Abstract
Four independent molecular data sets were sequenced in order to solve longstanding phylogenetic problems among Antarctic teleosts of the family Nototheniidae. The anatomical data of Balushkin (2000) were also coded into a matrix of 106 characters in order to test the parsimony of his taxonomic conclusions. Molecular results confirm Balushkin’s Pleuragrammatinae but not his Nototheniinae. Different genes used here found the “clade A” establishing the paraphyly of the Nototheniinae sensu lato; i.e. Lepidonotothen and Patagonotothen are more closely related to the Trematominae than to Notothenia. The genus Notothenia is paraphyletic and Paranotothenia should become Notothenia. Previously no molecular data set could assign a reliable position for the genus Gobionotothen. For the first time robust results are obtained for the phylogeny among the Trematominae. Trematomus scotti is the sister-group of all others, then Trematomus newnesi emerges, then Trematomus eulepidotus. Among the crown group, three clades emerge: 1: Trematomus hansoni + Trematomus bernacchii + Trematomus vicarius; 2: Trematomus pennellii + Trematomus lepidorhinus + Trematomus loennbergii; 3: Trematomus (Pagothenia) borchgrevinki + Trematomus nicolai. Pagothenia should become Trematomus to make the genus Trematomus monophyletic. The Trematomus tree found here did not match the topology obtained with Balushkin’s morphological matrix. The tree shows that the tendencies shown by some trematomines to secondarily colonize the water column are not gained through common ancestry.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Nototheniidae is the most speciose family of the Notothenioidei, the dominant taxonomic component of Antarctic teleosts. The suborder represents 35% of the “fish” species of the Southern Ocean, for which 97% are endemic and 46% of the fish species and 90% of the fish biomass of the continental shelf and upper slope (DeWitt 1971; Eastman and Clarke 1998). The family, in its traditional sense (DeWitt et al. 1990), was composed of the Eleginopinae, the Nototheniinae, the Trematominae and the Pleuragramminae, with various taxonomic contents according to different authors. One of the major recent advances concerning the delineation of the family was the exclusion of Eleginops based on morphological data (Balushkin 1992, 2000) as well as molecular data (Lecointre et al. 1997; Bargelloni and Lecointre 1998; Bargelloni et al. 2000; Near et al. 2004). Eleginops now appears as the sister-group of the rest of the non-bovichtid notothenioids. This is consistent with the fact that Eleginops is subantarctic and, interestingly, a basal nototheniid devoid of antifreeze proteins. Indeed, when mapped onto a tree (for instance in Lecointre and Ozouf-Costaz 2004 or Near et al. 2004), presence of antifreeze proteins imply that they have been acquired in the sister-group of Eleginops, in an ancestor of the rest of non-bovichtid notothenioids. In the present paper we will use the terms Nototheniidae or nototheniids in the restricted sense, i.e. without Eleginops (Balushkin 1992). The Nototheniidae sensu stricto are classically divided into three subfamilies (DeWitt et al. 1990; Balushkin 2000): the Nototheniinae (Notothenia, Paranotothenia, Gobionotothen, Lepidonotothen and Patagonotothen), the Trematominae (Trematomus, Pagothenia), and the Pleuragramminae (DeWitt et al. 1990) or Pleuragrammatinae (Balushkin 2000) (Dissostichus, Pleuragramma, Aethotaxis and Gvozdarus). With 50 known species, the nototheniids in that sense are the most morphologically diverse family of the suborder. Paradoxically, morphologists tended to consider them as monophyletic (Iwami 1985; Hastings 1993; Balushkin 2000) without characters supporting the monophyly of the group (except in Hastings 1993: 104, mentioning a single feature: branchiostegal membrane with a fold along the ithmus). Molecular phylogenies seemed, until recently, to challenge this monophyly (Bargelloni et al. 1994; Ritchie et al. 1997; Bargelloni et al. 2000). More specifically, molecular phylogenies encountered difficulties in recovering monophyletic nototheniids but without convincing alternatives; it was rather a lack of resolution in molecular trees than the clear demonstration of their paraphyly. For instance, the robustness of the node separating Pleuragramma from the rest of the family in Bargelloni et al. (2000) was rather poor (52%). This lack of resolution may be interpreted as an artefact due to a past sudden burst of nototheniid diversification just after the acquisition of antifreeze proteins, leaving too short time spans between cladogeneses for present molecular phylogenies to be resolved. The recent exception to these absences of resolution was the phylogeny published by Near et al. (2004) based on the complete 16S rDNA, where the nototheniids appeared monophyletic with the best support obtained to date [79% in maximum parsimony (MP) approach, 97% in maximum likelihood approach].
The present study starts from two considerations. First, there has been some disagreement about the monophyly of the family and the taxonomic content of each subfamily (Balushkin 2000; Bargelloni et al. 2000; Near et al. 2004). Second, in spite of the fact that the study of Near et al. (2004) provided answers to some of these questions, certain genera or species still have their phylogenetic position unresolved, either because discrepancies among studies, or because of a lack of resolution in those phylogenies, or lack of taxonomic sampling, without excluding the possibility of a mix of these causes. For instance, the phylogenetic position of the genus Gobionotothen is still unclear. None of the molecular phylogenies published to date recovered the position found by Balushkin (2000) for this genus, however, none of those phylogenies provided a clear (robust) answer. The phylogeny of the trematomine nototheniids is still unknown, in spite of the ecological importance of the group in coastal Antarctic environments (the most accessible to researchers), and in spite of some molecular attempts (Ritchie et al. 1996, 1997) that failed to provide reliable results. The Pleuragrammatinae (in the above sense of Balushkin 2000) is composed of nototheniids that secondarily acquired anatomical features allowing neutral buoyancy (at least in those taxa for which this parameter was measured). The monophyly of the group obtained from morphological data could have resulted from coding as homologuous some traits linked to neutral buoyancy independently acquired by convergence (Eastman 1993). Moreover, recovering the monophyly of the subfamily through molecular phylogenies has been highly problematic, even from the complete 16S gene data set of Near et al. (2004) where the node has one of the lowest bootstrap proportions. Last but not the least, interrelationships among subfamilies are far from clear from past molecular phylogenies (Bargelloni et al. 1994, 2000; Near et al. 2004), except for one feature: from the complete 16S data of Near et al. (2004) as well as from the partial 12S and 16S data of Bargelloni et al. (2000), the nototheniinae in the traditional sense are paraphyletic, with Lepidonotothen and Patagonotothen more closely related to the Trematominae than to other Nototheniinae like Notothenia.
The present work attempts to answer the above questions through the use of multiple data sets. First of all, we have transformed the morphological and anatomical data of Balushkin (2000) into a matrix for standard parsimony analysis, in order to check for the parsimony of the solution proposed by this author for the phylogeny of the nototheniids, as we already did with similar data (Iwami 1985) from channichthyids (Chen et al. 1998). Second, we sequenced in a collection of nototheniids three gene segments, chosen for their functional independence, and analysed them separately and simultaneously. The mitochondrial cytochrome b gene is a marker classically used in phylogenetic studies, specifically within teleostean families (Chen et al. 1998). The 5′ half of the gene (541 bp) has been sequenced. The MLL (Mixed Leukaemia-Like) gene is a teleostean nuclear orthologue of a gene that, in humans, encodes a protein of 4,498 amino acids involved in leukaemogenesis (Caldas et al. 1988a, b). Partial sequences from exon 26 was used as described in Dettaï and Lecointre (2005). The rhodopsin gene is a member of the opsin gene family that has five main paralogous genes in vertebrates involved in visual pigments. The protocol for obtaining orthologues is described in Chen et al. (2003). From the separate analyses of those four data sets, we retained clades repeated across independent trees as reliable, which is a more conservative approach to reliability than the “total evidence” approach (for a methodological justification, see Lecointre and Deleporte 2000, 2005; Chen et al. 2003; Dettaï and Lecointre 2004, 2005). As none of the above genes showed sufficient variability to provide phylogenetic resolution within the genus Trematomus, we sequenced 500 bp of the highly variable mitochondrial control region for all available species of the genus in order to obtain, for the first time, interrelationships for a significant sample of trematomine species. The present molecular results about nototheniid phylogeny are summarised in the form of a MRP (Matrix Representation with Parsimony) supertree, which is suitable for exploring taxonomic congruence among data sets even when taxonomic samplings are not exactly the same (Baum and Ragan 2004).
Materials and methods
Taxonomic sampling
Table 1 shows the samples available for the present study. The nomenclature is based on that of DeWitt et al. (1990). The taxonomic overlap among data sets is not complete because sequencings have been performed at different periods and, to a lesser extent, because of difficulties in obtaining reliable gene amplification for some taxa. More precisely, the overlap between nuclear and mitochondrial data sets is not complete because of the rather poor resolving power of the present nuclear genes. For example, confronted with the poor resolution provided by the MLL (Caldas et al. 1988a, b) and rhodopsin genes within the Nototheniidae, we decided to stop the sequencing at such a taxonomic scale. However, the sequences must be published for future use in other taxonomic contexts.
Molecular techniques
Samples were kept in 70% ethanol until DNA extraction following a classical protocol (Winnepenninckx et al. 1993). Sequence-specific amplifications were performed by PCR in a final 50 μl volume containing 5% DMSO, 300 μM of each dNTP, 0.3 μM of Taq DNA polymerase (Qiagen, Crawley, UK), 5 μl of 10× buffer (Qiagen) and 1.7 pM of each of the two primers (see Table 2); 0.01–0.1 μg of DNA were added depending on species. After denaturation for 2 min, the PCR was run for 40 cycles of (30 s, 94°C; 30 s, 50 or 54°C; 1 min, 72°C). The result was visualised on ethidium-bromide stained agarose gels, and purified with the Minelute PCR Purification kit (Qiagen). Sequencing was performed on a CEQ2000 Beckman sequencer, Version 4.3.9, with the products and according to the instructions of the manufacturer’s kit. Each sequence was obtained at least twice and checked against its chromatograms in Bioedit (Hall 1999). Potential contaminations and mix-ups were detected by pairwise sequence comparison and using Blast (Altschul et al. 1997) on GenBank (Benson et al. 2002) through the NCBI portal (http://www.ncbi.nlm.nih.gov/), and for dubious cases another sequencing was performed on a new extraction. All new sequences were deposited in GenBank (accession numbers listed in Table 1). Alignments were mainly performed by hand under BioEdit (Hall 1999). All the sequenced genes are partial: 573 positions in exon 26 for MLL, 759 for rhodopsin, 541 for cyt b, 480 for d-loop. The size of each dataset, number of taxa and number of informative positions for parsimony are given Table 4.
Morphological data
For the present work the morphological data of Balushkin (2000) have been recoded. In his original work, Balushkin considered sets of characters for each subfamily separately, each character being described in a binary manner “apomorphic state–plesiomorphic state”. As a consequence, a set of characters supporting the tree of a given subfamily (e.g. Pleuragrammatinae) was not the same as the set of characters used for another subfamily (e.g. Trematominae)–except for three characters. Nevertheless, we have chosen to employ a global matrix by coding “plesiomorphic state” as “0” for all plesiomorphic states of a given subfamily and for all taxa belonging to other subfamilies. Obviously such a coding may erase part of homoplasy. However, it should be stressed that what we are doing here is assessing through standard parsimony methods the parsimony of the phylogenetic hypotheses as published by Balushkin with non-standard methods. Introducing more information than contained in the original paper would create biases in this assessment. Table 3 shows the 106 characters used, with correspondences to Balushkin’s characters are as follows:
Characters 1–51: Balushkin’s characters 1–51 used for the Nototheniinae. Note that the present character 7 combines the seventh character of the Nototheniinae and the eighteenth character of the Trematominae (apomorphic state: operculum trapezoidal in shape, angle between the anterior and upper margins is more than 60°—plesiomorphic state: Operculum triangular in shape, angle between the anterior and upper margins less than 60°).
Characters 52–83: Balushkin’s characters 1–33 used for the Pleuragrammatinae. Note that Balushkin’s fourth character is transferred to character 84 corresponding to Balushkin’s first character used for the Trematominae (it is the same: scapular foramen located in scapula—scapular foramen between scapula and coracoid). Note that character 63 is the combination of Balushkin’s thirteenth character used for Pleuragrammatinae and fifteenth character used for Trematominae [it is the same: absence of two subocular bones (plcr1, plcr2)—presence of plcr1 and plcr2].
Characters 84–104: Balushkin’s characters 1–23 used for the Trematominae. Note that Balushkin’s character 15 is transferred to our character 63 (see above) and Balushkin’s character 18 is transferred to our character 7 (see above).
Characters 105 and 106: they are cited in Balushkin’s paper as characters uniting the Nototheniinae and the Trematominae: broad fusion of gill membranes to isthmus and decrease in number of branchiostegal rays to six.
Standard parsimony analysis was conducted using heuristic searches (TBR search, 1000 random addition sequences, characters unordered and unweighted) with PAUP*4.0b10 (Swofford 1999). Bootstrap proportions were calculated with 1,000 bootstrap pseudoreplicates.
Sequence data analysis
Saturation was evaluated for each data set separately according to classical techniques (Philippe et al. 1994; Hassanin et al. 1998, data not shown). Separate analyses have been conducted under MP to allow comparison between molecular and morphological results without running the risk of mixing taxonomic congruence with sensitivity effects. Heuristic searches [tree bisection reconnection (TBR) search, 1,000 random addition sequences, characters unordered and unweighted] were conducted with PAUP4.0b10 (Swofford 1999) as well as bootstrap values calculation with 1,000 pseudo-replicates. Lack of resolution in trees from nuclear genes reduced the number of clades of interest in the table of taxonomic congruence among trees (Table 4). As a result, empty cells in Table 4 are not due to contradiction but either to a lack of resolution because the gene is not sufficiently variable or to incomplete taxonomic sampling. For simplicity, a “yes” in cells of Table 4 means that the clade is present, a “no” means that it is contradicted by another hypothesis. As the adopted approach involves comparing trees obtained from independent datasets, the MP tree from Near et al. (2004) based on the complete 16S gene was included in Table 4 (but not in the supertree in Fig. 3). For simplicity we present only three trees: the tree based on morphological data (Fig. 1), the tree based on d-loop data because it is the most precise within the Trematominae (Fig. 2). Additionally, we provide here a “summary tree” which is actually a supertree (Fig. 3) obtained from coding in a matrix each node of each strict consensus tree from each molecular data set and calculating the summary tree (called “MRP” supertree) from that matrix using parsimony.
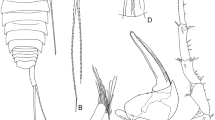
Strict consensus tree based on the morphological matrix of Table 3. For tree characteristics see Table 4. Letters refer to Table 4. Numbers are bootstrap proportions calculated from 1,000 pseudoreplicates. White bar: Pleuragrammatinae; black bar: Trematominae; dotted bar: Nototheniinae; sensu Balushkin (2000)
Most parsimonious tree based on d-loop data, on which nodes with a BP below 50% have been collapsed. For tree characteristics see Table 4. Letters refer to Table 4. Numbers are bootstrap proportions calculated from 1,000 pseudoreplicates. White bar Pleuragrammatinae; black bar Trematominae; dotted bar Nototheniinae; sensu Balushkin (2000). Numbers before names refer to the number of individuals sequenced for the species and present in the tree (as each species concerned is monophyletic, this presentation saves space)
Results
Table 4 summarises the number of trees and tree length obtained for each data set, and the presence of some clades of interest with regard to interrelationships among subfamilies: A: paraphyly of the Nototheniinae, Lepidonotothen and Patagonotothen being more closely related to the Trematominae than to other Nototheniinae sensu Balushkin like Notothenia; B: Lepidonotothen as the sister-group of Patagonotothen; C: monophyly of the Pleuragrammatinae (the “pelagic clade”); D: monophyly of the group Notothenia + Paranotothenia; E: monophyly of the genus Gobionotothen; F: monophyly of the Trematominae; G: Pagothenia embedded within Trematomus; H: Paranotothenia embedded within Notothenia (Paraphyletic Notothenia); I: monophyly of Lepidonotothen; J: monophyly of Patagonotothen; Z: monophyly of the Nototheniinae sensu Balushkin; X: monophyly of the Nototheniidae. Table 4 does not detail results within the Trematominae, because in each tree most of the clades are collapsed, except for the tree based on d-loop data. This is the reason why the tree based on d-loop data is shown Fig. 2. The only conclusion that could be extracted from other molecular data sets was that Trematomus scotti, Trematomus newnesi and Trematomus eulepidotus were found external to all other trematomines, which were all collapsed into a polytomy. Positions of the three basal trematomines are provided by rhodopsin and cyt b data, interrelationships among crown trematomines are provided by the d-loop sequences; then the supertree in Fig. 3 provides the overall picture one should retain.
Morphological data exhibited only 15 homoplastic characters (character 3 has a CI of 0.33 and characters 43–45, 48–51, 63, 78–83, have a CI of 0.5), which is a rather low level of homoplasy. The tree obtained (Fig. 1) is consistent with those shown in Balushkin’s publication. The “pelagic clade” (Balushkin’s Pleuragrammatinae) was found to be monophyletic and basal among the Nototheniidae. As a consequence, the monophyletic Trematominae (clade F) was found as the sister-group of the monophyletic Nototheniinae (sensu Balushkin, clade Z). Trematomus may be paraphyletic: if they are not basal, the genera Pagothenia and Cryothenia may have to be renamed Trematomus to obtain a monophyletic Trematomus. In the same way, if Indonotothenia was considered as a Notothenia, then one should rename Paranotothenia into Notothenia (clade H). Lepidonotothen sensu lato was paraphyletic and includes Gobionotothen.
Empty cells in Table 4 mean that there is no “signal”, neither for nor against the corresponding clade. Among molecular results, it is noticeable that the two nuclear genes were not sufficiently variable and should not be further exploited for nototheniid intra-relationships. Molecular results, when resolved, globally corroborated morphological results: there were only two points of conflict. First, morphological data supported the monophyly of the Nototheniinae sensu Balushkin (clade Z) while all molecular data providing resolution showed a sister-group relationship between clades B and F, i.e. between Lepidonotothen + Patagonotothen and the Trematominae. That clade, named A, was present in trees from MLL, cytochrome b, d-loop and the complete 16S gene from Near et al. (2004). Second, the monophyly of the genus Lepidonotothen sensu lato was challenged by morphological results, as Gobionotothen was embedded within it. The cytochrome b data set, however, supported the monophyly of the genus. Here the d-loop data set was of no help because there was only one available Lepidonotothen species. Nuclear data sets did not resolve those relationships. In Near et al.’s (2004) study, Lepidonotothen was not monophyletic, but for other reasons than in Balushkin’s (2000) study: Lepidonotothen larseni and Lepidonotothen nudifrons were more closely related to Patagonotothen than to other species of Lepidonotothen. However, monophyletic Lepidonotothen was not rejected in the author’s SH test based on 16S data. More data are needed to solve discrepancies about Lepidonotothen. A general feature of all molecular data collected to date is the impossibility to assign a stable and reliable position for the genus Gobionotothen. Here the genus was never included within clades B, C, F, D but was variably placed among them, according to genes and methods. The same result was found in Near et al.’s (2004) study, where it is clear that the nodes defining its position were not robust.
The d-loop data brought some additional information than other molecular data sets with regard to interrelationships among species of the genus Gobionotothen (i.e. within clade E) and among species of the crown Trematomus (clade F, Fig. 2). Three clades emerge: 1: Trematomus hansoni + Trematomus bernacchii + Trematomus vicarius; 2: Trematomus pennellii + Trematomus lepidorhinus + Trematomus loennbergii; 3: Trematomus (Pagothenia) borchgrevinki + Trematomus nicolai. For some Trematomus species, several individuals have been sequenced (numbers before names in Fig. 2).
Discussion
Figure 3 summarises the molecular phylogenetic conclusions with a supertree. The present molecular data could not solve interrelationships among nototheniid subfamilies, and could not establish the monophyly of the Nototheniidae. However, a number of conclusions emerge:
-
1.
Molecular results confirm Balushkin’s Pleuragrammatinae. This leads one to think that neutral buoyancy was gained by common ancestry. However, one should keep in mind that the anatomical features allowing neutral buoyancy are not the same in Dissostichus, Pleuragramma and Aethotaxis (Eastman 1993), rather suggesting parallelisms than common ancestry.
-
2.
Molecular results found the clade A several times independently (also present in Near et al.’s and in Bargelloni et al.’s), establishing the paraphyly of the Nototheniinae sensu lato. Lepidonotothen and Patagonotothen are more closely related to the Trematominae than to Notothenia. The term Nototheniinae may now be reserved for Notothenia. The reason of the discrepancy with Balushkin’s morphological data may be related to the fact that his data contain far more characters devoted to classifying species within each pre-defined subfamily than to characters covering several subfamilies. Such a structure in data collection may have prevented discovery of those characters that could legitimately challenge the monophyly of any subfamily, among them the Nototheniinae sensu lato. In Fig. 1 there are two synapomorphies supporting the clade Z without homoplasy: derived states for characters 1 and 46. Clade Z is characterised by the mesopterygoid overlapping the quadratum (character 1); and interruption of sensory canals of CPM and CT (character 46). Accepting the molecular hypothesis imply homoplastic changes for these two derived states.
-
3.
To date no molecular data set could assign a reliable position to the genus Gobionotothen.
-
4.
For Notothenia to be a monophyletic genus, Paranotothenia will have to be included in the genus Notothenia.
-
5.
As in previous studies, T. scotti (Ritchie et al. 1997; Bargelloni and Lecointre 1998; Bargelloni et al. 2000; Near et al. 2004), and T. newnesi (Bargelloni and Lecointre 1998; Bargelloni et al. 2000) are the most basal Trematominae. For the first time we present interrelationships among crown Trematominae (i.e. Trematominae less T. scotti, T. newnesi and T. eulepidotus). The present trematomine molecular phylogeny (Figs. 2, 3) is far better resolved than in Ritchie et al. (1996, 1997) or Bargelloni et al. (2000). Near et al. (2004) did not include sufficient taxa to allow such a comparison. However, as in Near et al. (2004), Ritchie et al. (1996), Bargelloni et al. (2000), the genus Trematomus was paraphyletic and Pagothenia should become Trematomus to make the genus monophyletic. The Trematomus tree found did not match the topology found from Balushkin’s morphological matrix (Fig. 1) where T. newnesi was the sister group of the clade Pagothenia + Cryothenia, and the remaining Trematomus species were placed in Pseudotrematomus. In our trees T. newnesi was clearly not the sister group of T. (Pagothenia) borchgrevinki. The tree found did not match to the ecomorphological index of Ekau (1988, 1991) describing the notothenioid mode of life from 1 (the most pelagic) to 10 (the most benthic) for a number of Trematominae including the present sample (Eastman 1993). Mapping values of this index onto the tree of Fig. 3 clearly showed that being benthic or pelagic was never attained by common ancestry, suggesting a high plasticity of the trematominae. If we map cryopelagic ecology, the same conclusion appears: cryopelagic taxa were not sister-groups but well separated (T. newnesi and T. borchgrevinki in Fig. 3). This is not congruent with Balushkin’s findings, where taxa with pelagic tendencies were clustered (Pagothenia, Cryothenia and T. newnesi, the only species in the genus Trematomus according to this author), and all the remaining species, benthic or epibenthic, being placed within Pseudotrematomus.
Better resolution within the nototheniid tree will probably be obtained in the future by using several variable nuclear markers, more variable than the ones used here. Other possibilities will be given from characters extracted from cytogenetics and genome structures, for instance by comparing among taxa the relative position of genes obtained from fluorescent in situ sequence hybridisations onto chromosomes. This could be used to study relative positions of clades A, C, D, E with regard to crown notothenioids.
References
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Balushkin AV (1992) Classification, phylogenetic relationships, and origins of the families of the suborder notothenioidei (Perciforms). J Ichthyol 32:90–110
Balushkin AV (2000) Morphology, classification, and evolution of notothenioid fishes of the Southern Ocean (Notothenioidei, Perciformes). J Ichthyol 40:S74–S109
Bargelloni L, Lecointre G (1998) Four years in notothenioid systematics: a molecular perspective. In: di Prisco G, Pisano E, Clarke A (eds) Fishes of Antarctica, a biological overview. Springer-Verlag, Milano, pp 259–273
Bargelloni L, Marcato S, Zane L, Patarnello T (2000) Mitochondrial phylogeny of notothenioids: a molecular approach to antarctic fish evolution and biogeography. Syst Biol 49:114–129
Bargelloni L, Ritchie PA, Battaglia B, Lambert DM, Meyer A (1994) Molecular evolution at subzero temperatures: mitochondrial and nuclear phylogenies of fishes from Antarctica (Suborder Notothenioidei), and the evolution of antifreeze glycopeptides. Mol Biol Evol 11:854–886
Baum BR, Ragan MA (2004) The MRP method. In: Bininda-Emonds RP (eds) Phylogenetic supertrees, Kluwer Academic publishers, Dordrecht, the Netherlands, pp 17–34
Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Rapp BA, Wheeler DL (2002) Genbank. Nucleic Acids Res 30:17–20
Caldas C, Kim MH, McGregor A, Cain D, Aparicio S, Wiedeman LM (1998a) Isolation and characterization of a pufferfish MLL (Mixed lineage leukemia-like) gene (fMll) reveals evolutionnary conservation in vertebrate genes related to Drosophila trithorax. Oncogene 16:3233–3241
Caldas C, So CW, McGregor A, Ford AM, McDonald B, Chan LC, Wiedemann LM (1998b) Exon scrambling of MLL transcripts occur commonly and mimic partial genomic duplication of the gene. Gene 208:167–176
Chen WJ, Bonillo C, Lecointre G (1998) Phylogeny of the Channichthyidae (Notothenioidei, Teleostei) based on two mitochondrial genes. In: di Prisco G, Pisano E, Clarke A (eds) Fishes of Antarctica, a biological overview. Springer-Verlag, Milano, pp 287–298
Chen WJ, Bonillo C, Lecointre G (2003) Reapeatability of clades as a criterion of reliability: a case study for molecular phylogeny of Acanthomorpha (Teleostei) with larger number of taxa. Mol Phylogenet Evol 26:262–288
Dettaï A, Lecointre G (2004) In search for notothenioid relatives. Antarct Sci 16:71–85
Dettaï A, Lecointre G (2005) Further support for the clades obtained by multiple molecular phylogenies in the acanthomorph bush. C R Biol 328:674–689
DeWitt HH (1971) Coastal and deep-water benthic fishes of the Antarctic. In: Bushnell VC (ed) Antarctic map folio series folio 15. American Geographical Society, New York, pp 1–10
DeWitt HH, Heemstra PC, Gon O (1990) Nototheniidae. In: Gon O, Heemstra PC (eds) Fishes of the Southern Ocean. JLB Smith Institute of Ichthyology, Grahamstown, South Africa, pp 279–331
Eastman JT (1993) Antarctic fish biology. Academic Press, San Diego
Eastman JT, Clarke A (1998) A comparison of adaptative radiations of Antarctic fishes with those of non-Antarctic fish. In: di Prisco G, Pisano E, Clarke A (eds) Fishes of Antarctica, a biological overview. Springer-Verlag, Milano, pp 3–26
Ekau W (1988) Ecomorphology of nototheniid fish from the Weddell Sea. Antarctica Ber Polarforsch 51:1–140
Ekau W (1991) Morphological adaptations and mode of life in High Antarctic fish. In: di Prisco G, Maresca B, Tota B (eds) Biology of Antarctic Fish. Springer-Verlag, Berlin, pp 23–39
Hall TA (1999) Bioedit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hassanin A, Lecointre G, Tillier S (1998) The “ Evolutionary Signal “ of homoplasy in protein coding gene sequences and its consequences for a priori weighting in phylogeny. C R Acad Sci 321:611–620
Hastings PA (1993) Relationships of the fishes of the perciform suborder Notothenioidei. In: Miller A (ed) A history and atlas of the fishes of the Antarctic Ocean. Foresta Institute for Ocean and Mountain Studies, Carson City, Nevada, pp 99–107
Iwami T (1985) Osteology and relationships of the family Channichthyidae. Mem Natl Inst Pol Res Tokyo Ser E 36:1–69
Lecointre G, Bonillo C, Ozouf-Costaz C, Hureau JC (1997) Molecular phylogeny of the Antarctic fishes: paraphyly of the Bovichtidae and no indication for the monophyly of the Notothenioidei (Teleostei). Polar Biol 18:193–208
Lecointre G, Deleporte P (2000) Le principe du “total evidence” requiert l’exclusion de données trompeuses. Biosystema 18:129–151
Lecointre G, Deleporte P (2005) Total evidence requires exclusion of phylogenetically misleading data. Zool Scripta 34:101–117
Lecointre G, Ozouf-Costaz C (2004) Les poissons à antigel de l’océan austral. Pour La Sci 320:48–54
Near TJ, Pesavento JJ, Cheng CC (2004) Phylogenetic investigations of Antarctic notothenioid fishes (Perciformes: Notothenioidei) using complete gene sequences of the mitochondrial encoded 16S rRNA. Mol Phylogenet Evol 32:881–891
Philippe H, Sorhannus U, Baroin A, Perasso R, Gasse F, Adoutte A (1994) Comparison of molecular and paleontological data in diatoms suggests a major gap in the fossil record. J Evol Biol 7:247–265
Ritchie PA, Bargelloni L, Meyer A, Taylor J, MacDonald JA, Lambert DM (1996) A mitochondrial phylogeny of the trematomid fishes (Nototheniidae, Perciformes) and the evolution of Antarctic fishes. Mol Phylogenet Evol 5(2):383–390
Ritchie PA, Lavoué S, Lecointre G (1997) Molecular phylogenetics and the evolution of Antarctic notothenioid nishes. Comp Biochem Physiol 118A 4:1009–1027
Swofford D (1999) PAUP*, Phylogenetic analysis using parsimony, version 4.0b10
Winnepenninckx B, Backeljau T, Wachter RD (1993) Extraction of high molecular weight DNA from molluscs. T I G 9:407
Acknowledgments
We thank Tom Near for having provided samples, Erwan Le Guilloux, François Vadurel and Philippe Koubbi for having sampled on the field in Terre Adélie. The present work was based on samples collected thanks to the ICEFISH 2004 cruise. Some of the samples were acquired thanks to the EPOS cruise for which we are grateful to the Alfred Wegener Institute and the European Science Foundation, along with the Centre National pour la Recherche Scientifique, the Muséum National d’Histoire Naturelle and the Terres Australes et Antarctiques Françaises. Samples from the Magellan Strait were collected thanks to the MNHN and the Instituto de la Patagonia, Punta Arenas. We thank the Institut Paul Emile Victor for support to the ICOTA program, and Philippe Koubbi for management. We thank Eric Pasquet and Annie Tillier for help in the “Service de Systématique moléculaire” of the IFR101. The ICEFISH cruise was supported by National Science Foundation grant OPP 01-32032 to H. William Detrich (Northeastern University). This is publication number 12 from the ICEFISH Cruise of 2004 (H.W. Detrich, Chief Scientist).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sanchez, S., Dettaï, A., Bonillo, C. et al. Molecular and morphological phylogenies of the Antarctic teleostean family Nototheniidae, with emphasis on the Trematominae. Polar Biol 30, 155–166 (2007). https://doi.org/10.1007/s00300-006-0170-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00300-006-0170-1