Abstract
Cone-rod dystrophy (CRD) is a form of inherited retinal degeneration (RD) causing blindness in man as well as in several breeds of dog. Previously, a 44 bp insertion in RPGRIP1 (retinitis pigmentosa GTPase regulator interacting protein-1) was associated with a recessive early-onset CRD (cone-rod dystrophy 1, cord1) in a Miniature longhaired dachshund (MLHD) research colony. Yet in the MLHD pet population, extensive range of the onset age has been observed among RD cases, with some RPGRIP1 −/− dogs lacking obvious clinical signs. Phenotypic variation has been known in human homologous diseases, including retinitis pigmentosa and Leber congenital amaurosis, indicating possible involvement of modifiers. To explore additional genetic loci associated with the phenotypic variation observed in MLHDs, a genome-wide association study was carried out using Canine SNP20 arrays in 83 RPGRIP1 −/− MLHDs with variable ages of onset or no clinical abnormality. Using these samples, comparison of 31 early-onset RD cases against 49 controls (15 late-onset RD and 34 normal dogs combined) identified a strong association (P = 5.05 × 10−13) at a single locus on canine chromosome 15. At this locus, the majority of early-onset RD cases but few of the controls were homozygous for a 1.49 Mb interval containing ~11 genes. We conclude that homozygosity at both RPGRIP1 and the newly mapped second locus is necessary to develop early-onset RD, whereas RPGRIP1 −/− alone leads to late-onset RD or no apparent clinical phenotype. This study establishes a unique model of canine RD requiring homozygous mutations at two distinct genetic loci for the manifestation of early-onset RD.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Inherited retinal diseases (RDs) leading to blindness are perhaps the most phenotypically and molecularly well-described inherited diseases in dogs (Aguirre and Acland 2006; Miyadera et al. 2012). Cone-rod dystrophy (CRD), a form of RD, is characterized by initial dysfunction of cone photoreceptors followed by that of the rods, while progressive retinal atrophy (PRA), more commonly known in the veterinary field and a homologue of retinitis pigmentosa (RP) in man, shows predominant dysfunction of the rods prior to progressive loss of both rods and cones. Unless examined by electroretinography (ERG) at an early stage of the disease, ophthalmoscopic findings at the time of presentation, which is usually prompted by altered behaviour indicating abnormal vision, are identical in CRD and PRA, and the two conditions are thus often being confused. In this article, the broader term RD will be used to describe the retinal condition unless ERG was performed. Based on similarities in phenotypic, etiologic, and genetic features, RDs in dogs have been recognized as excellent models for human homologous diseases, including RP and Leber congenital amaurosis (Acland et al. 2001, 2005; Annear et al. 2011; Bainbridge et al. 2003; Beltran 2009; Bennicelli et al. 2008). Many breeds are known to be susceptible to a form of RD showing breed-specific mode of inheritance (autosomal recessive, autosomal dominant, or X-linked), pathogenesis [dysplasia/degeneration affecting rod/cone photoreceptors or retinal pigment epithelium (RPE)], age of onset, and rate of progression. The unique and consistent phenotypes of RD within each breed correspond to distinct breed-specific mutations segregating within those breeds. All RDs previously investigated at the molecular level appear to show monogenic segregation caused by a single mutation. A diverse group of affected genes are often characterized by retina-specific expression or a retina-specific isoform disrupted by the mutation (Aguirre and Acland 2006).
A naturally occurring form of inherited RD has been recognized in Miniature longhaired dachshunds (MLHDs) as early as 1965 (Barnett 1965), while comprehensive studies to characterize its phenotype have been carried out using an MLHD research colony (Curtis and Barnett 1993; Mellersh et al. 2006). The colony was founded using a small number of affected MLHDs and developed by breeding closely related dogs where the disease segregated in an autosomal recessive manner. In affected dogs, ERG and histopathology indicated normal development of the retina followed by rapid degeneration, with the earliest sign of abnormalities detectable by ERG as early as 6 months old (Curtis and Barnett 1993). A more recent and extensive ERG study revealed initial dysfunction of the cone photoreceptors at 6 weeks of age followed by that of the rods by 40 weeks of age (Turney et al. 2007). The RD form observed in this research colony has been identified as CRD (Turney et al. 2007), and the disease in the wider population has been referred to as cone-rod dysplasia 1 (cord1) (Mellersh et al. 2006).
This research colony, described by Curtis and Barnett (1993) and Turney et al. (2007), was also used to investigate the molecular basis of cord1. Microsatellite-based homozygosity mapping followed by linkage analysis identified the cord1 locus as a 14.15 Mb interval on canine chromosome 15 (CFA15) (Mellersh et al. 2006). Sequencing of a strong positional–functional candidate gene, RPGRIP1 (RP ATPase regulator interacting protein 1) identified a 44 bp insertion (“RPGRIP1 insertion”) in exon 2. The resulting frameshift was predicted to lead to a premature stop codon, and, if unaffected by nonsense-mediated decay, give rise to markedly truncated protein lacking the C-terminus RPGR-interacting domain. As the RPGRIP1 insertion segregated completely with cord1 in the research colony (i.e., 15 RPGRIP1 −/−-affected cases and 14 RPGRIP1 +/−-obligate carriers), it was considered also to be causative of RD in the general MLHD population from which the founder of the research colony was derived.
However, by studying 264 MLHDs from the pet population, we have previously identified significant discordance between the RPGRIP1 genotype and the RD phenotype; 17% of clinically normal dogs aged 4 years or older were, unexpectedly, homozygous for the RPGRIP1 insertion (RPGRIP1 −/−) (Miyadera et al. 2009). The relative risk for clinical RD (in dogs of all ages) associated with the RPGRIP1 −/− genotype was 9.01 (CI95 = 5.06–16.04), but 20% of RD cases were not RPGRIP1 −/−, showing that forms of RD not associated with RPGRIP1 −/− are present. Furthermore, unlike the research colony, which uniformly showed an early onset, the age of clinical onset among cases from the pet population ranged broadly from 0.3 to 15 years among RPGRIP1 −/− dogs, showing a bimodal distribution. Hence it was likely that the population contained RPGRIP1 −/− individuals who were not clinically affected at the time of assessment but would show the disease signs later. Such reduced penetrance of the clinical signs of RD in RPGRIP1 −/− dogs as well as a striking variety in ages of RD onset was observed among 13 RPGRIP1 −/− dogs within a single extended MLHD family with a shared environment (Miyadera et al. 2009). Therefore, the cause of the phenotypic variation appeared to be genetic and with limited loci involved.
In the study described here, we sought to identify genetic factors controlling the variable age of onset as well as the presence or absence of RD among RPGRIP1 −/− dogs. Canine 22 K SNP array genotyping data from 83 RPGRIP1 −/− MLHDs were analysed. A genome-wide association study (GWAS) led to the identification of a single additional locus on CFA15 showing strong association with early-onset RD (EORD) while late-onset RD (LORD) and normal dogs above 4 years combined served as the best controls. Homozygosity mapping and haplotype analysis of the mapped region identified a 1.49 Mb critical interval. We have established that an early-onset form of RD or cord1 in MLHDs is caused by both RPGRIP1 −/− and homozygosity at this second locus, whereas RPGRIP1 −/− alone leads to either LORD or no clinically apparent phenotype when assessed by behaviour or ophthalmoscopy. This is the first evidence of a canine digenic disorder or that involving a modifier where two homozygous mutations at independent loci are required to lead to an EORD phenotype.
Materials and methods
Canine samples
DNA samples of 96 RPGRIP1 −/− MLHDs with either variable ages of RD onset or no clinical signs were used for microarray SNP genotyping (Table 1, Supplementary Table 1). For the clinically normal dogs, the oldest possible individuals (≥4.8 years) were included and younger dogs that could potentially develop RD later in life were excluded from the study. Of all the samples, 75 were unrelated Japanese pets according to three-generation pedigree records with an exception of one related pair (Supplementary Table 1).
Thirteen of the MLHDs from Japan were from a single extended family with shared environment (Family K). Eight samples were from the UK: six ophthalmologically normal pet dogs without pedigree information and two EORD cases from the research colony in the original cord1 studies (Curtis and Barnett 1993; Mellersh et al. 2006; Turney et al. 2007). To screen for polymorphisms within the interval mapped by GWAS, 13 RD-affected MLHDs from Japan that were not RPGRIP1 −/− (i.e., 5 RPGRIP1 +/− and 8 RPGRIP1 +/+) were used. All Japanese samples have been used in a previous study (Miyadera et al. 2009).
Diagnosis
Diagnostic methods and criteria have been detailed previously (Miyadera et al. 2009). General ophthalmic examination included menace response, pupillary light reflex, and dazzle reflex as initial assessment of visual function. Indirect ophthalmoscopy was performed using 14, 20, and 28 diopter lenses. Slit lamp biomicroscopy and intraocular pressure measurements were performed to exclude other ophthalmic conditions. Diagnosis of inherited RD was based on clinical histories of progressive visual impairment and ophthalmoscopic evidence of bilateral progressive retinal degeneration. The age of onset was defined as the age when ophthalmological abnormality was first observed or when the earliest signs of altered behaviour indicating abnormal vision were recognized. Dogs were determined to be normal by the absence of apparent ophthalmoscopic abnormality. In cases showing sudden onset of blindness with little ophthalmoscopic abnormality, ERG was performed at initial presentation; where ERG signals were lost, sudden acquired retinal degeneration (SARD) was diagnosed and such cases were not included in the study. All the affected MLHDs from Japan were examined by the same veterinary ophthalmologist (KK).
DNA preparation
DNA from the Japanese dogs was extracted from blood specimens as described previously (Miyadera et al. 2009). DNA from the UK pet and research colony dogs was extracted from cheek swabs and liver specimens, respectively. Blood-derived DNA was amplified by whole-genome amplification using either GenomePlex® Whole Genome Amplification Kit (Sigma-Aldrich, Dorset, UK) or Illustra™ GenomiPhi™ V2 DNA Amplification Kit (GE Healthcare Life Sciences, Little Chalfont, UK) for six samples used in the microarray study.
Microarray SNP genotyping
Genotyping was done using the Canine SNP20 BeadChip (Illumina, San Diego, CA, USA) on the BeadLab station (Illumina) at the Cambridge Genome Services (Department of Pathology, University of Cambridge) following the manufacturer’s protocol. The Canine SNP20 BeadChip types 22,362 SNPs genome-wide, of which, according to the manufacturer, 81.1% were polymorphic [minor allele frequency (MAF) > 0.05], whilst the average MAF was 0.23 in a panel of 32 control Dachshunds.
Genotypes were called using the BeadStudio 2.0 software (Illumina), using the cluster positions predetermined by the manufacturer for each SNP. Once the genotypes were automatically assigned, the calls were visually inspected to remove erroneous calls. Based on cluster separation, call frequency, Mendelian errors, heterozygote excess, minor allele frequency, and gender estimation, 2,189 monomorphic SNPs (9.8%) and 73 SNPs (0.3%) with visually bad genotyping quality were eliminated. Of the 96 samples genotyped, one sample (MLD001) showed a low genotyping rate of <98% and was discarded. Genotype reports were then converted to the appropriate format using the genetic data management software Progeny (Progeny Software, LLC., Delray Beach, FL, USA) before being imported to the PLINK program (version 1.07) (Purcell et al. 2007) for association analysis.
Genome-wide association analysis
One dog each from Family K and the original UK research colony was used in the analysis to avoid population stratification. Consequently, of the 96 dogs genotyped, 83 samples were analyzed in a series of different case–control GWAS analyses. These included 49 RD cases (0.3–16 years) and 34 ophthalmoscopically normal dogs (Normal, 4.8–14 years) (Table 1). Genome-wide association was calculated in various models of possible case–control combinations based on the SNP calls from these samples. SNPs with MAF > 0.01 and call rates of >90% were analyzed for association using the PLINK software, taking the data of each SNP independently (Purcell et al. 2007). Four different case–control combinations of the different phenotypic groups, Normal, LORD, MORD, and EORD, were studied (Table 2). Corrected genome-wide associations were calculated using a “max(T)” permutation procedure that corrects for multiple tests (100,000 permutations) and by the Bonferroni method, taking the number of independent tests as equal to the SNP number.
Sequencing of positional candidate genes
To find potential mutations associated with EORD, EORD cases (n = 1–5 for different amplicons) homozygous at the mapped 1.49 Mb interval and control cases (n = 1–5 for different amplicons) heterozygous at the same interval were screened. The exons and the exon–intron boundaries of four candidate genes, LRAT (lecithin retinol acyltransferase), GUCY1A3 (guanylate cyclase soluble subunit alpha-3), GUCY1B3 (guanylate cyclase soluble subunit beta-1), and MAP9 (microtubule-associated protein 9), were amplified by PCR and sequenced. For LRAT, the region 2.8 kb upstream of the translation initiation codon and the canine equivalent of human LRAT 3′-UTR were also sequenced. Primers are given in Supplementary Table 2 (LRAT) and Supplementary Table 3 (GUCY1A3, GUCY1B3, and MAP9).
Results
GWAS in search for loci controlling the age of RD onset
We hypothesized that the age of onset of RD in RPGRIP1 −/− MLHDs was controlled by one or more additional loci (Miyadera et al. 2009). To test this hypothesis, we used SNP microarray-based GWAS to map the potential gene modifiers in RPGRIP1 −/− dogs with different ages of clinical RD onset or whose eyes were normal. Due to the broad range of the age of onset, the affected dogs were sub-grouped as early-onset (EORD: n = 31, 0.3–3.4 years), mid-onset (MORD: n = 3, 4.0–5.2 years), or late-onset (LORD: n = 15, 5.6–16 years) (Fig. 1; Table 1). While the age of onset in the LORD cases could be as late as 16 years of age, had they been examined at a younger age they could have been Normal. In other words, the currently Normal dogs could potentially develop RD later in life. Therefore, it was not feasible to determine a definite boundary dividing the RPGRIP1 −/− population into cases and controls. To overcome this, four subgroups, EORD, MORD, LORD, and Normal, differing in age of clinical diagnosis or absence of disease phenotype, were combined in various ways to perform four alternative case–control analyses (Table 2, M1–M4).
Distribution of the age of RD onset in 49 RPGRIP1 −/− cases used in GWAS. EORD (<4 years), MORD (4–5.5 years), and LORD (>5.5 years) dogs are indicated in black, grey, and white bars, respectively. The vertical line with the hatched interval (11.6 ± 4.2 years) shows the reported lifespan in Dachshunds (mean ± SD; The Kennel Club 2004)
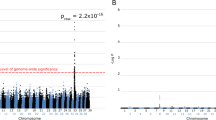
GWAS revealed a single strong association at CFA15 in all M1–M4 case-control models (Fig. 2a). The highest peak was found in the M1 model [p(raw) = 5.1 × 10−13] in which EORD cases were compared to LORD and Normal combined as controls. LORD only or Normal dogs only were used as controls in models M2 and M3, respectively. These models also showed strong association with the EORD cases [M2: p(raw) = 1.3 × 10−10, M3: p(raw) = 4.5 × 10−11] at the same locus. The association was weakest yet still noticeable in the M4 model [p(raw) = 1.5 × 10−6), where RD-affected dogs of any age were considered cases against the Normal controls. These observations suggest that the LORD group was genetically more similar to Normal rather than EORD or MORD and that LORD and Normal could be grouped together in this study. The second highest peak across all the case–control combinations could be found on CFA22 in the M2 model, among several other minor peaks.
Genome-wide association plots in different case–control models. The GWAS plots are compared in different case–control combinations (M1–M4) as defined in Table 2. a The level of significance for each SNP (−log10[p], y axis) is shown at the position on each chromosome (x axis). b Genome-wide significance of association was calculated using PLINK over 100,000 permutations. Each dot represents one of 19,506 SNPs passing quality control. Different coloured dots represent SNPs on different canine chromosomes
Data corrected for multiple hypotheses testing by permutation analysis (100,000 permutations) is shown in Fig. 2b. A single major peak at CFA15 is again prominent in all M1–M4 models, and the maximum –log[p] ≥ 5.0 value was reached in models M1 and M3. Bonferroni correction of the raw p values using number of SNPs gives for M1 [p(corr) = 1.07 × 10−8], and for M3 [p(corr) = 9.5 × 10−7]. Inspection of the association plot at CFA15 revealed consecutive SNPs showing strong association at around 55–60 Mb (Fig. 3). Interestingly, the interval mapped by GWAS fell on the same chromosome as the first cord1 locus containing RPGRIP1, identified in the research dogs (Mellersh et al. 2006). RPGRIP1 is located ~35 Mb further centromeric to the newly discovered second locus, and association plots (Fig. 3) and haplotype data (not shown) show that linkage disequilibrium (LD) does not extend across the two loci, suggesting that they are independent. As expected, the sample population, which was all RPGRIP1 −/−, showed no association (−log10[p] = 0) with the RPGRIP1 region (Fig. 3).
Regional CFA15 association plot in the M1 case–control study [31 EORD cases against 49 controls (15 LORD + 34 Normal)]. The y axis represents the level of significance for each SNP (−log10[p]) at the position on CFA15 along the x axis. Each dot represents a SNP on CFA15 passing quality control. The arrow indicates the location of RPGRIP1
Quantile–quantile plot
To evaluate the likelihood of false-positive genome-wide association findings caused by population stratification from some other source, a trend test of association was conducted for the 19,506 SNPs that passed quality control. The observed p values were compared to the expected values under the null hypothesis in a quantile–quantile (Q–Q) plot. The comparison of observed and expected p values in the M1 model showed significant deviation in SNPs at the higher end of −log10[p] (genomic inflation factor λ = 1.32) (Fig. 4). Based on the bottom 90% of SNPs, excluding the strong association and a small number of false-positive SNPs or possible additional contributing loci, the genomic inflation factor was λ = 0.96, implying a possibility of false-positive associations resulting from population stratification or cryptic relatedness.
Q–Q plot showing the observed p values against the expected p values in the M1 case–control study for all 19,506 SNPs (genomic inflation factor λ = 1.32). The ascending line represents the null model where the observed p value matches the expected value. Calculation based on the bottom 90% of SNPs of the Q–Q plot indicates background of minimal genomic inflation with low possibility of population stratification or cryptic relatedness (λ = 0.96), while the higher SNPs represent true association
Homozygosity mapping and haplotype analysis
Investigation of the top-hit SNP and its flanking SNPs on CFA15 in EORD cases led to the identification of a homozygosity region of 13 consecutive SNPs spanning 1.49 Mb (CFA15: 55,411,377–56,901,029 bp) (Supplementary Fig. 1). Haplotypes for the region were inferred using the PHASE program to interpret genotyping data from the various controls (Supplementary Fig. 2) where the haplotype predominant in the EORD cases is shown in red (EORD haplotype). While 90.3% of EORD (n = 28/31) were homozygous for the EORD haplotype, all MORD (n = 3), 13.3% of LORD (n = 2/15), and 2.9% of Normal (n = 1/34) were homozygous for the EORD haplotype (Supplementary Fig. 1). The only Normal dog homozygous for the EORD haplotype was 4.9 years old, younger than any of the LORD dogs. Of the 31 EORD dogs, three with the age of onset at 0.3, 2, and 3 years were not homozygous for the region of the EORD haplotype and carried part of or whole haplotype blocks present in the control chromosomes (shown as blue, green, or yellow blocks in Supplementary Fig. 2). The haplotype patterns of these three EORD dogs appeared to be substantially dissimilar to the rest of the EORD dogs.
Among the extended Japanese MLHD family whose 12 of 13 members were not used for GWAS (Family K: 2 EORD, 1 MORD, 5 LORD, and 5 Normal), RD cases with the four earliest ages of onset (<6.6 years) were homozygous for the EORD haplotype. The rest of the LORD and the Normal dogs carried only one or no EORD haplotype.
Sequencing of positional–functional candidate genes
According to the public genome databases (NCBI and Ensembl), the mapped 1.49 Mb interval (CFA15: 55.41–56.90 Mb) syntenic to human chromosome 4 contained two known (GUCY1A3 and GUCY1B3), seven predicted (LRAT, RBM46, NPY2R, MAP9, ACCN5, TDO2, and CTSO), and two hypothetical (LOC612503 and LOC475558) genes (Fig. 5). Inspection of these genes revealed a strong positional and functional candidate gene, LRAT (lecithin retinol acyltransferase). LRAT encodes a microsomal enzyme that catalyses the esterification of all-trans-retinol into all-trans-retinyl ester (Saari and Bredberg 1989). Sequencing of the coding and the noncoding LRAT regions identified a synonymous substitution in exon 1 (329G>A) and several polymorphisms in the noncoding regions as compared with the CanFam 2.0 reference sequence of the Boxer (Table 3). In addition, there were changes in copy number of three repeat elements in the EORD chromosome compared with the control chromosomes whilst both differ in copy number from the Boxer reference. Sequencing of three other positional candidate genes, GUCY1A3, GUCY1B3, and MAP9, in EORD cases and controls showed additional polymorphisms in all dogs compared with CanFam 2.0, but did not reveal any mutation segregating with the EORD phenotype.
Regional map of association with EORD on CFA15 (10,000 permutations) and the genes at the locus. The region highlighted in grey encompasses the 1.49 Mb EORD critical interval defined by homozygosity mapping and haplotype analysis (CFA15: 55.41–56.90 Mb, CanFam2.0). The locations of the corresponding known and predicted genes (Ensembl release 50, NCBI) are shown below the association plot. The four positional–functional candidate genes resequenced are underlined
Discussion
Until the current study, all recognized canine RDs were considered monogenic with high, if not complete, penetrance (Aguirre and Acland 2006; Miyadera et al. 2012). Cord1, a form of autosomal recessive RD in MLHDs, was characterized phenotypically in a research colony (Curtis and Barnett 1993), and a 44-bp insertion in RPGRIP1 has been associated with the disease (Mellersh et al. 2006). A DNA test to screen for this mutation allows breeders to avoid producing RPGRIP1 −/− puppies. Consequently, the proportion of RPGRIP1 −/− MLHDs undergoing DNA testing has decreased from 18 to 5% in the last 4 years (Animal Health Trust, UK). Meanwhile, we previously reported substantial variation in the age of onset and genotype–phenotype discordance among the pet MLHD population (Miyadera et al. 2009). The current study adds to the molecular picture of RD in MLHDs in the phenotypically and genetically heterogeneous pet population.
The 44-bp insertion in RPGRIP1 appears to be tightly linked to RD in the pet population of MLHDs (Miyadera et al. 2009), but penetrance of the RPGRIP1 −/− genotype is partial, and the genotype-specific relative risk for homozygotes is only around 9. This relative risk is low compared with most monogenic diseases, and the age of RD onset differs markedly in different RPGRIP1 −/− dogs. Here we have shown that a single additional locus could distinguish EORD from other RPGRIP1 −/− dogs (i.e., LORD and Normal). In the latter group, pathology may develop too slowly to become clinically apparent during life, thus manifesting as LORD in some dogs and as Normal in others. On the other hand, we have previously observed loss of cone ERG signals in apparently normal Beagles that were homozygous for a longer variant of the RPGRIP1 insertion (RPGRIP1 −L/−L) (Miyadera et al. 2009). Furthermore, cone ERG was found to be grossly reduced in apparently normal RPGRIP1 −/− MLHDs from the UK pet population (Busse et al. 2011). Hence, the consequence of RPGRIP1 −/− alone could be proposed as subclinical cone degeneration progressing into LORD with age-related incomplete penetrance.
It should be noted that the clinical subgroup LORD defined in the current study is not the direct homolog of the human condition, L-ORD (late-onset retinal degeneration), which has been molecularly characterized (Hayward et al. 2003; Jacobson et al. 2001; Milam et al. 2000).
There are human and murine diseases initially characterized as monogenic and later proven to be modulated or caused by two or more loci (Badano and Katsanis 2002). Depending on the level of the influence that the second locus exerts on the phenotype, it can be regarded as either a modifier or a synergistic second allele of a digenic disease. A modifier could be an extra variant that may not be pathogenic by itself but alters the primary phenotype caused by mutations in other genes. Examples of RDs include a mouse model of autosomal dominant RP caused by a Rho mutation where sequence variants in RPE65 either reduce or accelerate disease progression (Samardzija et al. 2006); a common RPGRIP1L variant modifies the phenotypic expressivity of ciliopathies caused by mutations in other genes (Khanna et al. 2009); and an AHI1 polymorphism increases the risk of retinal degeneration by sevenfold in human nephronophthisis patients caused by mutations in NPHP1 and other genes (Louie et al. 2010). Digenic RP has been reported in man where heterozygous mutations in both ROM1 and RDS are required to cause a seemingly dominant RP (Kajiwara et al. 1994). In some families with Bardet-Biedl syndrome (Fauser et al. 2003) or nephronophthisis (Hoefele et al. 2007), a mutation in a second gene may be required together with a homozygous mutation in the first gene to exhibit a clinical phenotype. Whether the mapped 1.49 Mb locus in the current study is a modifier of RPGRIP1 −/− or a second allele of a digenic condition remains to be confirmed. However, the delayed onset in the LORD dogs that fits into the “control” group indicates that the mapped locus is an accelerating modifier for a late to subclinical RD caused by RPGRIP1 −/−.
Only one of 13 RD-affected MLHDs that are not RPGRIP1 −/− (age = 0.3–9.6 years; mean ± SD = 5.9 ± 3.5 years) was homozygous for the EORD haplotype at the second locus (data not shown). This indicates that few, if any, RD cases may be caused by the mutation at the second RD locus alone. The high frequency of the EORD haplotype among the normal dogs suggests that the pathogenic effect of the second RD locus alone could be mild. As such, the causative mutation at this locus could be in the regulatory or noncoding region, or in microRNAs, which could regulate the expression of genes at a distance (Visel et al. 2010). Given the large distance between the two loci (35.2 Mb apart on CFA15), long-range cis effects may be unlikely.
Other than the strong hit at CFA15, several minor peaks of association were observed in other chromosomes in different case–control combinations. None of these peaks reached statistical significance after correction for multiple testing, so that none of the loci can be regarded as having proven involvement. It is possible that one or more of them account for a small proportion of the phenotypic variation that cannot be explained by RPGRIP1 −/− and the second locus. Further study of animals with both major loci fixed could identify additional loci accounting for an additional part of the variation.
In general, identification of loci involved in multigenic traits would not be as straightforward as in monogenic traits. With hindsight, the initial use of the inbred MLHD research colony facilitated mapping of the first RD locus (RPGRIP1) (Mellersh et al. 2006) helped by the phenotypic, genetic, and environmental uniformity. Based on the uniform EORD phenotype observed in the research colony and the non-identification of the second locus in the initial whole-genome scan (Mellersh et al. 2006), it is likely that all the dogs in the research colony were homozygous or “fixed” at the second locus. Indeed, typing of the founder dogs of the original research colony revealed that the three founders representing all the chromosomal variants in the colony were each homozygotes for the EORD haplotype (data not shown). Given that the entire colony is fixed for the EORD haplotype, the second locus does not seem to be evidently pathogenic on its own since the non-RPGRIP −/− dogs in the colony did not show clinical signs within the limit of the study (Curtis and Barnett 1993). In the subsequent GWAS we report in this article, using MLHDs from a genetically heterogeneous pet population and fixing the RPGRIP1 locus as RPGRIP1 −/−, we were able to map a second locus involved in RD.
Geographical mixing of samples may cause population stratification (Karlsson et al. 2007; Quignon et al. 2007). In our study, admixture of UK Dachshund DNA with a mainly Japanese sample group did not cause evident population stratification as assessed by Q–Q plot or Eigenstrat (data not shown), whilst increasing power. The relatively recent introduction of MLHDs to Japan and occasional gene flow from the UK to Japan may account for the lack of observable genetic divergence between the two groups.
Of the genes within the second RD interval, LRAT is a strong candidate gene based on its involvement in severe EORDs in human patients (Thompson et al. 2001). LRAT is expressed in the RPE as well as in the liver and converts all-trans-retinol (vitamin A) to all-trans-retinyl ester, which is an essential intermediate compound in the visual cycle (Saari et al. 1993). Lrat knockout mice (Lrat −/−) showed rapid cone degeneration after eye opening with normal rod trafficking (Zhang et al. 2008); RPE was devoid of all-trans-retinol or all-trans-retinyl esters; functional rhodopsin of the photoreceptors was absent; and ERG responses were highly reduced (Batten et al. 2004). Although RPGRIP1 and LRAT are not known to colocalise or interact directly, mutations in each gene may have a cumulative effect causing a disease phenotype. Human LRAT, but not that of mice, has an upstream noncoding exon. No equivalent canine exon is predicted in the current Ensembl, UCSC, and NCBI databases. An upstream sequence in the dog does align with both this exon and the proximal part of the human promoter. One difference between EORD and LORD alleles at LRAT is the number of 86 bp repeats in the putative intron 1 of this model. EORD alleles have one more repeat than LORD and two more than the CanFam2.0 reference sequence. The significance of this is not clear.
Other genes in the mapped interval could also be candidates. GUCY1A3 and GUCY1B3 are involved in cGMP biosynthesis and are members of the same membrane guanylyl cyclase family as GUCY2D (guanylate cyclase 2D). GUCY2D encodes a retina-specific guanylate cyclase causing Leber congenital amaurosis and cone-rod dystrophy-6 (Kelsell et al. 1998; Payne et al. 2001; Perrault et al. 1996), but GUCY1A3 and GUCY1B3 are not specific to retina and have known functions in response to nitric oxide. MAP9 is required for bipolar spindle assembly, mitotic progression, and cytokinesis (Saffin et al. 2005). As MAP9 could have a role in stabilizing microtubules, it may also affect the photoreceptor’s connecting cilium. The absence of candidate mutations in the coding and noncoding regions of LRAT and in the coding regions of three other candidate genes indicates that the causative mutation could lie outside of known genes and alter gene regulation, as happens in many human complex traits (Altshuler and Daly 2007). While the actual mutation at the second locus remains to be identified, ongoing studies include resequencing of the entire 1.49 Mb region by targeted next-generation sequencing. Furthermore, the functional consequence of polymorphisms identified in the genes involved in the pathogenesis of RD in MLHDs is being investigated at the RNA and protein levels.
Some RD-affected MLHDs are neither RPGRIP1 −/− nor homozygous for the EORD haplotype at the second locus, indicating the presence of further independent RD loci. As in human RDs, phenotypic and genotypic heterogeneities may complicate the clinical and molecular diagnoses (den Hollander et al. 2008; Hartong et al. 2006; Kennan et al. 2005) so much as to be described as complex traits (Wright et al. 2010). Environmental factors could also play a role as the dose of light acting on the photoreceptor is known to enhance the progression of RD in animals and humans (Sanyal and Hawkins 1986; Taylor et al. 1990; Wang et al. 1997). In autosomal dominant RD dogs with RHO mutation, the extent of light exposure has also dramatically accelerated neurodegeneration (Cideciyan et al. 2005).
Once the second mutation is identified, a direct DNA test for the two mutations will allow accurate prediction of the onset of RD. Dogs homozygous for both the RPGRIP1 insertion and the second locus would develop EORD. RPGRIP1 −/− dogs that are not homozygous for the second mutation should fit in the spectrum ranging from LORD to Normal. Yet, when applying DNA testing results to breeding, it is important that the risk levels and the frequencies of each locus be considered. More work at the population level will be required to measure these accurately.
Elucidating the molecular basis for RD in MLHDs showing either early or late to no onset has established a reliable canine model for the understanding of phenotypic and genetic variability observed in human RDs (Acland et al. 2001, 2005; Annear et al. 2011; Bainbridge et al. 2003; Bennicelli et al. 2008). The accurate prediction of the RD phenotype expressing either EORD or LORD to Normal according to the combination of the genotypes of two RD loci is essential in the assessment of gene therapy.
In conclusion, a GWAS in MLHD dogs homozygous for a previously found RD locus (RPGRIP1 −/−) and showing variable ages of RD onset or no clinical signs has identified an independent second RD locus that modifies the age of onset. Homozygosity at both the first (RPGRIP1 −/−) and the newly identified second locus on CFA15 is required to develop EORD, establishing the condition as the first canine RD involving at least two loci and a unique model for the heterogenic RDs in humans. Further analysis of the second locus, which is currently defined as a 1.49-Mb interval, is expected to identify the gene and the mutation and its pathogenic involvement in relation with RPGRIP1. Initially considered a simple Mendelian disease, we have established the involvement of a second genetic locus in EORD or cord1 in MLHDs.
References
Acland GM, Aguirre GD, Ray J, Zhang Q, Aleman TS, Cideciyan AV, Pearce-Kelling SE, Anand V, Zeng Y, Maguire AM, Jacobson SG, Hauswirth WW, Bennett J (2001) Gene therapy restores vision in a canine model of childhood blindness. Nat Genet 28:92–95
Acland GM, Aguirre GD, Bennett J, Aleman TS, Cideciyan AV, Bennicelli J, Dejneka NS, Pearce-Kelling SE, Maguire AM, Palczewski K, Hauswirth WW, Jacobson SG (2005) Long-term restoration of rod and cone vision by single dose rAAV-mediated gene transfer to the retina in a canine model of childhood blindness. Mol Ther 12:1072–1082
Aguirre GD, Acland GM (2006) Models, mutants and man: searching for unique phenotypes and genes in the dog model of inherited retinal degeneration. In: Ostrander EA, Giger U, Lindblad-Toh K (eds) The dog and its genome. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, pp 291–325
Altshuler D, Daly M (2007) Guilt beyond a reasonable doubt. Nat Genet 39:813–815
Annear MJ, Bartoe JT, Barker SE, Smith AJ, Curran PG, Bainbridge JW, Ali RR, Petersen-Jones SM (2011) Gene therapy in the second eye of RPE65-deficient dogs improves retinal function. Gene Ther 18:53–61
Badano JL, Katsanis N (2002) Beyond Mendel: an evolving view of human genetic disease transmission. Nat Rev Genet 3:779–789
Bainbridge JW, Mistry A, Schlichtenbrede FC, Smith A, Broderick C, De Alwis M, Georgiadis A, Taylor PM, Squires M, Sethi C, Charteris D, Thrasher AJ, Sargan D, Ali RR (2003) Stable rAAV-mediated transduction of rod and cone photoreceptors in the canine retina. Gene Ther 10:1336–1344
Barnett KC (1965) Retinal atrophy. Vet Rec 77:1543–1560
Batten ML, Imanishi Y, Maeda T, Tu DC, Moise AR, Bronson D, Possin D, Van Gelder RN, Baehr W, Palczewski K (2004) Lecithin-retinol acyltransferase is essential for accumulation of all-trans-retinyl esters in the eye and in the liver. J Biol Chem 279:10422–10432
Beltran WA (2009) The use of canine models of inherited retinal degeneration to test novel therapeutic approaches. Vet Ophthalmol 12:192–204
Bennicelli J, Wright JF, Komaromy A, Jacobs JB, Hauck B, Zelenaia O, Mingozzi F, Hui D, Chung D, Rex TS, Wei Z, Qu G, Zhou S, Zeiss C, Arruda VR, Acland GM, Dell’Osso LF, High KA, Maguire AM, Bennett J (2008) Reversal of blindness in animal models of leber congenital amaurosis using optimized AAV2-mediated gene transfer. Mol Ther 16:458–465
Busse C, Barnett KC, Mellersh CS, Adams VJ (2011) Ophthalmic and cone derived electrodiagnostic findings in outbred miniature long-haired dachshunds homozygous for a RPGRIP1 mutation. Vet Ophthalmol 14:146–152
Cideciyan AV, Jacobson SG, Aleman TS, Gu D, Pearce-Kelling SE, Sumaroka A, Acland GM, Aguirre GD (2005) In vivo dynamics of retinal injury and repair in the rhodopsin mutant dog model of human retinitis pigmentosa. Proc Natl Acad Sci USA 102:5233–5238
Curtis R, Barnett KC (1993) Progressive retinal atrophy in miniature longhaired dachshund dogs. Br Vet J 149:71–85
den Hollander AI, Roepman R, Koenekoop RK, Cremers FP (2008) Leber congenital amaurosis: genes, proteins and disease mechanisms. Prog Retin Eye Res 27:391–419
Fauser S, Munz M, Besch D (2003) Further support for digenic inheritance in Bardet-Biedl syndrome. J Med Genet 40:e104
Hartong DT, Berson EL, Dryja TP (2006) Retinitis pigmentosa. Lancet 368:1795–1809
Hayward C, Shu X, Cideciyan AV, Lennon A, Barran P, Zareparsi S, Sawyer L, Hendry G, Dhillon B, Milam AH, Luthert PJ, Swaroop A, Hastie ND, Jacobson SG, Wright AF (2003) Mutation in a short-chain collagen gene, CTRP5, results in extracellular deposit formation in late-onset retinal degeneration: a genetic model for age-related macular degeneration. Hum Mol Genet 12:2657–2667
Hoefele J, Wolf MT, O’Toole JF, Otto EA, Schultheiss U, Deschenes G, Attanasio M, Utsch B, Antignac C, Hildebrandt F (2007) Evidence of oligogenic inheritance in nephronophthisis. J Am Soc Nephrol 18:2789–2795
Jacobson SG, Cideciyan AV, Wright E, Wright AF (2001) Phenotypic marker for early disease detection in dominant late-onset retinal degeneration. Invest Ophthalmol Vis Sci 42:1882–1890
Kajiwara K, Berson EL, Dryja TP (1994) Digenic retinitis pigmentosa due to mutations at the unlinked peripherin/RDS and ROM1 loci. Science 264:1604–1608
Karlsson EK, Baranowska I, Wade CM, Salmon Hillbertz NH, Zody MC, Anderson N, Biagi TM, Patterson N, Pielberg GR, Kulbokas EJ 3rd, Comstock KE, Keller ET, Mesirov JP, von Euler H, Kampe O, Hedhammar A, Lander ES, Andersson G, Andersson L, Lindblad-Toh K (2007) Efficient mapping of mendelian traits in dogs through genome-wide association. Nat Genet 39:1321–1328
Kelsell RE, Gregory-Evans K, Payne AM, Perrault I, Kaplan J, Yang RB, Garbers DL, Bird AC, Moore AT, Hunt DM (1998) Mutations in the retinal guanylate cyclase (RETGC-1) gene in dominant cone-rod dystrophy. Hum Mol Genet 7:1179–1184
Kennan A, Aherne A, Humphries P (2005) Light in retinitis pigmentosa. Trends Genet 21:103–110
Khanna H, Davis EE, Murga-Zamalloa CA, Estrada-Cuzcano A, Lopez I, den Hollander AI, Zonneveld MN, Othman MI, Waseem N, Chakarova CF, Maubaret C, Diaz-Font A, Macdonald I, Muzny DM, Wheeler DA, Morgan M, Lewis LR, Logan CV, Tan PL, Beer MA, Inglehearn CF, Lewis RA, Jacobson SG, Bergmann C, Beales PL, Attie-Bitach T, Johnson CA, Otto EA, Bhattacharya SS, Hildebrandt F, Gibbs RA, Koenekoop RK, Swaroop A, Katsanis N (2009) A common allele in RPGRIP1L is a modifier of retinal degeneration in ciliopathies. Nat Genet 41:739–745
Louie CM, Caridi G, Lopes VS, Brancati F, Kispert A, Lancaster MA, Schlossman AM, Otto EA, Leitges M, Grone HJ, Lopez I, Gudiseva HV, O’Toole JF, Vallespin E, Ayyagari R, Ayuso C, Cremers FP, den Hollander AI, Koenekoop RK, Dallapiccola B, Ghiggeri GM, Hildebrandt F, Valente EM, Williams DS, Gleeson JG (2010) AHI1 is required for photoreceptor outer development and is a modifier for retinal degeneration in nephronophthisis. Nat Genet 42:175–180
Mellersh CS, Boursnell ME, Pettitt L, Ryder EJ, Holmes NG, Grafham D, Forman OP, Sampson J, Barnett KC, Blanton S, Binns MM, Vaudin M (2006) Canine RPGRIP1 mutation establishes cone-rod dystrophy in miniature longhaired dachshunds as a homologue of human Leber congenital amaurosis. Genomics 88:293–301
Milam AH, Curcio CA, Cideciyan AV, Saxena S, John SK, Kruth HS, Malek G, Heckenlively JR, Weleber RG, Jacobson SG (2000) Dominant late-onset retinal degeneration with regional variation of sub-retinal pigment epithelium deposits, retinal function, and photoreceptor degeneration. Ophthalmology 107:2256–2266
Miyadera K, Kato K, Aguirre-Hernandez J, Tokuriki T, Morimoto K, Busse C, Barnett K, Holmes N, Ogawa H, Sasaki N, Mellersh CS, Sargan DR (2009) Phenotypic variation and genotype–phenotype discordance in canine cone-rod dystrophy with an RPGRIP1 mutation. Mol Vis 15:2287–2305
Miyadera K, Acland GM, Aguiree GD (2012) Genetic and phenotypic variation of inherited retinal diseases in dogs: the power of within- and across-breed studies. Mamm Genome. doi:10.1007/s00335-011-9361-3
Payne AM, Morris AG, Downes SM, Johnson S, Bird AC, Moore AT, Bhattacharya SS, Hunt DM (2001) Clustering and frequency of mutations in the retinal guanylate cyclase (GUCY2D) gene in patients with dominant cone-rod dystrophies. J Med Genet 38:611–614
Perrault I, Rozet JM, Calvas P, Gerber S, Camuzat A, Dollfus H, Chatelin S, Souied E, Ghazi I, Leowski C, Bonnemaison M, Le Paslier D, Frezal J, Dufier JL, Pittler S, Munnich A, Kaplan J (1996) Retinal-specific guanylate cyclase gene mutations in Leber’s congenital amaurosis. Nat Genet 14:461–464
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575
Quignon P, Herbin L, Cadieu E, Kirkness EF, Hedan B, Mosher DS, Galibert F, Andre C, Ostrander EA, Hitte C (2007) Canine population structure: assessment and impact of intra-breed stratification on SNP-based association studies. PLoS One 2:e1324
Saari JC, Bredberg DL (1989) Lecithin:retinol acyltransferase in retinal pigment epithelial microsomes. J Biol Chem 264:8636–8640
Saari JC, Bredberg DL, Farrell DF (1993) Retinol esterification in bovine retinal pigment epithelium: reversibility of lecithin:retinol acyltransferase. Biochem J 291(Pt 3):697–700
Saffin JM, Venoux M, Prigent C, Espeut J, Poulat F, Giorgi D, Abrieu A, Rouquier S (2005) ASAP, a human microtubule-associated protein required for bipolar spindle assembly and cytokinesis. Proc Natl Acad Sci USA 102:11302–11307
Samardzija M, Wenzel A, Naash M, Reme CE, Grimm C (2006) Rpe65 as a modifier gene for inherited retinal degeneration. Eur J Neurosci 23:1028–1034
Sanyal S, Hawkins RK (1986) Development and degeneration of retina in rds mutant mice: effects of light on the rate of degeneration in albino and pigmented homozygous and heterozygous mutant and normal mice. Vision Res 26:1177–1185
Taylor HR, Munoz B, West S, Bressler NM, Bressler SB, Rosenthal FS (1990) Visible light and risk of age-related macular degeneration. Trans Am Ophthalmol Soc 88:163–173 discussion 173–168
The Kennel Club/British Small Animal Veterinary Association Purebred Dog Health Survey (2004) http://www.thekennelclub.org.uk/item/549. Accessed 29 Sep 2011
Thompson DA, Li Y, McHenry CL, Carlson TJ, Ding X, Sieving PA, Apfelstedt-Sylla E, Gal A (2001) Mutations in the gene encoding lecithin retinol acyltransferase are associated with early-onset severe retinal dystrophy. Nat Genet 28:123–124
Turney C, Chong NH, Alexander RA, Hogg CR, Fleming L, Flack D, Barnett KC, Bird AC, Holder GE, Luthert PJ (2007) Pathological and electrophysiological features of a canine cone-rod dystrophy in the miniature longhaired dachshund. Invest Ophthalmol Vis Sci 48:4240–4249
Visel A, Zhu Y, May D, Afzal V, Gong E, Attanasio C, Blow MJ, Cohen JC, Rubin EM, Pennacchio LA (2010) Targeted deletion of the 9p21 non-coding coronary artery disease risk interval in mice. Nature 464:409–412
Wang M, Lam TT, Tso MO, Naash MI (1997) Expression of a mutant opsin gene increases the susceptibility of the retina to light damage. Vis Neurosci 14:55–62
Wright AF, Chakarova CF, Abd El-Aziz MM, Bhattacharya SS (2010) Photoreceptor degeneration: genetic and mechanistic dissection of a complex trait. Nat Rev Genet 11:273–284
Zhang H, Fan J, Li S, Karan S, Rohrer B, Palczewski K, Frederick JM, Crouch RK, Baehr W (2008) Trafficking of membrane-associated proteins to cone photoreceptor outer segments requires the chromophore 11-cis-retinal. J Neurosci 28:4008–4014
Acknowledgments
The authors thank Emily Clemente at Cambridge Genomic Services, Department of Pathology, University of Cambridge, for microarray genotyping and Oliver Forman for helpful discussions. The many veterinary clinicians and dog owners across Japan, the UK, and elsewhere are gratefully acknowledged for their participation in the study. This study was supported by the Kennel Club Charitable Trust (RG55218).
Disclosure
Cathryn S. Mellersh is affiliated with the Animal Health Trust, UK, and a charitable organization offering DNA testing for RD in MLHDs.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Miyadera, K., Kato, K., Boursnell, M. et al. Genome-wide association study in RPGRIP1 −/− dogs identifies a modifier locus that determines the onset of retinal degeneration. Mamm Genome 23, 212–223 (2012). https://doi.org/10.1007/s00335-011-9384-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00335-011-9384-9