates for microorganisms) might have been possible stresses for micorbiota. These conditions might have influenced the composition of microbial communities in the rice field ecosystem.
Conclusions
Rice soils were characterized by the predominance of actinomycetes and Gram-positive bacteria in comparison with other habitats. In contrast, the microbial communities of the floodwater and percolating water were characterized by the predominance of Gram-negative bacteria and eukaryotes (presumably algae), and Gram-negative bacteria, respectively. The community structure of RS materials was characterized by the predominance of fungi. Gram-positive bacteria became predominant in RS after application to flooded rice soils, while RS placed in a drained rice field after rice harvest was characterized by the predominance of Gram-negative bacteria and fungi. PLFA patterns of composting RS were similar to those of soil samples. The community structures of biota at respective habitats were stable and specific, irrespective of the season of sampling and the duration of decomposition, which was estimated to contribute to the biological diversity of the rice field ecosystem as a whole.
References
Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37:911–917
Cahyani VR, Watanabe A, Matsuya K, Asakawa S, Kimura M (2002) Succession of microbiota estimated by phospholipid fatty acid analysis and changes in organic constituents during the composting process of rice straw. Soil Sci Plant Nutr 48:735–743
Cahyani VR, Matsuya K, Asakawa S, Kimura M (2003) Succession and phylogenetic composition of bacterial communities responsible for the composting process of rice straw estimated by PCR-DGGE analysis. Soil Sci Plant Nutr 49:619–630
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Herrmann RF, Shann JF (1997) Microbial community changes during composting of municipal solid waste. Microb Ecol 33:78–85
Hirota T (2001) Statistical macros for Excel. http://www.tuat.ac.jp/~ethology/Columbo/Stat/
Ito Y, Sato K (2002) Problems around the indices of species diversity for comparison of different communities. Seibutsukagaku 53:204–220
Kimura M (2000) Anaerobic microbiology in waterlogged rice fields. In: Bollag JM, Stotzky G (eds) Soil biochemistry, vol 10. Marcel Dekker, New York, pp 35–138
Kimura M, Kishi H, Okabe A, Maie N (2001a) Phospholipid fatty acid composition of microbiota in the percolating water from a rice paddy microcosm. Soil Sci Plant Nutr 47:533–545
Kimura M, Miyaki M, Fujinaka K, Maie N (2001b) Microbiota responsible for the decomposition of rice straw in a submerged paddy soil estimated from phospholipid fatty acid composition. Soil Sci Plant Nutr 47:569–578
Kroppenstedt RM (1992) The genus Nocardiopsis. In: Balows A, Trüper HG, Dworkin M, Harder W, Schleifer K-H (eds) The prokaryotes, 2nd edn, vol 2. Springer, Berlin Heidelberg New York, pp 1139–1156
Lechevalier H, Lechevalier MP (1988) Chemotaxonomic use of lipids — an overview. In: Ratledge C, Wilkinson SG (eds) Microbial lipid, vol 1. Academic Press, London, pp 869–902
Lösel DM (1988) Fungal lipids. In: Ratledge C, Wilkinson SG (eds) Microbial lipids, vol 1. Academic Press, London, pp 699–806
Lueders T, Friedrich M (2000) Archaeal population dynamics during sequential reduction processes in rice field soil. Appl Environ Microbiol 66:2732–2742
Nakamura A, Tun CC, Asakawa S, Kimura M (2003) Microbial community responsible for the decomposition of rice straw in a paddy field: estimation by phospholipid fatty acid analysis. Biol Fertil Soils 38:288–295
Nannipieri P, Acher J, Ceccherini MT, Landi L, Pietramellara G, Renella G (2003) Microbial diversity and soil functions. Eur J Soil Sci 54:655–670
Okabe A, Oike H, Toyota K, Kimura M (2000a) Comparison of phospholipid fatty acid composition in floodwater and plow layer soil during the rice cultivation period in a Japanese paddy field. Soil Sci Plant Nutr 46:893–904
Okabe A, Toyota K, Kimura M (2000b) Seasonal variations of phospholipid fatty acid composition in the floodwater of a Japanese paddy field under a long-term fertilizer trial. Soil Sci Plant Nutr 46:177–188
O'Leary WM, Wilkinson SG (1988) Gram-positive bacteria. In: Ratledge C, Wilkinson SG (eds) Microbial lipids, vol 1. Academic Press, London, pp 117–202
Shimizu M, Hayashi M, Matsuya K, Murase J, Kimura M (2002a) Comparison of microbial communities in percolating water from plow layer and subsoil layer estimated by phospholipid fatty acid (PLFA) analysis. Soil Sci Plant Nutr 48:877–881
Shimizu M, Nakajima Y, Matsuya K, Kimura M (2002b) Comparison of phospholipid fatty acid composition in percolating water, floodwater, and the plow layer soil during the rice cultivation period in a Japanese paddy field. Soil Sci Plant Nutr 48:595–600
Takai Y, Kamura T (1966) The mechanism of reduction in waterlogged paddy soil. Folia Microbiol 11:304–313
Tanahashi T, Murase J, Matsuya K, Asakawa S, Kimura M (2004) Microbial communities responsible for the decomposition of rice straw compost in a Japanese rice paddy field determined by phospholipid fatty acid (PLFA) analysis. Soil Sci Plant Nutr 50:1229–1236
Vestal JR, White DC (1989) Lipid analysis in microbial ecology: quantitative approaches to the study of microbial communities. BioScience 39:535–541
White DC, Davis WM, Nickels JS, King JD, Bobbie RJ (1979) Determination of the sedimentary microbial biomass by extractable lipid phosphate. Oecologia 40:51–62
Wilkinson SG (1988) Gram-negative bacteria. In: Ratledge C, Wilkinson SG (eds) Microbial lipids, vol 1. Academic Press, London, pp 299–488
Zelles L (1999) Fatty acid patterns of phospholipids and lipopolysaccharides in the characterisation of microbial communities in soil: a review. Biol Fertil Soils 29:111–129
Acknowledgements
We thank the coworkers in our laboratory for their valuable comments and also the staff members of Aichi-ken Anjo Research and Extension Center for allowing us to use the experimental fields for soil sampling. We also appreciate A. Nakanishi, T. Torimaru, and N. Tomaru of the Graduate School of Bioagricultural Sciences, Nagoya University, for their help in the AMOVA analysis. This study was supported in part by a Grant-in Aid for Scientific Research (B; No. 11460031) from the Japan society for the Promotion of Science.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kimura, M., Asakawa, S. Comparison of community structures of microbiota at main habitats in rice field ecosystems based on phospholipid fatty acid analysis. Biol Fertil Soils 43, 20–29 (2006). https://doi.org/10.1007/s00374-005-0057-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00374-005-0057-2