Abstract
Purpose
To assess the role of the MR radiomic signature in preoperative prediction of lymph node (LN) metastasis in patients with esophageal cancer (EC).
Patients and methods
A total of 181 EC patients were enrolled in this study between April 2015 and September 2017. Their LN metastases were pathologically confirmed. The first half of this cohort (90 patients) was set as the training cohort, and the second half (91 patients) was set as the validation cohort. A total of 1578 radiomic features were extracted from MR images (T2-TSE-BLADE and contrast-enhanced StarVIBE). The lasso and elastic net regression model was exploited for dimension reduction and selection of the feature space. The multivariable logistic regression analysis was adopted to identify the radiomic signature of pathologically involved LNs. The discriminating performance was assessed with the area under receiver-operating characteristic curve (AUC). The Mann-Whitney U test was adopted for testing the potential correlation of the radiomic signature and the LN status in both training and validation cohorts.
Results
Nine radiomic features were selected to create the radiomic signature significantly associated with LN metastasis (p < 0.001). AUC of radiomic signature performance in the training cohort was 0.821 (95% CI: 0.7042-0.9376) and in the validation cohort was 0.762 (95% CI: 0.7127-0.812). This model showed good discrimination between metastatic and non-metastatic lymph nodes.
Conclusion
The present study showed MRI radiomic features that could potentially predict metastatic LN involvement in the preoperative evaluation of EC patients.
Key Points
• The role of MRI in preoperative staging of esophageal cancer patients is increasing.
• MRI radiomic features showed the ability to predict LN metastasis in EC patients.
• ICCs showed excellent interreader agreement of the extracted MR features.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The presence of lymph node metastasis is an important prognostic factor for curable esophageal cancer (EC) [7, 13, 19]. Lymphatic spread of EC is highly variable and unpredictable because of the unique submucosal lymphatic drainage system of the esophagus [9]. The presence of lymph node involvement is generally associated with worse overall survival [24]. All positive lymph nodes should be removed together with the tumor to improve long-term survival. Extended lymph node resection might increase the incidence of postoperative recurrence and worsen the prognosis of EC patients [4]. Lymph node staging is also important for the decision to administer neoadjuvant therapy [23]. Therefore, lymph node involvement should be assessed before treatment. However, the detection accuracy of positive lymph nodes on preoperative CT still remains controversial, and the reported sensitivity, specificity and accuracy ranged between 37.3%-67.2%, 63.9%-96.4% and 85.8%-87.2%, respectively [12]. MRI has been shown to be more accurate in detecting positive lymph nodes in other cancers [6, 14, 20]. However, conventional MRI has limited image quality in the chest. Recently, contrast-enhanced StarVIBE has been applied in the detection of EC because of its high image quality [18], especially in patients who are unable to suspend respiration [2]. Meanwhile, T2-TSE-BLADE involves an acquisition scheme similar to the periodically rotated overlapping parallel lines with an enhanced reconstruction (BLADE) technique [17], and it may decrease motion artifacts in non-cooperative patients.
Radiomics is gaining momentum in cancer research [11]. High-throughput mining extracts quantitative image features from digitally encrypted medical images, and this is coupled with powerful image-based signatures that could potentially enhance precision diagnosis and treatment. Radiomic research recently revealed the potential of MRI to substantially improve the ability to detect or predict lymph node metastases [5, 11].
In this study, we aim to build and validate a MR radiomic-based model based on T2-TSE-BLADE and contrast-enhanced StarVIBE for predicting LN metastasis in preoperative EC patients.
Materials & method
Patients
This prospective study was approved by the institutional review board, and the patients’ informed consents were obtained. In the present study, 181 patients with EC were enrolled between April 2015 and September 2017. Pathological confirmation of lymph node metastasis status was obtained in all cases.
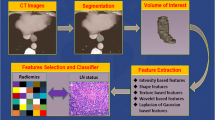
Inclusion criteria were: (1) patients with endoscopically biopsy-proven potentially resectable EC and T1/T2/T3/T4a staging by CT; (2) patients who received lymph node dissection within 7 days after the baseline MRI study; (3) patients who had pathologically confirmed LN status after surgery. Exclusion criteria were: (1) patients who were under 18 years of age; (2) patients who received treatment (radiotherapy or chemotherapy) before surgery; (3) patients who had received prior treatment in other institutions; (4) histological grade was not confirmed; (e) patients who could not tolerate the MR examination. The flowchart of patients included this study is shown in Fig. 1.
Image acquisition
All MR images were reconstructed with a standard kernel. These MR images were retrieved from the picture archiving and communication system (PACS) (Neusoft v5.5.60801).
MRI examination was performed on a 3-T MR scanner (MAGNETOM Skyra, Siemens Healthcare) with routine sequences, T2-TSE-BLADE and contrast-enhanced StarVIBE. The total scanning time was about 40 min. T2-TSE-BLADE was performed with the following parameters: TR/TE = 5000 ms/97 ms; voxel = 0.9 mm × 0.9 mm × 3.0 mm; FOV = 240 mm × 240 mm, scan time = 240 s-360 s. StarVIBE was performed for the whole chest during free breathing at 20 s post-contrast media administration with the following parameters: TR/TE = 3.98 ms/1.91 ms; FOV = 300 mm × 300 mm × 72 mm; resolution = 1.0 mm × 1.0 mm × 3.0 mm; flip angle = 12 degrees; radial views = 1659; acquisition time = 309 s. Gadopentetate dimeglumine [0.2 ml/kg of body weight, (Consun)] was injected at a rate of 2.5 ml/s by a MR-compatible automated double-tube high-pressure injector (Spectris Solaris EP, Medrad) immediately followed by an equal volume of normal saline solution to flush the tube.
Tumor segmentation
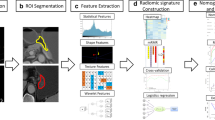
Manual segmentation of the EC was performed on each patient's MR images utilizing “ITK-SNAP” (www.itksnap.org), which is an open-source and free software application (Fig. 2). Two radiologists (reader 1 and 2), both with 5 years of chest radiology experience, carefully contoured the tumor on all T2-TSE-BLADE and StarVIBE images to generate two 3D segmentations of the entire tumor. A senior (reader 3) with > 15 years of chest radiology experience examined all segmented tumors and selected the best segmentation to include in the analysis. These regions of interests (ROIs) were performed for subsequent feature extraction for further analysis.
Flowchart of this study: (I) Original MR images: T2-TSE-BLADE and contrast-enhanced StarVIBE. (II) Segmentation was performed on both T2-TSE-BLADE and contrast-enhanced StarVIBE images to define the tumor region. (III) Radiomic features were extracted from the tumor region, including shape, first-order histogram, texture and wavelet group analysis. (IV) Several features were selected to build the radiomic signature using the least absolute shrinkage and selection operator (LASSO) method. (V) Finally, the classification ability of the radiomic signature was tested by the receiver-operating characteristics (ROC) curves in both the training and validation cohort
Radiomic feature calculation, selection, and signature building
Patients were allocated into a training and validation cohort depending on the date of surgery. MR images of the first half of this cohort (90 patients with surgery performed between April 2015 and September 2017) were set as the training cohort. The remaining half (91 patients with surgery performed by September 2017) were set as the validation cohort.
Radiomic features were calculated based on the segmentation results from the training cohort using a homemade program in the Matlab (Matlab 2014b). Features were categorized into three primary types: (1) 8 shapes and 1 size feature; (2) 14 first-order histogram statistics and (3) 116 texture-based features. The following procedure was to add five additional wavelet filters on MR images and re-calculate type 2 and type 3 features; hence, a total of 789 features of each tumor were obtained (9 + 14 + 116 + 5 * 130 = 789). For each MRI sequence, the 789 total extracted features covered the major feature pool in recent radiomic studies. The details of the radiomic feature calculation is shown in the Appendix [10]. Since two MR sequences were utilized, there were 1578 radiomic features in total for each patient.
The proper feature selection procedure was used to simplify the building model and avoid over-fitting issues. The “elastic net” approach was used to select the calculated features, which could be considered a combination the LASSO (least absolute shrinkage selection operator) and the ridge regression approaches. Ten-fold cross validation was used in the parameter tuning of the “elastic net.” For tuning coefficient λ and α, the criterion of minimum standard deviation and maximum AUC was followed, respectively.
The logistic regression model was exploited in the “elastic net” approach to build the radiomic signature for each patient. After the training procedure, a linear combination of the selected features was extracted. The sums of those linear combinations formed the radiomic signatures of each patient.
Statistics analysis
Statistical analysis was performed in R (version 3.3.0; http://www.Rproject.org). The package used is in Appendix Table S1. The statistical significance levels were all set as two sided at p < 0.05 in the current study. Interobserver reproducibility of radiomic feature extraction was assessed by inter-class correlation coefficients (ICCs). An ICC > 0.75 was considered good agreement. A kappa test was used to evaluate the differences between the features generated by reader 1 and 2.
A two-sample t-test was conducted for common comparisons of patients’ characteristics for continuous variables, and Fisher’s exact test and the χ2 test were used for categorical variables. The Mann-Whitney U test was adopted for testing the potential correlation of the radiomic signature and the LN status in both the training and validation cohorts.
Results
Patients characteristics
Table 1 shows the demographic statistics of patients in the training and validation cohorts. There were no significant differences between the training and validation cohorts in terms of gender, age, position of the tumor (upper, middle and lower parts of the thoracic esophagus), post-treatment T stage and post-treatment N stage.
Radiomic features
ICCs showed the excellence of 1578 extracting features from the two MRI sequences as assessed by the two radiologists, and the kappa value was 0.943. The features were selected with non-zero coefficients, based on the elastic net approach in the training cohort. As a result, 9 out of 1578 radiomic features were included. These were the included t2_length, t2_Sphericity, t2_ a2_GLRL_GLN_90, t2_hd_GLCM_CONTRAST_0, t2_vd_GLRL_RLN_0, dyn_a1_GLCM_PROBABILITY_90, dyn_a1_GLRL_SRE_135, dyn_hd_GLRL_HGRE_90 and dyn_dd_ENTROPY. These features included two shape and size features, six texture features and one wavelet filter feature. The parameter-tuning procedure of the regression model and the feature space reduction are illustrated in Fig. 3, and the name and description of the selected features are listed in Table 2. Distribution of nine features that could distinguish between positive and negative lymph node metastasis was analyzed by t-test. Of those, the five features (t2_length, t2_Sphericity, t2_hd_GLCM_CONTRAST_0, t2_vd_GLRL_RLN_0, dyn_hd_GLRL_HGRE_90) had a p value < 0.05.The p value is shown in Fig. 4.
Feature selection using the elastic net method with a logistic regression model. (a) Tuning parameter λ in the elastic net model and λ were selected under the minimum criteria. The vertical line was drawn at the value chosen according to 10-fold cross-validation, including nine optimized nonzero coefficients. (b) The model coefficient trend lines of the 1578 radiomic features. A coefficient profile plot was performed by coefficients against the L1 norm (inverse proportional to log λ)
Radiomic signature discrimination
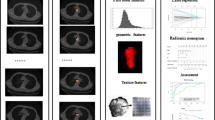
The radiomic signature was built by employing the selected features in the last section, which is the linear combination of the logistic regression model of those features. The radiomic signature’s discriminative power of the LN metastasis was assessed by two ROCs in the training and validation cohorts (Fig. 5). Radiomic scores (Rad scores) of EC patients in the training and validation cohorts were calculated through the elastic net model with selected features with their corresponding weights. Each patient’s Rad scores in both the training and validation cohorts are shown in Fig. 6.
ROCs were employed to assess the MRI radiomic signature discriminative performance of the LN metastasis in preoperative esophageal cancer patients. AUC in the training cohort with 0.821 (95% CI: 0.7042-0.9376, sensitivity = 68.0%, specificity = 92.3%) and AUC in the validation cohort with 0.762 (95% CI: 0.7127-0.812, sensitivity = 80.7%, specificity = 70.3%)
Discussion
The present study showed that the proposed MRI radiomics-based model involving the radiomic signature and radiological observation factors has potential ability in predicting LN metastasis preoperatively in EC patients. These observations are based on high-quality T2-TSE-BLADE and contrast-enhanced StarVIBE.
Lymph node status is the single most important prognostic factor in EC [1, 21]. CT is the most commonly utilized imaging modality in staging patients with EC. However, the accuracy of CT in detecting lymph node metastasis in EC is still controversial, because detection of pathological lymph nodes on CT depends primarily on size criteria [12]. MRI has higher soft tissue resolution than CT, and a prior study demonstrated its superiority to CT in T staging for EC patients [18].
The present study showed the feasibility of the MR radiomic feature for predicting LN metastasis in EC patients. We extracted 1578 quantitative image features of tumors using both T2-TSE-BLADE and contrast-enhanced StarVIBE. These sequences can provide high image quality and anatomic details in EC with the ability to accurately delineate the different layers of the esophageal wall. Hence, both T2-TSE-BLADE and contrast-enhanced StarVIBE were feasible in texture analysis. The image features assessed included shape, first order histogram, texture and wavelet group analysis and the elastic net method with a logistic regression model that was employed to reduce dimensionality. The current study showed that a stable classification mode with a similar AUC value in both the primary and validation cohorts based on the MR images and radiomic features obtained from both T2-TSE-BLADE and contrast-enhanced StarVIBE are useful to differentiate metastatic from non-metastatic lymph nodes.
Gray-level histograms and texture features have been useful in feature extraction and in discriminating between benign and malignant lesions [3, 25]. Texture features analysis on MR imaging was also proven to discriminate between benign and malignant masses of the breast [16]. In the present study, nine radiomic features, which included two shape and size features, six texture features and one wavelet filter feature, were selected to build the MR model, which was useful in assessing lymph node status in EC, and 9 features of the 181 cases had a proper ratio for building a predicting model that could avoid overfitting. It is still challenging to identify other radiomic features related to lymph node status in EC patients. Features of t2_length and t2_Sphericity, which are shape and size features, are highly consistent with the radiologists’ experience, and they describe the external contour information of the tumor. The longer length and larger sphericity indicate more tumor invasions; hence, this leads to higher risk of LN metastasis. Although these two features can be captured subjectively, additional features were extracted from MR images of EC patients, and these can be quantified and statistically analyzed. These six texture features and one wavelet filter feature include t2_ a2_GLRL_GLN_90, t2_hd_GLCM_CONTRAST_0, t2_vd_GLRL_RLN_0, dyn_a1_GLCM_PROBABILITY_90, dyn_a1_GLRL_SRE_135, dyn_hd_GLRL_HGRE_90 and dyn dd_ENTROPY and mainly represent the texture complexity of tumors, which was highly associated with the tumors’ heterogeneity, and prognosis [15, 26].
There are some limitations to this study. First, it did not analyze multimodality medical images, especially CT images. Second, MR images did not include DWI, which may expand the feature pool and show more valuable radiomic features because the image quality of DWI at 3.0 T may be associated with significant artifacts, although DWI has shown powerful ability in differentiating benign and malignant lymph nodes in some cancers [8, 22]. Third, a larger sample size will improve the confidence and performance of this EC model. Moreover, a larger multicenter database combining genomic and radiomic information could potentially improve the confidence and performance of the current model.
In conclusion, our study showed that MR radiomic features have the potential to predict lymph node status in EC patients. They could be used clinically to assess lymph node status for EC patients preoperatively.
Abbreviations
- AUC:
-
Area under receiver operating characteristic curve
- CT:
-
Computed tomography
- EC:
-
Esophageal cancer
- ICC:
-
Interclass correlation coefficient
- LASSO:
-
Least absolute shrinkage selection operator
- LN:
-
Lymph node
- MRI:
-
Magnetic resonance imaging
- ROI:
-
Regions of interest
- T2WI:
-
T2-weighted imaging
References
Akutsu Y, Matsubara H (2011) The significance of lymph node status as a prognostic factor for esophageal cancer. Surg Today 41:1190–1195
Azevedo RM, de Campos RO, Ramalho M, Heredia V, Dale BM, Semelka RC (2011) Free-breathing 3D T1-weighted gradient-echo sequence with radial data sampling in abdominal MRI: preliminary observations. AJR Am J Roentgenol 197:650–657
Bayanati H, R ET, Souza CA et al (2015) Quantitative CT texture and shape analysis: can it differentiate benign and malignant mediastinal lymph nodes in patients with primary lung cancer? Eur Radiol 25:480–487
Booka E, Takeuchi H, Nishi T et al (2015) The impact of postoperative complications on survivals after esophagectomy for esophageal cancer. Medicine (Baltimore) 94:e1369
Coroller TP, Agrawal V, Huynh E et al (2017) Radiomic-based pathological response prediction from primary tumors and lymph nodes in NSCLC. J Thorac Oncol 12:467–476
Dappa E, Elger T, Hasenburg A, Duber C, Battista MJ, Hotker AM (2017) The value of advanced MRI techniques in the assessment of cervical cancer: a review. Insights Imaging 8:471–481
Hofstetter W, Correa AM, Bekele N et al (2007) Proposed modification of nodal status in AJCC esophageal cancer staging system. Ann Thorac Surg 84:365–373 discussion 374-365
Holzapfel K, Gaa J, Schubert EC et al (2016) Value of diffusion-weighted MR imaging in the diagnosis of lymph node metastases in patients with cholangiocarcinoma. Abdom Radiol 41:1937–1941
Hosch SB, Stoecklein NH, Pichlmeier U et al (2001) Esophageal cancer: the mode of lymphatic tumor cell spread and its prognostic significance. J Clin Oncol 19:1970–1975
Huang YQ, Liang CH, He L et al (2016) Development and validation of a radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J Clin Oncol 34:2157–2164
Lambin P, Leijenaar RTH, Deist TM et al (2017) Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 14:749–762
Liu J, Wang Z, Shao H, Qu D, Liu J, Yao L (2017) Improving CT detection sensitivity for nodal metastases in oesophageal cancer with combination of smaller size and lymph node axial ratio. Eur Radiol. https://doi.org/10.1007/s00330-017-4935-4
Mariette C, Piessen G, Briez N, Triboulet JP (2008) The number of metastatic lymph nodes and the ratio between metastatic and examined lymph nodes are independent prognostic factors in esophageal cancer regardless of neoadjuvant chemoradiation or lymphadenectomy extent. Ann Surg 247:365–371
Moreno CC, Sullivan PS, Mittal PK (2017) MRI Evaluation of rectal cancer: staging and restaging. Curr Probl Diagn Radiol 46:234–241
Ng F, Ganeshan B, Kozarski R, Miles KA, Goh V (2013) Assessment of primary colorectal cancer heterogeneity by using whole-tumor texture analysis: contrast-enhanced CT texture as a biomarker of 5-year survival. Radiology 266:177–184
Nie K, Chen JH, Yu HJ, Chu Y, Nalcioglu O, Su MY (2008) Quantitative analysis of lesion morphology and texture features for diagnostic prediction in breast MRI. Acad Radiol 15:1513–1525
Ohgiya Y, Suyama J, Seino N et al (2010) MRI of the neck at 3 Tesla using the periodically rotated overlapping parallel lines with enhanced reconstruction (PROPELLER) (BLADE) sequence compared with T2-weighted fast spin-echo sequence. J Magn Reson Imaging 32:1061–1067
Qu J, Zhang H, Wang Z et al (2018) Comparison between free-breathing radial VIBE on 3-T MRI and endoscopic ultrasound for preoperative T staging of resectable oesophageal cancer, with histopathological correlation. Eur Radiol 28:780–787
Rice TW, Chen LQ, Hofstetter WL et al (2016) Worldwide Esophageal Cancer Collaboration: pathologic staging data. Dis Esophagus 29:724–733
Shen G, Zhou H, Jia Z, Deng H (2015) Diagnostic performance of diffusion-weighted MRI for detection of pelvic metastatic lymph nodes in patients with cervical cancer: a systematic review and meta-analysis. Br J Radiol 88:20150063
Twine CP, Lewis WG, Morgan MA et al (2009) The assessment of prognosis of surgically resected oesophageal cancer is dependent on the number of lymph nodes examined pathologically. Histopathology 55:46–52
Vag T, Heck MM, Beer AJ et al (2014) Preoperative lymph node staging in patients with primary prostate cancer: comparison and correlation of quantitative imaging parameters in diffusion-weighted imaging and 11C-choline PET/CT. Eur Radiol 24:1821–1826
van Rossum PS, van Hillegersberg R, Lever FM et al (2013) Imaging strategies in the management of oesophageal cancer: what's the role of MRI? Eur Radiol 23:1753–1765
Visser E, van Rossum PSN, Ruurda JP, van Hillegersberg R (2017) Impact of lymph node yield on overall survival in patients treated with neoadjuvant chemoradiotherapy followed by esophagectomy for cancer: a population-based cohort study in the Netherlands. Ann Surg 266:863–869
Woods BJ, Clymer BD, Kurc T et al (2007) Malignant-lesion segmentation using 4D co-occurrence texture analysis applied to dynamic contrast-enhanced magnetic resonance breast image data. J Magn Reson Imaging 25:495–501
Yip C, Davnall F, Kozarski R et al (2015) Assessment of changes in tumor heterogeneity following neoadjuvant chemotherapy in primary esophageal cancer. Dis Esophagus 28:172–179
Funding
This study has received funding from the National Natural Science Foundation of China (nos. 81501549, 81772012), the National Key Research and Development Plan of China under grant nos. 2017YFA0205200 and 2016YFC0103001, Beijing Municipal Science & Technology Commission (no. Z171100000117023) and special funding from the Henan Health Science and Technology Innovation Talent Project (no. 201004057).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Guarantor
The scientific guarantor of this publication is Hailiang Li.
Conflict of interest
The authors of this manuscript declare relationships with the following companies: Siemens. Three authors from Siemens provided the prototype sequence and reviewed the paper without any conflict of interest.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• retrospective
• case-control study/observational/experimental
• performed at one institution
Electronic supplementary material
ESM 1
(DOCX 28 kb)
Rights and permissions
About this article
Cite this article
Qu, J., Shen, C., Qin, J. et al. The MR radiomic signature can predict preoperative lymph node metastasis in patients with esophageal cancer. Eur Radiol 29, 906–914 (2019). https://doi.org/10.1007/s00330-018-5583-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-018-5583-z